Gonadal transcriptomic analysis and differentially expressed genes between the testes and ovaries in Trachinotus ovatus
Pingping He, Peng Zhu, Pinyun Wei, Xiofei Zhuo, Yun M,, Xiohn Chen,Yong Lin, Youhou Xu, Hui Luo, Jinxi Peng,*
a Guangxi Key Laboratory of Aquatic Genetic Breeding and Healthy Aquaculture, Guangxi Academy of Fisheries Sciences, Nanning, 530021, China
b Beibu Gulf University, Qinzhou, 535000, China
c College of Animal Science, Southwest University, Chongqing, 402460, China
Keywords:Transcriptome Trachinotus ovatus Gonad High-throughput sequencing RT-qPCR
ABSTRACT The Trachinotus ovatus is a popular aquaculture species in China. There are no obvious morphological differences between male and female fish, even during maturity, prompting research studies on sex-related features of this fish. To examine sex determination- and gonadal development-related genes, we conducted transcriptome analysis of the ovaries and testes of T. ovatus. A total of 345,972,132 high-quality clean reads were obtained from 12 libraries. In addition, 28,137 gonad-expressed unigenes were obtained by mapping the clean reads to the T. ovatus reference genome. A total of 8,237 differentially expressed genes (DEGs) were identified between stage I ovaries and testes, including 3,235 testicular upregulated and 5,002 ovarian upregulated genes. Furthermore,13,448 DEGs were obtained between stage III ovarian and testicular libraries, including 7,576 testicular upregulated and 5,872 ovarian upregulated genes. The DEGs included some sex-determining genes such as sry, dmrt1,and amh. DEGs between ovarian and testicular libraries were significantly enriched with KEGG pathways that are involved in gonadal development, sex determination, and gonadal function. Then, 10 DEGs (seven testicular upregulated and three ovarian upregulated unigenes) were selected for quantitative real-time PCR analysis. Four genes (zinc-binding protein A33-like, pro-opiomelanocortin-like, leucine-rich repeat and transmembrane domain-containing protein 2-like, and forkhead boxC1) showed a specific testicular expression pattern. The gonadally expressed as well as testicular and ovarian DEGs provide useful information for further research on the reproductive biology of T. ovatus.
1. Introduction
Trachinotus ovatusis widely distributed in Southeast Asia, Australia,and eastern Africa. In recent years,T. ovatusaquaculture has rapidly developed along the southern China coast (Li, Peng, & Zhou, 2014), due to its fast growth and high-quality flesh (Tan et al., 2017).T. ovatusreaches gonadal maturity in at least four years, and their sexes are generally hard to distinguish based on appearance, even at maturity(Jiang et al., 2015). This makes the identification of parents for selective breeding difficult. Thus, studies on sex-related features such as sex-specific markers and gene mining, gonadal development, and maturity mechanism analyses have become vital in this fish. Although the genome ofT. ovatushas been assembled (Zhang et al., 2019a), to date, reports onT. ovatussex remain limited Xie et al. (2014); Zhang et al., 2019b.
Transcriptome sequencing can efficiently reveal gene expression in specific tissues or organs (Afonso et al., 2019; Krüger et al., 2019; Ma et al., 2019; Meyer et al., 2019; Tang et al., 2019), and gonad transcriptome analysis has been commonly applied in seeking gonadal differentiation genes in various aquatic animals, includingAcipenser schrenckii(Jin et al., 2015),Scophthalmus maximus(Hu et al., 2016)Patinopecten yessoensis(Zhou et al., 2019), musselPerumytilus purpuratus
(Carolina et al., 2018),Macrobrachium rosenbergii(Jiang et al., 2019),Sillago sihama(Tian et al., 2019), andAcipenser dabryanus(Chen et al.,2018). To date, some sex-determining genes have been identified (Li &Gui, 2018; Raymond, Murphy, OSullivan, Bardwell, & Zarkower, 2000;Mei & Gui, 2015; Peng et al., 2015; Tian et al., 2019), such as sex-determining region of Y (sry),doublesex- and mab-3-related transcription factor 1 (dmrt1),doublesex (Dsx), and anti-Müllerian hormone(amh) in males, as well as forkhead box l2 (foxl2)in females. In addition,many genes involving gonadal development and gametogenesis were also identified. In rainbow trout,gsdfregulates the proliferation of primordial germ cells (PGCs) and spermatogonia (Sawatari, Shikina,Takeuchi, & Yoshizaki, 2007). Gnrhr is related to testicular function maintenance inSparus aurataandS. sihama(Forner-Piquer et al., 2019;Tian et al., 2019).
