Tilapia Lake Virus (TiLV) disease: Current status of understanding
Nilv Aih, Anirn Pul, Tnmoy Gon Choudhury, Himdri Sh
a Department of Fisheries, Government of Tripura, Tripura, 799006, India
b ICAR-Central Institute of Freshwater Aquaculture, Kausalyaganga, Bhubaneswar, 751002, India
c Department of Aquatic Health & Environment, College of Fisheries, Central Agricultural University (Imphal), Lembucherra, Tripura, 799210, India
Keywords:Co-habitation Diagnosis Tilapinevirus Syncytial Tilapia lake virus
ABSTRACT Tilapia Lake Virus (TiLV) disease is an emerging and transboundary disease of tilapia cultures, causing mortality up to 90% globally. TiLV is a negative sense single stranded RNA virus belongs to family Amnoonviridae, genus Tilapinevirus and species Tilapia tilapinevirus. The first TiLV outbreak to fishes was reported from Israel followed by other countries viz., Ecuador, Colombia, Egypt, Thailand, Chinese Taipei, India, Malaysia, Bangladesh,Uganda, Tanzania, Peru, Mexico, Philippines, Indonesia, and USA. All the life stages of Tilapia (belonging to the family Cichlidae) are vulnerable to TiLV infection. However, river barb and giant gourami have also been found susceptible to TiLV infection. The virus infects the vital organs of the fish, including eyes, brain, and liver. The notable pathological finding of this disease includes syncytial cell formation and massive hepatocellular necrosis with pyknotic and karyolytic nuclei in the liver cells of infected fish. The disease is very contagious and spreads through both horizontal and vertical transmission. Several sensitive and rapid molecular diagnostic tools like reverse transcriptase polymerase chain reaction (RT-PCR), RT-quantitative PCR (RT-qPCR), loop-mediated isothermal amplification (LAMP) have been developed for early detection of the virus. Till date, no comprehensive control measures have been developed throughout the globe, although aggressive work on this line is going on. Implementations of strict good management practices, including quarantine protocols, are the only available option to combat the outbreak and spread of the disease. This review emphasizes the etiology,occurrence and distribution, mode of transmission, pathology and pathogenesis, diagnosis, possible control measures, and challenges of TiLV disease.
1. Introduction
Tilapia culture is one of the fastest-growing and lucrative trades in the aquaculture industry. Due to high protein content, large size, rapid growth and palatability tilapia culture has taken a significant acceleration in the recent past. The worldwide production of tilapia and other cichlids was estimated at 6.3 million metric tons in 2018 (FAO, 2019).This species is being cultured in many countries, and some of the largest producers are China, Thailand, Ecuador, Egypt, and Indonesia, whereas the largest importer is United States (Harvey, 2016). Tilapia cultivation provides a major avenue of earning for the farming community (Cleasby et al., 2014; Gomna, 2011). Besides their food values, they are also used for controlling algae and mosquito larvae in the aquatic environment.However, the intensification of the culture system is making them prone to different emerging pathogens (Abowei, Briyai, & Bassey, 2011; Bigarr´e et al., 2009). Until 2009, no viral diseases were reported in Tilapia.However, during the summer of 2009, enormous mortalities of both wild and farmed hybrid tilapia (O. niloticus×O. aureus) were observed in different parts of Israel, and the etiological agent was subsequently identified in 2013 as Tilapia Lake Virus (TiLV) (Eyngor et al., 2014).Later on, the disease was reported from other Asian, African, and American countries (Ahasan et al., 2020; Amal et al., 2018; Behera et al.,2018; Dong et al., 2017; ; Ferguson et al., 2014; Nicholson et al., 2017;OIE, 2017a,b,cOIE, 2018a,b,c; Pulido, Mora, Hung, Dong, & Senapin,2019; Surachetpong et al., 2017; Tsofack et al., 2017).
A similar type of disease was also observed in the fingerings of Nile tilapia (O. niloticus) in Ecuador named syncytial hepatitis of tilapia(SHT) (Ferguson et al., 2014). After the first report of TiLV from Israel,the virus associated with SHT found to have close genetic similarity with TiLV reported by Eyngor et al. (2014). In Thailand, a disease with unknown etiology, initially called ‘tilapia one-month mortality syndrome’,was subsequently identified as TiLV disease (Surachetpong et al., 2017;Tattiyapong, Dachavichitlead, & Surachetpong, 2017). The virus has initially been classified as a novel Orthomyxo-like virus despite its variability in the histopathological responses (Bacharach et al., 2016).The outbreaks of TiLV disease during the summer season were reported to cause massive mortality in different life stages of tilapia and can transmit from one pond to the other (Eyngor et al., 2014). The outbreak of TiLV has not been reported to affect any other species in polyculture system incorporating tilapia, suggesting the specificity of the disease only to tilapia. However, recently in Malaysia, TiLV disease has been reported from river carp (Abdullah et al., 2018). High mortalities (80%–90%) were observed in Israel, Ecuador, and Colombia, however, no second outbreak was observed once mortalities were ceased in a particular pond (Bacharach et al., 2016; Del-Pozo et al., 2017; Eyngor et al., 2014; Ferguson et al., 2014; Tsofack et al., 2017). Thus, there is an urgent requirement for the researchers to understand the current emergence of this viral disease. This review highlights the etiology,occurrence, distribution, mode of transmission, pathology, pathogenesis, diagnosis, and the possible control measures and challenges for this newly emerged disease and its way forward.
2. Etiology
The aetiological agent of Tilapia Lake Virus disease (TiLVD) is Tilapia Lake Virus (TiLV), confirmed by Koch’s postulates (Tattiyapong et al., 2017). It is an enveloped, negative-sense ssRNA virus with ten segments in the genome; genome length is 10,323 kb with diameter varies between 55 and 100 nm (Bacharach et al., 2016; Del-Pozo et al.,2017; Eyngor et al., 2014; Surachetpong et al., 2017; Tattiyapong et al.,2017). Each of the ten segments has open reading frames (ORF), which may have code for 14 different proteins (Acharya et al., 2019). All the ten segments of TiLV show some sort of genetic closeness with different viruses of the family Orthomyxoviridae. Among all the segments,segment 1 shows weak sequence homology with PB1 subunit of influenza C virus, and it is presumed to encode for the polymerase of TiLV(Bacharach et al., 2016). Segment 1 is the largest in TiLV genome, with a nucleotide size of 1.641 kb. Segment 1, 2, 4 and 5 shows very strong homology with Dhori virus, Segment 6, 7 and 8 shows evolutionary closeness with the influenza virus whereas segment 3 has some weak homology with the infectious salmon anemia virus (Acharya et al.,2019). Sequence alignment of segments using Geneious software’s revealed a unique feature, which is common with influenza viruses. The virus was initially categorized under Orthomyxo-like virus (Eyngor et al., 2014) but later on classed into family Amnoonviridae and genusTilapinevirusand speciesTilapia tilapinevirus(Adams et al., 2017; Chengula, Mutoloki, Evensen, & Munang’andu, 2019). The complete genome sequences of TiLV from Israel, Thailand, Bangladesh, USA, Peru and Ecuador have been reported (Ahasan et al., 2020; Al-Hussinee, Subramaniam, Ahasan, Keleher, & Waltzek, 2018; Bacharach et al., 2016;Chaput et al., 2020; Pulido et al., 2019; Subramaniam, Ferguson,Kabuusu, & Waltzek, 2019; Surachetpong et al., 2017; Surachetpong,Roy, & Nicholson, 2020).