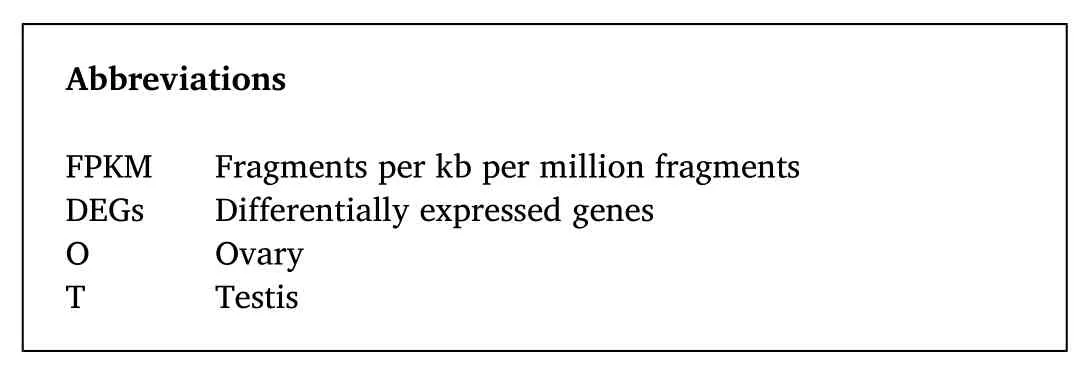
?
To obtain more information on the molecular mechanism of sex determination, gonadal differentiation, and gonadal development inT. ovatus,we conducted a transcriptome analysis of ovaries and testes ofT. ovatus. The gonadal transcriptome data will provide useful information for studying gonad differentiation and development mechanisms inT. ovatus.
2. Materials and methods
2.1. Ethics statement
All of the procedures involved in the treatment ofT. ovatuswere executed with the approval of the Animal Care and Use Committee of the Guangxi Academy of Fishery Sciences.
2.2. Sample collection
Fifty-five one-year-old and nine three-to four-year-oldT. ovatusindividuals were provided by the Guangxi Academy of Fisheries Sciences(Nanning, China). After anaesthetization, the fish were sacrificed, and their gonads were collected. Parts of the gonad of each fish were immediately immersed in RNAlater (Jierui Biotechnology Co., Ltd.,Shanghai, China), and the rest were kept in 4% paraformaldehyde (PFA)for sectioning and HE staining.
Gonadal tissue sections were used to determine the sex and developmental stage of the fish. The fish with gonads mainly containing oogonia or spermatogonia were classified as stage I. The fish with gonads mainly containing stage II oogonia or spermatogonia and primary spermatogonia were identified as stage II. Ovaries mainly consisted of stage III oocytes and the testes comprised of spermatogonia, primary spermatogonia, secondary spermatocytes, and spermatids were identified as stage III. Ovaries mainly consisted of stage Ⅳ oocytes and the testes containing some sperms were identified as stage Ⅳ. The ovary mainly consisted of stage Ⅴ oocytes and the testes full of sperms were identified as stage Ⅴ. The ovary with a few oocytes remaining and testes with a few sperm were identified as stage Ⅳ. The gonadal development stages of the collected fishes were mainly at stages I–III.
TwelveT. ovatusindividuals were used for Solexa sequencing (six at stage I, and six at stage III) and 16 individuals (eight at stage I, and eight at stage III) for RT-qPCR. Among these, 14 fish were at stage I, with mean body weight of 416.67 ±98.32 g and mean body length of 22.58± 1.36 cm, whereas the others were at stage III, with a mean body weight of 2274.00 ±1911.00 g and mean body length of 37.35 ±15.00 cm.
2.3. RNA extraction, libraries construction, and sequencing
Total RNA was extracted from the gonad samples with TRIzol(Invitrogen, CA, USA). Concentrations of the separated RNA weredetected with a NanoDrop 2000 spectrophotometer, and the quality was determined with an Agilent 2100 Bioanalyzer. The cDNA libraries were sequenced on an Illumina HiSeq™ 2500.

Table 1 Primer sequences used in RT-real-time quantitative PCR (RT-qPCR).
2.4. Sequence assembly and functional annotation
Clean data were obtained after removal of adapters and reads with>10% unknown nucleotides, and >50% low-quality bases (Q < 10)excluded from the raw reads. The clean reads were then mapped to the reference genome (the reference genome is unpublished data that were kindly provided by Xiaohan Chen) using TopHat2 (Kim et al., 2013).Then, the mapped reads were assembled using Cufflinks software. The unigenes were then annotated using NR, Swiss-Prot, KOG, COG, GO, and KEGG databases with BLAST2GO (https://www.blast2go.com).
2.5. Analysis of differentially expressed unigenes and GO/KEGG enrichment analysis
To distinguish DEGs between testis and ovaries, the expression levels of unigenes were determined using the fragments per kb per million fragments method (FPKM). The DEseq2 software was used to distinguish DEGs between the two sexes. The threshold of theP-value was determined by the false-discovery rate (FDR). Unigenes with an FDR ≤0.01 and fold changes >2 were considered as DEGs in our study.
Moreover, functional enrichment analysis was conducted to identify which GO terms and KEGG pathways were significantly enriched by DEGs. GO functional enrichment and KEGG pathway analyses were performed by Goatools (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do).
2.6. Real-time quantitative PCR (RT-qPCR)
To validate the significantly DEGs in the testes and ovaries transcriptome ofT. ovatus, 10 DEGs were selected for real-time quantitative PCR (RT-qPCR). The sequences of seven testicular upregulated and three ovarian upregulated genes were selected for primer design. The primer sequences are shown in Table 1β-Actinwas used as internal reference gene. Total RNAs were reverse-transcribed with a reverse transcription system (TaKaRa). QPCR was conducted on an ABI 7500 PCR system (CA,USA) with qPCR reagents (TaKaRa). The qPCR program was as follows:20 s at 95◦C; followed by 40 cycles of 15 s at 95◦C and 30 s at 55◦C.Each group included two pooled samples as biological replicates, and each sample was tested in triplicate. The relative expression levels of genes were calculated with the 2−ΔΔCTmethod. Data were analyzed with SPSS 23.0, and differences withP<0.05 were considered to be significant.
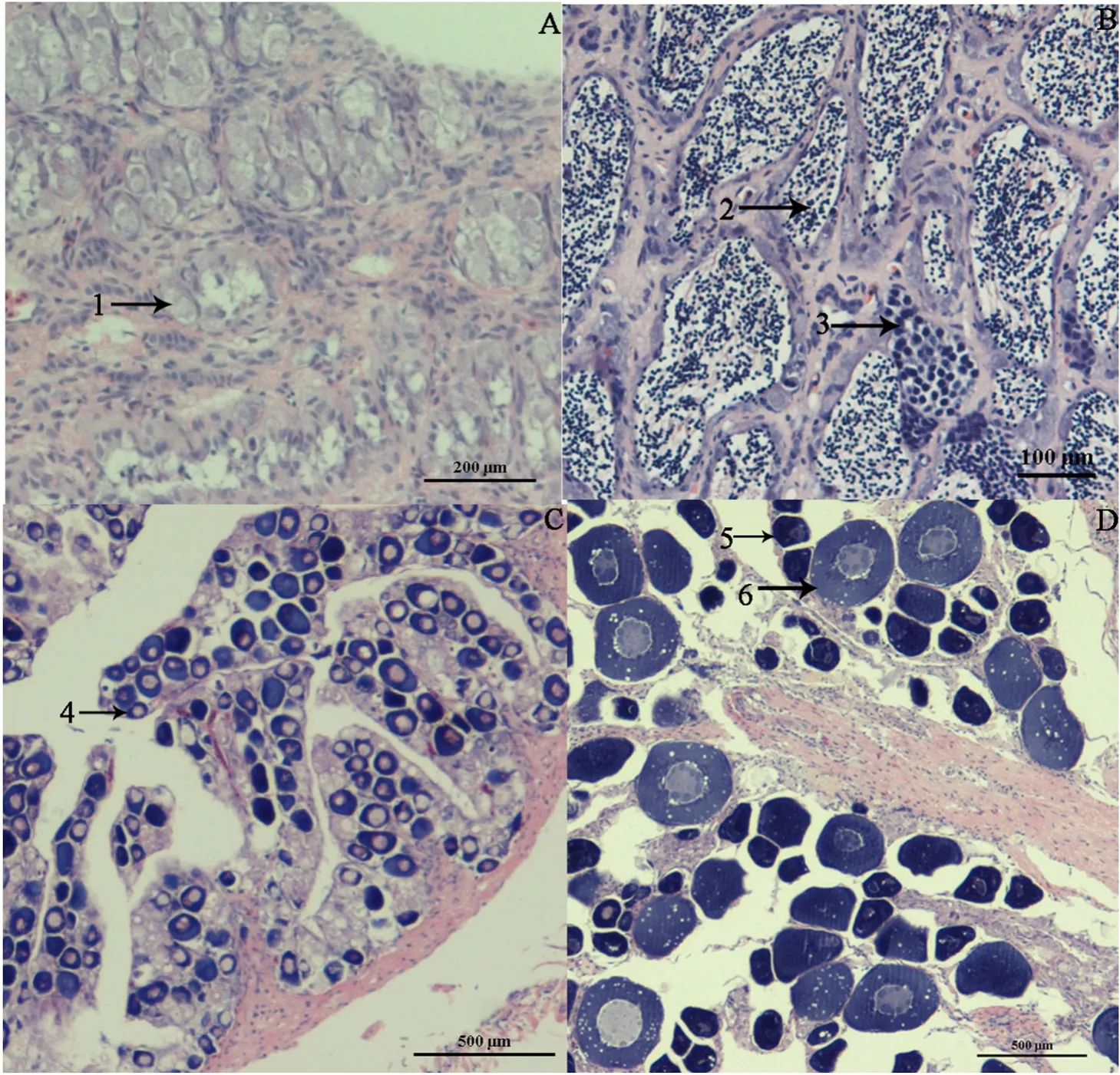
Fig. 1. Gonadal sections of T. ovatus. A. Representative section of the stage I testes. B. Representative section of the stage III testes. C. Representative section of the stage I ovary. D. Representative section of the stage III ovary. Arrows: 1. Spermatogonium 2. Spermatocyte, 3. Spermatid, 4. Oogonia, 5. Oocyte at stage II, 6. Oocyte at stage III.
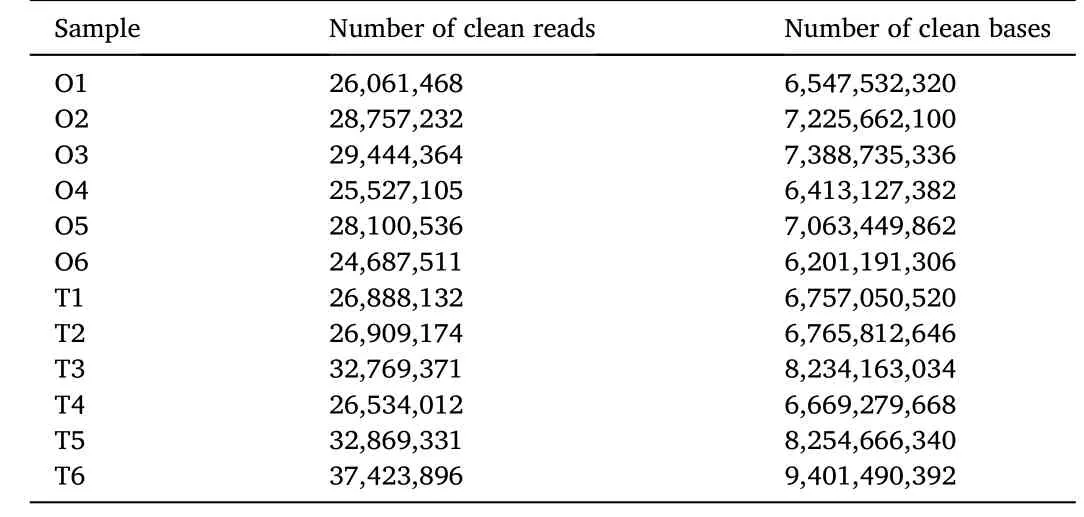
Table 2 Sequencing data statistics.