There are also reports of co-infection with different bacterial pathogens in the TiLV infected fishes (Nicholson et al., 2017; Surachetpong et al., 2017). Different species ofAeromonas viz A. veronii, A. ichthiosmia,A. enteropaelogenes, A. hydrophilia, Flavobacteriumspp.andStreptococcusspp, were detected from TiLV-infected fishes (Abdullah et al., 2018;Nicholson et al., 2017). Recently, a case of natural co-infection of TiLV andA. veroniiin Malaysian red hybrid tilapia has been reported (Amal et al., 2018). Co-infection study revealed that the TiLV withAeromonasspp.was more frequent than with other bacterial species. The reason explained to be the availability of impressive virulence factors and their ability to adapt to the adverse condition (Nicholson, Mon-on, Jaemwimol, Tattiyapong, & Surachetpong, 2020). This information indicates the requirement of further studies to confirm the possible involvement of other pathogens during TiLV infection.
3. Occurrence and distribution
During the summer of 2009, mass mortality of farmed tilapia all over Israel raises the eyebrow of the scientific community. However, the first scientific reporting of the disease as TiLV was from Israel in the year 2013 (Eyngor et al., 2014), and in the same year, it was reported from Ecuador (Ferguson et al., 2014). However, the genomic epidemiology study by Thawornwattana et al. (2020) suggests the origin of TiLV be between 2003 and 2009, 5–10 years before the first report of the virus in Israel. TiLV is widely distributed in Asian countriesviz. Thailand (Dong et al. 2017a; Surachetpong et al., 2017), Chinese Taipei (OIE, 2017b),India (Behera et al., 2018), Malaysia (Abdullah et al., 2018; Amal et al.,2018), Indonesia (Koesharyani et al., 2018), Philippines (OIE, 2017a)and Bangladesh (Debnath et al., 2020; Chaput et al., 2020). The disease has also been reported from Colombia (Tsofack et al., 2017), Peru (OIE,2018c), USA (Ahasan et al., 2020), Mexico (OIE, 2018b), and African countries like Egypt (Fathi et al., 2017; Nicholson et al., 2017), Uganda and Tanzania (parts of Lake Victoria) (Mugimba et al., 2018) (Fig. 1).Genome sequence analysis of viral isolates from different countries shows 97.2%–99% nucleotide sequence identity between Ecuador and Israel strains, 95.38%–95.68% in Thailand-Peru, 96.32%–96.71% in Thailand-Israel, and 96.89%–97.13% in Israel-Peru strain (Pulido et al.,2019). The sequences obtained from the Ecuador strain were compared with Thailand and Peru isolates and found 95.77% and 97.24% similarity, respectively. However, this study revealed that isolates from Thailand and Peru are more closely related to Israel but show genetic distant between themselves, which suggests both the countries may have infected with the TiLV virus of Israel origin. More than 45 countries are believed to be at high risk of TiLV, including different southeast Asian countries and Sub-Saharan African countries (Dong, Ataguba, et al.,2017; Hounmanou et al., 2017). The widespread occurrence of this virus across the tropical region of the world from America to Asia and Africa poses a significant threat of serious pandemic to tilapia farming. Many African countries imports tilapia seeds from TiLV reported countries,and this possesses the risk of cross-country and potential cross-continents spread of the virus (Dong, Siriroob, et al., 2017). As there are many reports of mortality of tilapias worldwide, samples should be screened for the TiLV with the advanced diagnostic tools to understand the spread, geographic distribution, and transboundary nature of the disease. Infected populations of Tilapia are the sole reported source of TiLV, but the source of the virus is not known (Dong, Ataguba,et al., 2017; Ferguson et al., 2014).
Numbers of tilapia species, namely Nile tilapia (O. niloticus), red tilapia (Oreochromissp.), and hybrid tilapia (O. niloticus×O. aureus)have been reported to be affected by TiLV, causing mortality up to 90%(Amal et al., 2018; Behera et al., 2018; Dong, Ataguba, et al., 2017;Eyngor et al., 2014; Ferguson et al., 2014; Mugimba et al., 2019; Surachetpong et al., 2017). Besides, TiLV has been identified from various species of wild tilapias (Tilapia zilli, O. aureus, Sarotherodon galilaeus, and Tristamellasimonis intermedia) in Israel (Eyngor et al., 2014) (Table 1).Furthermore, all the life stages of Tilapia viz., fertilized eggs, yolk-sac larvae, fries, fingerlings, and adults are susceptible to TiLV (Dong,Ataguba, et al., 2017; Dong, Siriroob, et al., 2017; Ferguson et al., 2014;Surachetpong et al., 2017). Upon co-habitation with Tilapia in the culture pond, fishes like Indian Major Carps (Labeo rohita, Catla catlaandCirrhinus mrigala), milkfish (Chanos chanos), pearl spot (Etroplus suratensis),Mugil cephalus,Cyprinus carpioandLiza ramadadid not show any mortality during the outbreak of disease (Behera et al., 2018; Eyngor et al., 2014; Fathi et al., 2017; Pradhan et al., 2020). Another study by Jaemwimol et al. (2018) showed resistance to TiLV infection by warm water species likeTrichogaster pectoralis,Pangasianodon hypophtthalmus,Clarius macrocephalus,Channa striata,Anabas testudineus,C. carpio,Barbodes gonionotus Lates calcarifer. However, for the first time other than tilapia, the river barb (Barbonymus schwanenfeldii) and giant gourami (Osphronemus goramy) were found infected by TiLV (Abdullah et al., 2018; Jaemwimol et al., 2018).
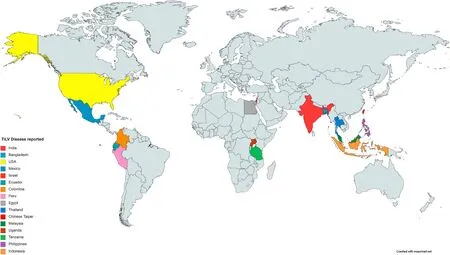
Fig. 1. Global distribution of Tilapia Lake Virus disease (*Tanzania and Uganda detected TiLV RNA in healthy Tilapia).