3. Results
3.1. Confirmation of sex and developmental stages using gonadal sections
Based on the results of histological assessment of gonadal sections, in total of 28 fishes were selected for transcriptome sequencing and RTqPCR. Twelve fishes were used in transcriptome sequencing, including six males and six females (each stage/sex using three libraries), and 16 fishes for RT-qPCR, including eight males and eight females, each stage/sex using four fishes. Among the 28 fishes, 14 were at stage I, with gonads mainly containing oogonia or spermatogonia, whereas the others were at stage III, where ovary mainly consisted of secondary oocytes,whereas the testes largely comprised spermatocytes and spermatids(Fig. 1).
3.2. Illumina sequencing, sequence assembly, and annotation
A total of 12 libraries were constructed, including stage I ovary, stage III ovary, stage I testis, and stage III testis (each stage/sex consisting of three libraries). After filtering low-quality reads, a total of 345,972,132(162,578,216 and 183,393,916 from ovarian and testicular libraries,respectively) clean reads were obtained from the 12 libraries (Table 2).The clean reads data can be downloaded from the NCBI Sequence Read Archive [SRA] with accession number PRJNA609165. We map the clean reads to theT. ovatusreference genome with TopHat2 software, and the percentage of mapped reads ranged from 83.38% to 86.84% among samples. A total of 28,137 unigenes were expressed in the gonads.Functional annotation was conducted by searching the NR, Swiss-Prot,KOG, COG, GO, and KEGG databases. A total of 23,678 unigenes were annotated, including NR (23,655), Swiss-Prot (16,129), KOG (16,017),COG (6,998), GO (12,503), and KEGG (13,692).

Fig. 2. Volcano plot of differences in gene expression between ovaries and testes. Red: testis-upregulated genes; Green: testis-downregulated genes. A. stage III, B.stage I. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3. Distribution of differentially expressed genes (DEGs) in gonad among GO terms in biological processes, molecular functions, and cellular components. A.stage III, B. stage I.
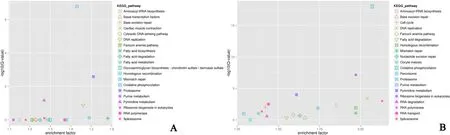
Fig. 4. The top 20 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched by the DEGs. A. stage III, B. stage I.

Table 3 Differentially expressed genes involved in gonadal differentiation and gonadal development.
3.3. Identification and functional annotation of DEGs between ovaries and testes
The expression levels of unigenes were determined with FPKM in Cufflinks software. Subsequently, we identified DEGs between ovaries and testes based on gene expression levels with the DEseq2 software. A total of 13,448 DEGs were identified between stage III ovaries and testes,including 7,576 testis upregulated and 5,872 ovary upregulated genes(Fig. 2A; Table S1). A total of 8,237 DEGs were identified between stage I ovaries and testes, including 3,235 testis upregulated and 5,002 ovary upregulated genes (Fig. 2B; Table S2). The distribution of DEGs in the GO terms is shown in Fig. 3A (stage III) and Fig. 3B (stage I). DEGs between ovaries and testes were significantly enriched in the functional categories of biological processes (BP), molecular function (MF), and cellular component (CC). Within the term BP, the DEGs significantly enriched in the functional categories of single-organism developmental processes and cellular process. Within the term MF, the DEGs (stage III or stage I) significantly enriched in the functional categories of binding and catalytic activity. Within the term CC, the DEGs (stage III or stage I)significantly enriched in the functional categories of cell parts and cell.
The top 20 KEGG pathways enriched by the DEGs (Fig. 4A, stage III;Fig. 4B, stage I) included fatty acid biosynthesis, fatty acid degradation,pyrimidine metabolism, proteasome, and oxidative phosphorylation,etc.
3.4. Identification of sex determination, gonadal development, and gametogenesis-related genes
In this study, we identified some male-biased or specific sexdetermining genes in stage III DEGs, including thesry(Scaffold_169.38),dmrt1(Scaffold_169.588),amh(Scaffold_256.343). Some female-biased genes were also identified in the stage IIIT. ovatusgonad transcriptome, such as cytochrome P450 2K1-like (Scaffold_782.536).Among these,sry,dmrt1, andamhwere also detected in stage I DEGs.
In addition, some genes involving gonadal development and gametogenesis were identified in the stage III DEGs, including gonadal somaderived factor 1 (gsdf) (Scaffold_271.330), gonadotrophin-releasing hormone receptor (gnrhr) (Scaffold_187.212), zona pellucida spermbinding proteins (zps) (Scaffold_1381.107, Scaffold_169.387), and other potential candidate genes (Table 3). Among these,gsdfandzpswere also detected in stage I DEGs.
3.5. RT-qPCR confirmation of DEGs
Ten most significant DEGs between stage III ovaries and testes from transcriptome analysis were selected for RT-qPCR validation (Table 4),including seven testicular-upregulated (Scaffold 1151.20, Zinc-binding protein A33-like; Scaffold 745.327, Pro-opiomelanocortin-like;Scaffold105.41, leucine-rich repeat and transmembrane domaincontaining protein 2-like; and Scaffold 671.293, Forkhead box C1;Scaffold 1068.175, G0/G1 Switch protein 2; Scaffold 1068.793, Chromosome C1orf74 homolog (cunh1orf74); and Scaffold 134.49, homeobox protein Zampogna-like) and three ovarian upregulated unigenes(Scaffold 169.213, forkhead box protein H1-like; Scaffold 671.306,insulin-like growth factor-binding protein 1; and Scaffold 671.234,aquaporin-1-like) (Fig. 5). Most of the RT-qPCR results were consistent with the high-throughput sequencing data, except for Scaffold 671.23.
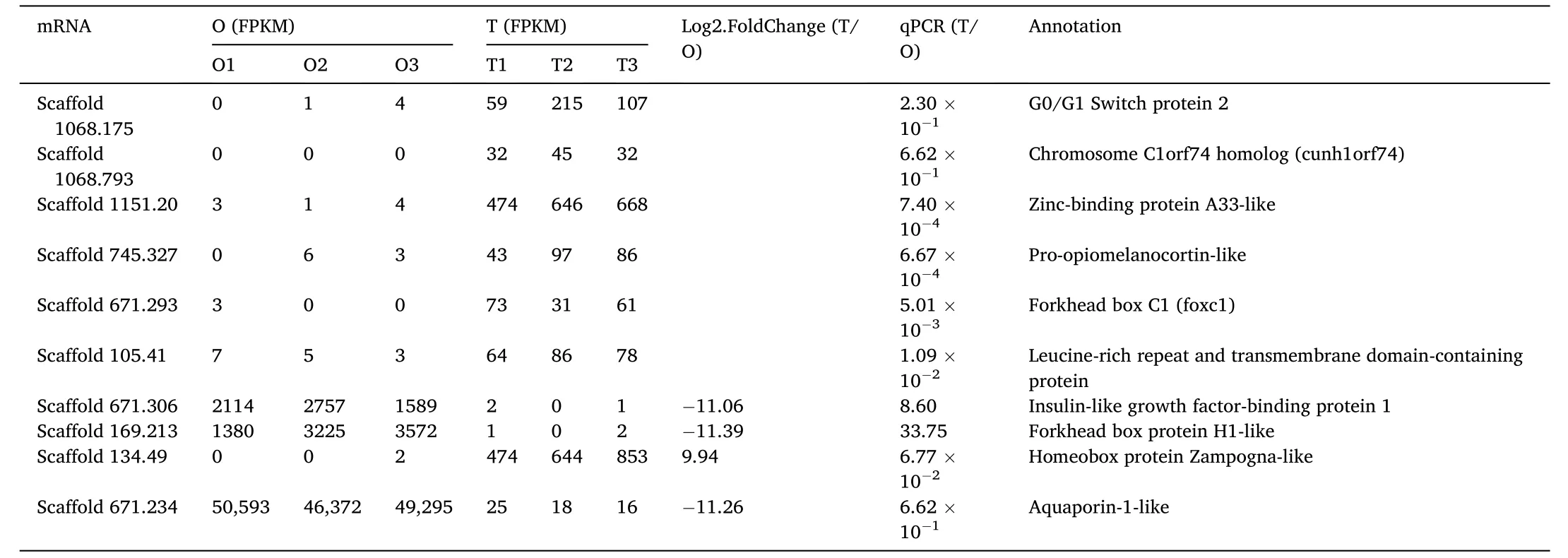
Table 4 DEGs identified using Solexa sequencing and validated by RT-qPCR.

Fig. 5. Real-time quantitative PCR (RT-qPCR) validation of the DEGs identified by transcriptome sequencing.