The mortality rate in TiLV infected farmed fishes varies in different geographical locations. In Israel, above 80% mortality been observed in TiLV affected farms (Eyngor et al., 2014). In the case of Ecuador tilapia culture systems, mortality in a tune of 10%–80% was observed within 4–7 days of post-transfer to grow-out ponds (Ferguson et al., 2014). In Thailand, 20%–90% mortality was observed within the first month of grow-out cage cultures (Dong, Ataguba, et al., 2017; Surachetpong et al.,2017). In India, 80%–90% (Behera et al., 2018), in Chinese Taipei 6.4%(OIE, 2017b) and in Malaysia 15% (OIE, 2017c) mortality were observed in TilV infected tilapia. However, the average mortality level was 9.2% (range 5%–15%) in TiLV infected farms of Egypt (Fathi et al.,2017). In Timah lake of Malaysia, mortality stopped after 50 days of infection, whereas peak mortality observed between 8 and 13 days post-infection (Abdullah et al., 2018). The wide range and diverse pattern of mortality for TiLV infected fish in different countries may be due to the status of fish and farm and geographical locations. Further study is required to conclude the diverse pattern of mortality for TiLV disease.
TiLV outbreaks were mostly reported during the hot summer in Israel, Egypt, Thailand, especially during May to October (Eyngor et al.,2014; Fathi et al., 2017) and even up to November in Ecuador (Ferguson et al., 2014). However, TiLV positive samples were also reported in Thailand during October and May (Surachetpong et al., 2017). Clinical outbreaks of the disease were reported from the above countries within the water temperature range of 22–32◦C. Predisposing factors like low initial body weight at the time of transfer to on-growing ponds, low water temperatures, high dissolved oxygen (DO), high stocking density,and more numbers of production cycles per year increase the natural incidence and severity of TiLV disease in farmed tilapia (Kabuusu, Aire,Stroup, Macpherson, & Ferguson, 2018).
4. Mode of transmission
The TiLV infection can be transmitted from one fish to another(waterborne and physical transfer), which has been experimentally demonstrated. In the co-habitation study, cumulative mortality of 50%with the clinical disease was observed (Eyngor et al., 2014). Adult Tilapia may have asymptomatic infections as the apparently healthy adults were also found positive for TiLV (Mugimba et al., 2018; Senapin,Shyam, Meemetta, Rattanarojpong, & Dong, 2018). It may be due to the immune system that makes them resistant to disease, and they may act as asymptomatic carriers that pass the virus to their offspring. The vertical transfer potential of this virus has been demonstrated by several authors (Dong et al., 2020; Yamkasem, Tattiyapong, Kamlangdee, &Surachetpong, 2019). Post IM-injection challenge with TiLV, the viral particle was detected in the reproductive organs, serum, and eggs (both fertilized and unfertilized) of Tilapia. Due to the aggressive feeding habit of tilapia, it is believed that intragastric route is the prime route of infection as intra peritoneal route needs to pass the first line of defence before they get entry into the body (Pierezan, Yun, Surachetpong, &Soto, 2019). However, whether it is intragastrically inoculated or intracoelomically injected the presence of TiLV observed in vital organs like brain and liver (Pierezan et al., 2019). Detection of virus in the mucus of Tilapia suggests mucus as a possible mode of horizontal transmission through co-habitation (Eyngor et al., 2014; Liamnimitr,Thammatorn, Sonicha, Tattiyapong, & Surachetpong, 2018). A co-habitation study revealed that healthy Tilapia might acquire infection within 1–2 days after exposure to infected fish (Liamnimitr et al.,2018). The transfer of the virus may be directly correlated with the stress status of the animal, as stress is the most crucial risk factor for the disease (Ferguson et al., 2014).
5. Pathology and pathogenesis
Many generalized clinical symptoms are associated with TiLV disease; however, high mortality associated with corneal opacity may be one of the overt clinical signs during outbreaks though the subsequent confirmatory diagnosis is must (Eyngor et al., 2014). Fishes were identified in both natural and cultured conditions with symptoms like anorexia, poor body condition, severe anemia, bilateral exophthalmia,skin abrasion and congestion, scale protrusion, and abdominal swelling(Jaemwimol et al., 2018; Surachetpong et al., 2017; Tsofack et al.,2017). Furthermore, symptoms like loss of appetite, pale coloration of the body, gathering in the bottom, sluggish movement, abnormal swimming, and avoidance of schooling before death were also observed during the outbreak (Dong, Ataguba, et al., 2017). There are also reports of in-apparent infection with TiLV in farmed Tilapia (Senapin et al.,2018), demonstrating that even clinically healthy adult and fingerlings of Tilapia with no signs of diseases or mortality can be diagnosed with TiLV positive.
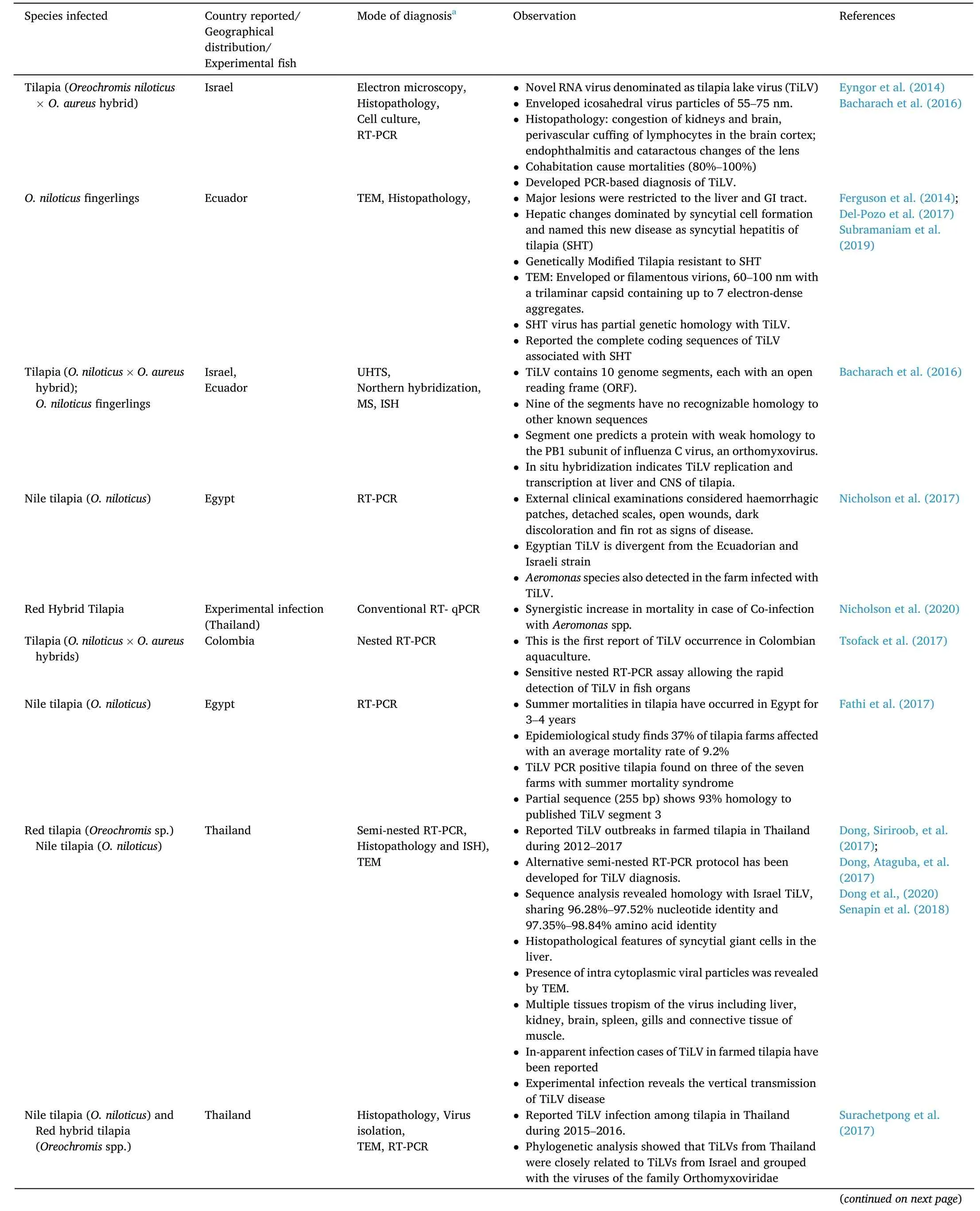
Table 1 Incidence of Tilapia Lake Virus (TiLV) disease.
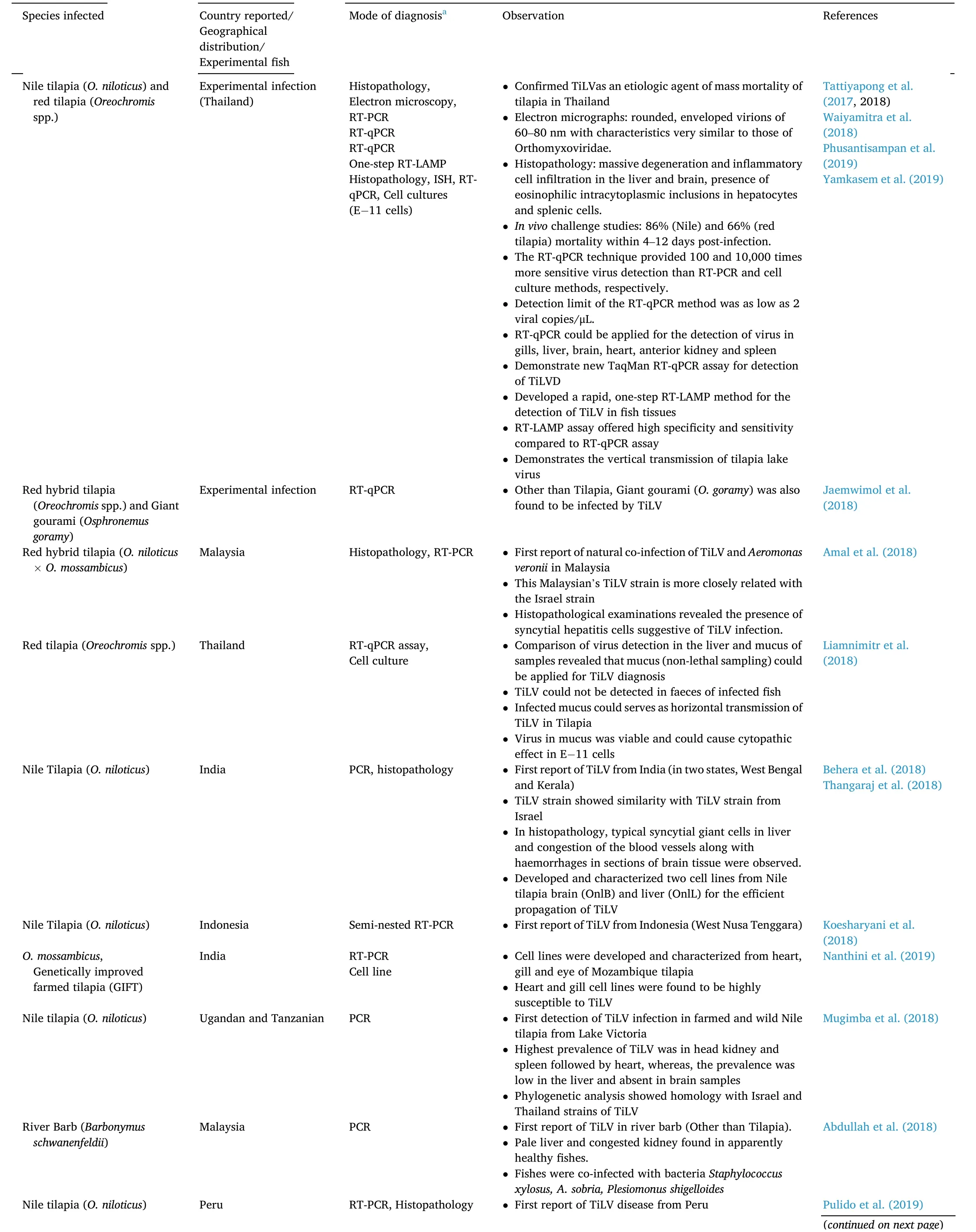
Table 1 (continued)
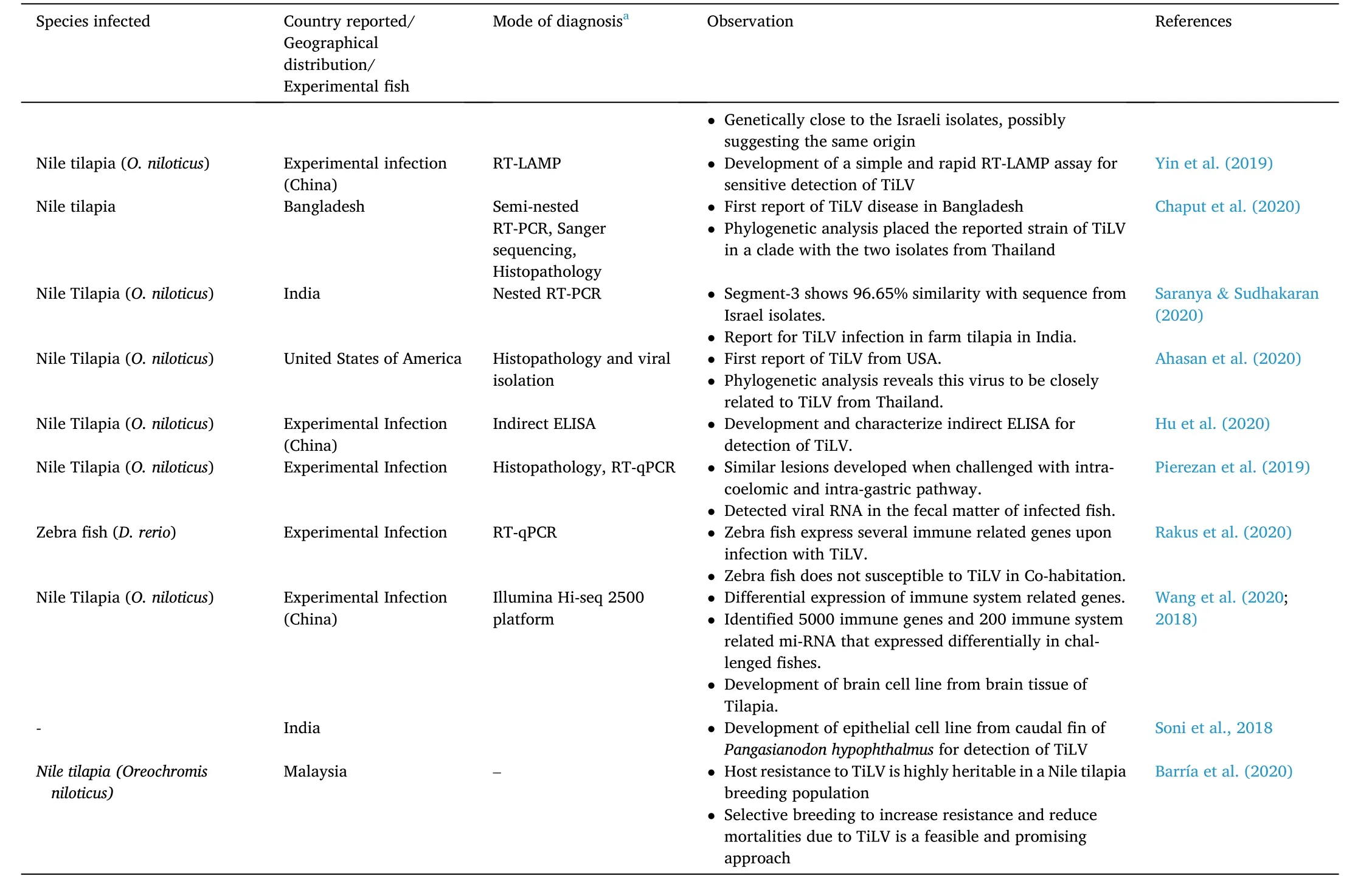
Table 1 (continued)
Histopathological examination of brains of TiLV infected fishes revealed multifocal haemorrhages, edema, proliferative glial cells and capillary congestion (Amal et al., 2018; Eyngor et al., 2014; Tsofack et al., 2017). In the liver of infected fish, syncytial cell formation and massive hepatocellular necrosis with pyknotic and karyolitic nuclei most commonly observed (Behera et al., 2018; Dong, Siriroob, et al., 2017;Ferguson et al., 2014; Jaemwimol et al., 2018; Senapin et al., 2018;Tsofack et al., 2017). In addition, eosinophilic intracytoplasmic inclusion bodies were present in the liver cells and splenic cells of infected fish. Multiple necrotic foci were observed in the anterior kidney with increased melanomacrophage centers (MMCs), and in spleen dispersion of melanin granules were observed (Tattiyapong et al., 2017). Mild congestion and pale liver were observed in apparently healthy river barb in Malaysia (Abdullah et al., 2018).
RNA-Seq analysis of separately prepared mRNA and miRNA library from TiLV infected fish reveals that Tilapia initiates a robust immune response to TiLV infection, around 5000 genes, and 200 miRNA expressed differently, which are associated with the immune system(Wang et al., 2020). It is further required to study the interaction of mRNA and mi RNA to understand the mechanism of pathogenesis of TiLV fully.
Regardless of the route of transmission, Tilapia developed identical clinical and histopathological lesions, when challenged with both intracoelomic and intragastric pathway (Pierezan et al., 2019). Artificial infection by intraperitoneal injection of TiLV in Tilapia suggests developing protective immunity and triggers rapid antibody response within seven days (Tattiyapong, Dechavichitlead, Waltzek, & Surachetpong,2020). The