4. Discussion
To date, gonad transcriptomes have been widely used to identify sex determination- and development-related genes in fish species (Chen et al., 2018; Li & Gui, 2018; Mei & Gui, 2015; Tian et al., 2019), and several sex-determining genes have been identified, such assry,dmrt1,SoxH,Dsx,β-catenin,wnt 4,amh, andfoxl2(Li & Gui, 2018; Mei & Gui,2015; Peng et al., 2015; Tian et al., 2019). To date, only a few sex-related genes have been reported inT. ovatus(Xie et al., 2014). Here, we conducted transcriptome analysis on the ovaries and testes ofT. ovatus,identified numerous genes and important pathways that are involved in gonadal development and gametogenesis, and their differential expression between ovaries and testes. Providing vital data for the reproductive biology study inT. ovatus.
In this study, we identified several male sex-determining genes in theT. ovatusgonad transcriptome, includingsry,dmrt1, andamh.Sryis a sex-determining gene in mammals (Larney, Bailey, & Koopman, 2014),and decreasedSryexpression bring testicular defects (Larney et al.,2014).Srypromotes testis development by activating the downstream geneSox9(Sekido & Lovell-Badge, 2008). As a sex-determining gene in fish,dmrt1encodes an important transcription factor for testis differentiation (Webster et al., 2017).Dmrt1has also been identified as a testicular upregulated gene in a previous transcriptome analysis ofT. ovatus(Xie et al., 2014).Amhregulates male germ cell accumulation in several fish species (Yan et al., 2019).Amhis a target of gonadotropic follicle-stimulating hormone (fsh) in some fish species (Pfennig,Standke, & Gutzeit, 2015; Zhai et al., 2017).Fem-1is male sex-determining gene inC. elegans(Doniach & Hodgkin, 1984; Hodgkin,1986).Dsxis a male sex-determining gene in invertebrate, and in vertebrates (Burtis & Baker, 1989; Raymond, Murphy, O; Sullivan, Bardwell, & Zarkower, 2000; Kato, Kobayashi, Watanabe, & Iguchi, 2011). In this study, thesry, dmrt1, andamhgenes all showed upregulated expression in the testis. Their function in sex determination and gonad development requires further investigation.
Some female-biased genes were also identified inT. ovatusgonad transcriptome, such as cytochrome P450 2K1-like. Cytochrome P450 family 19 subfamily A (cyp19a) is a female-related gene inPelodiscus sinensiandCarassius auratus. Cyp 19 catalyzes the conversion of androgens to estrogens and is critical in sex differentiation (Chen, Jiang,Gu, & Shi, 2014; Liang, Meng, Cao, Li, & Zou, 2019). Cytochrome P450 2K1-like gene (Scaffold_782.536), another member of the P450 family,is also upregulated in the ovary ofT. ovatus, indicating its potential role in ovary differentiation. Foxl2 is a female sex-determining gene in vertebrates (Li & Gui, 2018; Mei & Gui, 2015; Peng et al., 2015; Tian et al.,2019). InAndrias davidianus, 21 forkhead box (Fox) genes were identified, of which 18 genes exhibited sexually dimorphic expression (15 ovary-biased and 3 testis-biased genes) (Hu, Xiao, Wang, Tian, & Meng,2018).
Some genes involved in gonadal development and gametogenesis were also identified in ourT. ovatusgonad transcriptome analysis,includinggsdf,gnrhr, andzps(Tian et al., 2019). In rainbow trout,gsdfwas identified to regulate the proliferation of primordial germ cells(PGCs) and spermatogonia (Sawatari et al., 2007), and the overexpression ofgsdfin Nile tilapia lead to testis differentiation in females(Kaneko et al., 2015). InS. sihama,gsdfshowed a male-biased pattern of expression (Tian et al., 2019). InT. ovatus, thegsdfgene was up-regulated in the testis, which is concordant with these reports.
Gnrhris related to testicular function maintenance in goats,Sparus aurata, andS. sihama(Forner-Piquer et al., 2019; Han et al., 2019; Tian et al., 2019). InT. ovatus,gnrhrwas upregulated in the testis, which is concordant with these previous studies.
Zpsis involved in ovarian function maintenance in mice (Hu et al.,2012). InS. aurata, all of the fourZPmRNA isoforms are transcribed in the ovary (Modig et al., 2006). InT. ovatus,Zpsis upregulated in the ovaries, which was similar to the findings of other studies.