intraperitonial injection of TiLV in zebrafish induced series of histopathological changes in liver, kidney and spleen of fishes and turn on expression of several immune genes, but less mortality rate than tilapia (Rakus et al., 2020). Thus, zebrafishes can be used as future model fish for studying the pathogenicity and host-pathogen interaction.
Electron microscope analysis revealed the presence of 75–80 nm viral particles in the intra-cytoplasmic membrane and cytoplasmic area(most abundantly) (Eyngor et al., 2014). The presence of TiLV within the diseased hepatocytes was confirmed by transmission electron microscopy that in turn also confirmed the earlier reports of syncytial hepatitis(Del-Pozo et al., 2017).In-situhybridization revealed that TiLV replication and transcription occurs at the central nervous system, liver,kidney, gills, spleen, gastrointestinal tract, and connective muscle tissue(Dong, Siriroob, et al., 2017; Tsofack et al., 2017) (Table 1).
6. Diagnosis
Although many authors described various pathological lesions and clinical signs of TiLV infected fish, many of these clinical signs are similar to other viral and bacterial diseases. Besides that, subclinical infection of TiLV also has been reported (Senapin et al., 2018), which makes it further challenging to diagnose the presence of virus with only observing clinical lesions (Hu et al., 2020). However, high level of mortality in Tilapia population with mostly corneal opacity can be considered one of the signs of this disease. Other clinical symptoms include severe anemia, skin erosion, pale coloration of gills, and swollen abdomen (Dong, Siriroob, et al., 2017; Ferguson et al., 2014). Many authors have described specific and sensitive PCR based diagnostic of TiLV disease. Several Reverse-transcriptase PCR (RT-PCR), Nested RT-PCR, RT-quantitative PCR (RT-qPCR), and TiLV sensitive cell lines are also developed for precise diagnosis of this threat.
For PCR diagnosis purposes, brain, kidney, liver, and spleen of the infected fish can be used, however; brain and liver are the most targeted organ for the sampling of TiLV (Amal et al., 2018; Dong, Siriroob, et al.,2017; Fathi et al., 2017; Nicholson et al., 2017; Surachetpong et al.,2017; Tsofack et al., 2017). Mucus and faeces are used as non-lethal mode of sampling for viral detection in several other finfishes. Mucus from the TiLV infected moribund fishes can be kept up to 30 days at−20◦C and use for detecting viral RNA (Liamnimitr et al., 2018). Other non-destructive screening includes blood & liver biopsy for TiLV detection (Chiamkunakorn et al., 2019). RNA detected in faeces of Tilapia, suggesting faces as a possible source of horizontal transmission and non-lethal mode of sampling for TiLV detection (Pierezan et al.,2019).
Recently a very sensitive nested RT-PCR assay developed, which detects segment 3 of the virus (Tsofack et al., 2017). Nested PCR with outer primer (Nested ext-1’ and Nested ext-2) for 491 bp long PCR fragment from TiLV clone 7450 (GenBank Accession No. KJ605629) was amplified, and a set of inner primer (ME1 and 7450/150R/ME2) were used to re-amplify the diluted product derived from external primer to produce a 250 bp product (Eyngor et al., 2014) (Table 2). The different products from external and internal primer alone, and the combination of both the primer was compared, and it was found that nested PCR was much more specific with the detection level as low as seven numbers of copies of TiLV as compared to 70,000 in case of single PCR reaction(Tsofack et al., 2017). However, out of the 20 nucleotides of primer,13–18 nucleotides were matched with fish genes resulted in the false positive outcome. To counter this problem, a semi-nested PCR was developed (Dong, Siriroob, et al., 2017) by using only Nested ext 1 and ME1 in first step PCR, omitting primer Nested ext-2, followed by second step PCR using primer 7450/150R/ME2 and ME1. This has resulted in increasing the analytical sensitivity of the method, which has been able to detect 7.5 viral copies in a reaction. Thus the heavily infected individual produces two bands of 415 bp and 250 bp size and lightly infected fish produces a single 250 bp (Dong, Siriroob, et al., 2017). But, nested PCR is prone to contaminations, sometimes difficult for interpretation.Several primers used for the amplification of different segments of TiLV are tabulated in Table 2.
Tattiyapong, Sirikanchana, & Surachetpong (2017) developed SYBR green-based RT-qPCR technique, which was more sensitive than RT-PCR and virus isolation in cell culture methods. As low as two viral copies/μl can be detected by this RT-qPCR method. Besides, fish tissues, including gills, liver, brain, heart, anterior kidney and spleen could be used for TiLV detection by this RT-qPCR technique. In addition, the RT-qPCR showed high specificity against TiLV without amplification of important fish pathogens and fish genes. TaqMan probe-based RT-qPCR method was developed and validated for TiLV (Waiyamitra et al., 2018).Waiyamitra et al. (2018) compared the TaqMan probe-based and SYBR green-based RT-qPCR for detection of TiLV. Though, as per principal,TaqMan qPCR considered to be more specific due to the inclusion of the probe, but, both the methods surprisingly performed equally well for detection of TiLV (Waiyamitra et al., 2018) which also reported by Tajadini et al. (2014). Nicholson, Rawiwan & Surachetpong (2018)documented the whole process of virus detection, starting from sample collection, RNA isolation, c-DNA synthesis etc. for conventional PCR and SYBR green-based RT-qPCR.
Despite the fact that TaqMan qPCR is generally considered to be more specific due to the inclusion of the probe, we have found that our TiLV TaqMan and SYBR green RT-qPCR assays perform equally well, as reported by others comparing TaqMan and SYBR green assays (Tajadini et al., 2014).
Loop-Mediated isothermal amplification (LAMP) technique was developed by Phusantisampan, Tattiyapong, Mutrakulcharoen, Sriariyanun, & Surachetpong, (2019) based on the primer designed against the segment-3 of the virus isolated from Thailand. The sensitivity and specificity were compared between the newly developed LAMP method with previously developed RT-qPCR (Tattiyapong et al., 2017) technique. The specificity of both the methods was likely to be same, but the sensitivity of RT-qPCR was ten times more than the RT-LAMP method.But the RT-LAMP technique has the additional advantage of detecting virus through colorimetric changes in the tube, unlike RT-qPCR, which is an expensive and tedious technique. In another study, RT-LAMP technique is developed by designing primer against the Segment 1 of TiLV genome (Yin et al., 2019). Carefully designed primers achieved 100% specificity and sensitivity against the conserved region of the S1 gene of TiLV. Still, none of the PCR diagnosis techniques of TiLV have been fully validated and it solely depends on detection of viral nucleic acids (Hu et al., 2020).
Few cell lines were found to be suitable for TiLV with a different level of visible cytopathic effect (CPE) (Eyngor et al., 2014; Soni, Pradhan,Swaminathan, & Sood, 2018; Tsofack et al., 2017). Only E−11 out of the eight established cell line (E−11, CHSE-214, BF-2, BB, EPC, KF-1, RTG-2 and FHM) was found to be permissive to the TiLV virus (Eyngor et al.,2014; Tsofack et al., 2017). In E−11 cell line, CPE was visible after five to seven days of inoculation and after nine to ten days, the disintegration of cell monolayer was observed (Eyngor et al., 2014). A comparative study between tilapia ovary (TO-2), brain (OmB and TmB) shows TO-2 does not support replication of TiLV virus (Tsofack et al., 2017). It was found that E−11 cells were more convenient in detecting the CPE post infection in relatively short time and OmB cells could help us monitor CPE in longer time post-infection. However, monitoring CPE in TmB cells is difficult as it does not support clear plaque formation. In another study, infectingPuntius fasciatuscaudal fin (CFF) cell line with TiLV showed cytopathic effects on 3 days post-infection (DPI), and by 6–7 DPI more than 50% of the cells were detached (Behera et al., 2019). Two cell lines of Nile tilapia brain (OnlB) and liver (OnlL) were developed, which recorded maximum TiLV titer of 107.3±0.05and 107.0±0.96tissue culture infectious doses (TCID)50/ml, in OnlL and OnlB respectively. Experimental infection (in vivo) of Tilapia with infected cell culture supernatant (OnlL and OnlB) reproduced symptoms of disease and mortality in 10 days post-infection and virus re-isolate from challenged fish(Thangaraj et al., 2018). Nanthini et al. (2019) developed three cell lines from heart, gill and eye ofO. mossambicusand from these, heart and gill cell lines were highly susceptible to TiLV and found to be useful for its isolation. Recently brain cell lines developed from Tilapia and hybrid of snakeshead fishes (Channa argus(♂) ×Channa maculate(♀)) show potential for replicating TiLV virus (Wang et al., 2018, 2019; 2019). Above findings suggests the potential of using the cell lines for detection and studying the epidemiology of the disease.