In this study, the pro-opiomelanocortin-like (Scaffold 745.327) was identified to be a testis-specific gene by RT-qPCR. In the eel pituitary,the most abundant transcript is codes for pro-opiomelanocortin, which is the precursor of hormones of the melanocortin system, and interacts with the reproductive system (Ager-Wick et al., 2013).Pro-opiomelanocortin was found to be highly expressed before spawning compared with post-spawning inL. rohita, indicating possible roles in gonad maturation (Sahu et al., 2013). Pro-opiomelanocortin is related to testicular function maintenance in monkeys (EI, Ramaswamy, Sahu,& Plant, 2000).
Fox c1 is related to cancer and stress responses (Gong et al., 2019;Zhang, 2019).Let-7bacts downstream ofHif-1αto and promotes hypoxia-mediated cell proliferation through the downregulation of Forkhead box H1 (Fox h1) in zebrafish (Huang et al., 2017). In this study,Foxc1(Scaffold 671.293) was identified to be a testis-specific gene, andFoxh1(Scaffold 169.213) was identified to be an ovarian upregulated gene by RT-qPCR. These results suggest that various genes within one family have different functions.
Aquaporin 1 overexpression in the tunica vaginalis probably cause adult primary hydrocele testis (Hattori et al., 2014). In goose, aquaporin 1 is specifically expressed in the testis, which suggests that aquaporin 1 is related to water homeostasis regulation in the reproductive system of male (Skowronski, Skowronski, Leska, Robak, & Nielsen, 2009). Aquaporin 1b, which is synthesized in oocytes, is essential for mediating water uptake into eel oocytes (Kagawa et al., 2011), indicating different roles between aquaporin 1 and aquaporin 1b. In this study, aquaporin-1 was identified to be upregulated in the testis by RT-qPCR, which was consistent with the findings of previous research.
Of the top 20 KEGG pathways enriched by the DEGs between ovaries and testes ofT. ovatus, fatty acid biosynthesis is associated with the gonadal development in the sea urchinStrongylocentrotus(Wang, Ding,Ding, & Chang, 2019). The fatty acid degradation pathway appears to be associated with ovarian development of the swimming crab (Yu et al.,2015). The proteasome pathway is related to gonadal development and sex differentiation inSpodoptera litura(Sun et al., 2019), and is also implicated in testicular function in mice (Hou et al., 2014), as well as Chinese mitten crab (Wang et al., 2012). The pyrimidine metabolism pathway is involved in ovarian function of humans (RoyChoudhury et al., 2016; Wei, Xin, Wang, & Hao, 2018). The glycosaminoglycan biosynthesis pathway is associated with ovarian follicle activation in zebrafish (Zhu, Pardeshi, Chen, & Ge, 2018). These previous studies thus support our results that the DEGs between ovaries and testes ofT. ovatusare significantly enriched in those pathways involved in gonadal development, sex decision, and gonadal function.
Credit author statement
Pingping He: Experimental operation, Data curation, Writing-Original draft preparation, Peng Zhu: Funding Acquisition, Pinyuan Wei: Software, Xiaofei Zhuo: Sample collection, Wei Li: Data curation,Wenbin Zhao: Formal analysis, Xiaohan Chen: Genome Resources,Yong Lin: Project administration, Jinxia Peng: Writing- Review &Editing.
Declaration of competing interest
The authors declare that there is no conflicts of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant No. 31660740), the Fundamental Research Funds for nonprofit research institutes under Guangxi Zhuang Autonomous Region (Grant No. GXIF-2016-19), Science and Technology Major Project of Guangxi [Grant number AA18242031], and Guangxi Key Laboratory of Aquatic Genetic Breeding and Healthy Aquaculture Opening Fund (Grant No. 16-380-45-B-3). We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Appendix B. Supplementary data
Supplementary data related to this article can be found at htt ps://doi.org/10.1016/j.aaf.2020.09.007.
 Aquaculture and Fisheries2022年1期
Aquaculture and Fisheries2022年1期
- Aquaculture and Fisheries的其它文章
- A review of gynogenesis manipulation in aquatic animals
- Tilapia Lake Virus (TiLV) disease: Current status of understanding
- Classification and morphology of circulating haemocytes in the razor clam Sinonovacula constricta
- Potential probiotic and health fostering effect of host gut-derived Enterococcus faecalis on freshwater prawn, Macrobrachium rosenbergii
- Utility of gillnets for selectively targeting penaeids off Iran
- Refining tickler chains for penaeid trawls