New, low cost and improved diagnostic methods like enzyme-linked immune sorbent assay (ELISA), rapid antigen strip test should also be encouraged (Hu et al., 2020; Jansen, Dong, & Mohan, 2019; Tattiyapong et al., 2020). Antibody-based diagnosis methods are a cost-effective option in field-level diagnosis, but it is also proved to be very effective in studying the history and status of infection of the virus in the host cell.Indirect enzyme-linked immunosorbent assay (iELISA) can be used to monitor the changes in the production of antibodies in the cell that canhelp to develop commercial vaccines (Tattiyapong et al., 2020). Indirect ELISA technique shows more relative specificity compared to IFA and semi-nested PCR but less in relative sensitivity compared to semi-nested PCR (Hu et al., 2020). Hu et al. (2020) successfully produced and characterized anti- IgM antibody in the lab, which is found to be very specific and a key to further improvise ELISA based diagnosis technique of TiLV at commercial level.
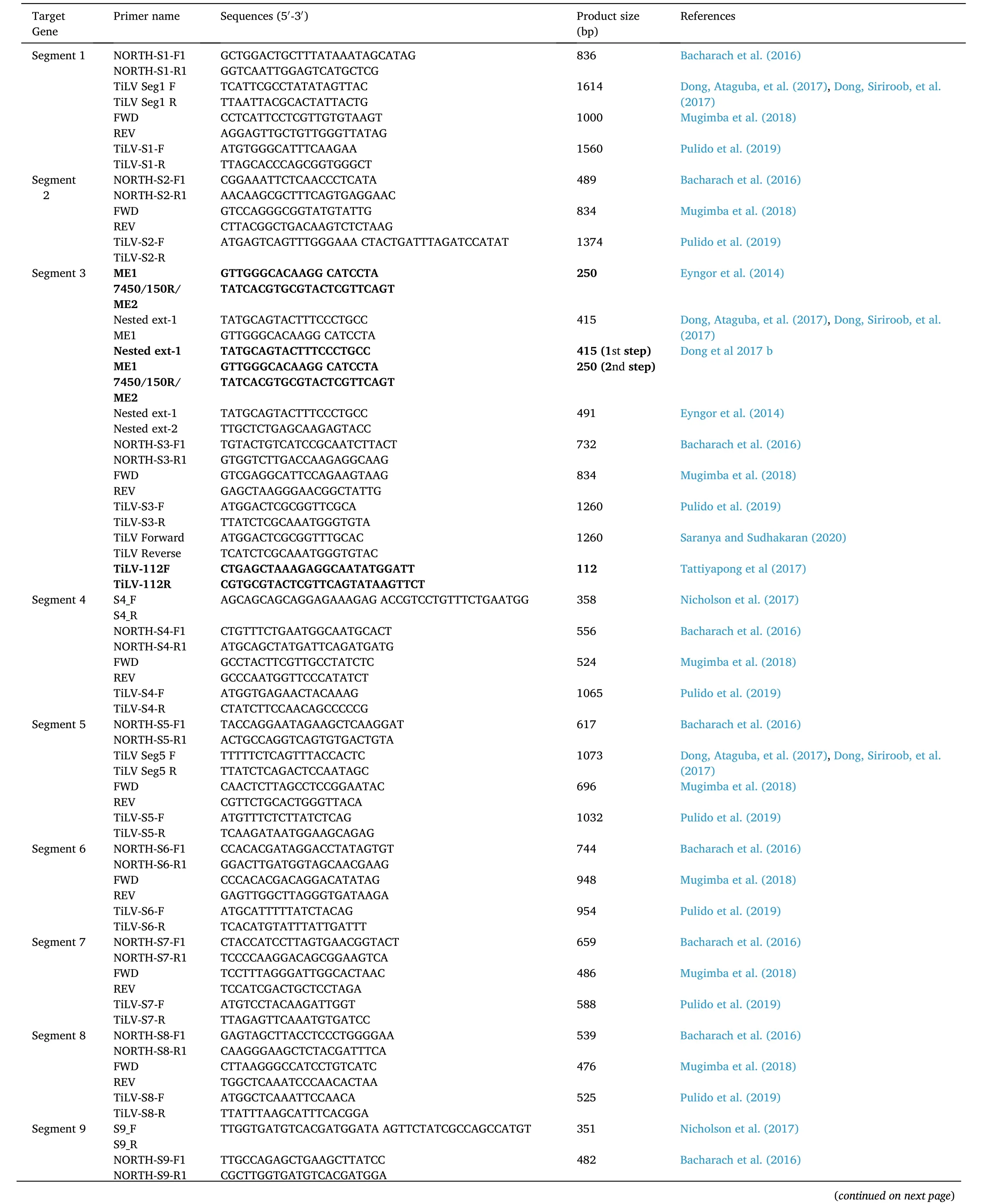
Table 2 Primers used for detection of different segments of Tilapia Lake Virus (TiLV).
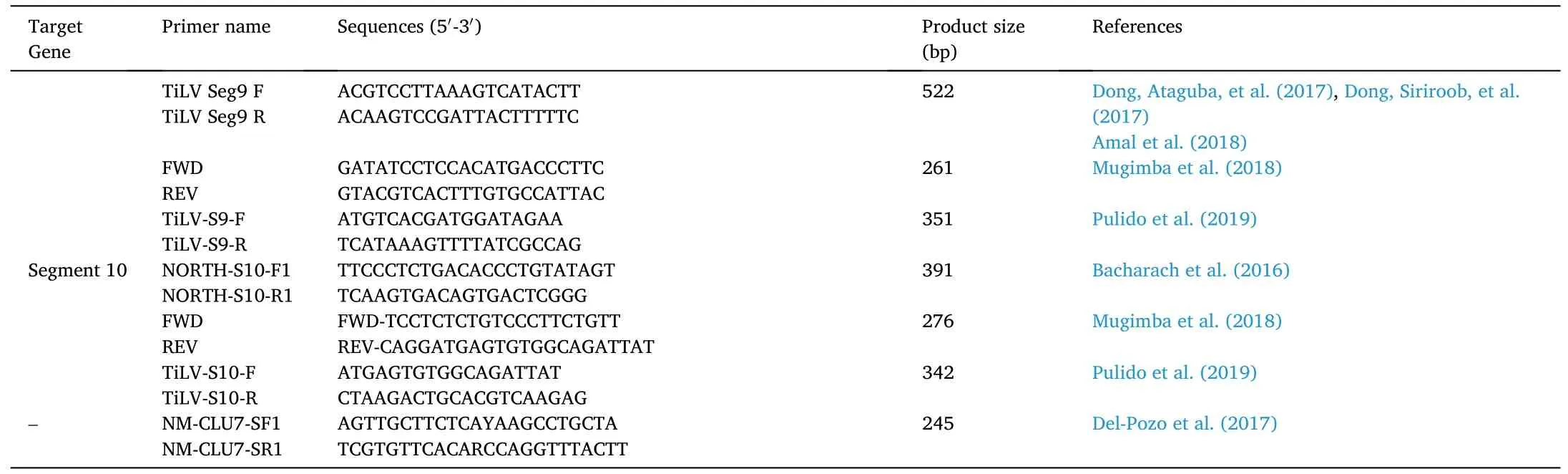
Table 2 (continued)
In-situ hybridization of segment-1 for brain and segment-3 for liver tissue was carried out for genomic RNA and mRNA. The analysis found that, in brain, hybridization signals were confined to leptomeninges,adjacent to the blood vessels, whereas in liver, signals were observed in hepatocytes, and the formation of some multinucleated cells was also observed. Dong, Siriroob, et al. (2017) found strongest signals in both peripheral membrane of liver and hepatocytes. Primary and secondary lamellae of gill and hematopoietic tissue of kidney showed the strong positive result.
7. Control measures and challenges
Till date, no effective means of therapeutic measures have been discovered for controlling TiLV. Hence, the only option left is the implementation of good management practices, bio-security, and exhaustive quarantine protocols. Affordable disease prevention and control measures including good farming practices, good water quality management, nutrition and sanitation should be followed. FAO and OIE provide control measures and bio-security procedures for TiLV disease management. Farmers should use seeds (larvae and fingerlings) of Tilapia of their farms or collect from local or regional farmers or hatcheries where no record of abnormalities, mass mortality, or diseases reported earlier. Also, Tilapia from TiLV vulnerable places/countries should be avoided to restrict the movement of virus from infected places to the uninfected regions. However, under unavoidable circumstances strict screening and quarantine should be followed (OIE, 2017d).Research found 14 days of post freezing fillet from subclinically infected Tilapia with TiLV contains no virulent viral particles (Thammatorn,Rawiwan, & Surachetpong, 2019). Such freezing procedure may be followed by commercial exporters from different countries to prevent further spread of this virus.
In the recent development, few authors have claimed the viral infection can be prevented (in vitro) by using common disinfectant(Jaemwimol, Sirikanchana, Tattiyapong, Mongkolsuk, & Surachetpong,2019; Soto, Yun, & Surachetpong, 2019). Furtherin vivostudy is required to establish the possible impact of these disinfectants in fish and environment. The dose, effective temperature and efficacy of the disinfectant are mentioned in the Table 3.
Bacharach and Eldar (2019) experimentally developed a live attenuated vaccine against TiLV in Israel. Upon administration through bathing or intraperitoneal injection of this live attenuated TiLV vaccine to wild TiLV challenged Tilapia (through cohabitation) resulted in 56%–58% relative percent survival within 21 days of post-vaccination. Author patented this vaccine (US20160354458A1) but yet to commercialize.
A literature survey revealed that the fish survived from an outbreak of TiLV were relatively resistant to the virus in the next phase of culture,which makes the researchers hopeful regarding successful control of this particular disease in the near future (Eyngor et al., 2014). Different strains of Tilapia showed different level of susceptibility towards this virus (Ferguson et al., 2014), which gives hope of existence of TiLV resistant strains (OIE, 2017d). Such evidence encourages opting for gene profiling to promote genetically improved Tilapia strains that are resistant to TiLV. Eyngor et al. (2014) and Barría et al. (2020) have also suggested selective breeding of resistant tilapia species or the fish survived during the TiLV outbreak can be a very effective measure to control this devastating disease in the future. However, till then, every country that is expecting the disease in their culture system should immediately go for a very extensive surveillance programme for this virus, and a country without the virus report can be declared as a disease-free zone. Yang, Lu, Lin, Chen, Liao (2018) suggested valuable models/tools for assessing the population transmission dynamics of TiLV disease that allows a more rigorous evaluation of different control strategies to reduce the disease.
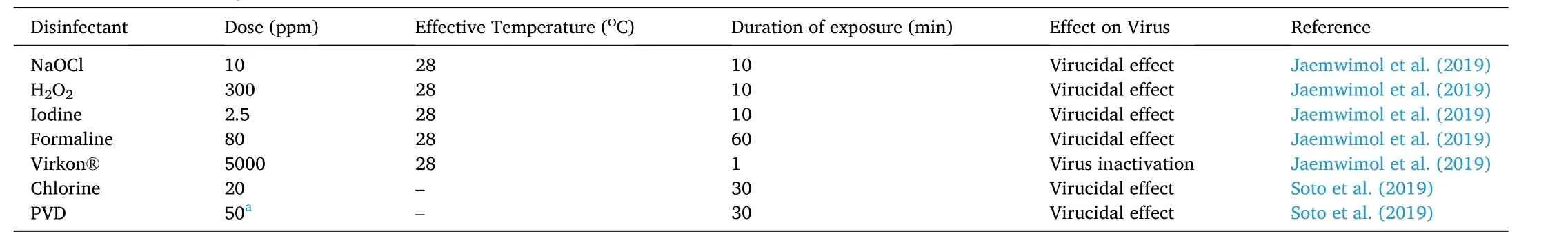
Table 3 Common disinfectants against tilapia lake virus.
However, there are many challenges for the available control measures of TiLV disease as the bio-security and quarantine procedures provided by FAO and OIE are for commercially larger scale farm and less applied for small-scale level. But, in many countries fish farmers have too little facilities and infrastructures with no/less knowledge for quarantine of fish, hence not able to implement the same. Additionally,many countries have minimum or inadequate opportunities to improve farmers’ disease management skills (Hounmanou et al., 2017). The available vaccine are still in process of development which requires some levels of technology, infrastructure, and financial assistance.Moreover, the efficiency of the vaccine as prophylactic measures is a matter of concern as it mostly causes disease among young fish whose immune system is not well developed.
8. Conclusion and future perspectives
Undoubtedly, TiLV is the most devastating disease for the tilapiabased aquaculture systems known to date. The virus has been discovered very recently; however, it is expected that the virus may be present in the system from long back and persisted unidentified. Furthermore,after its first report from Isreal, many countries started reporting the same in a very short span of time, which indicates the vulnerability of Tilapia culture system globally in the future. As the disease seems to spread very fast and affects a huge number of fish in different countries in a short period of time, the disease can be considered as a transboundary animal disease (TAD). There is a fear of the global spread of TiLV disease, as many countries import fish products/seeds from commercial hatcheries where TiLV has been reported. If the TiLV disease(with the known mortalities of up to 90%) spread globally, that will impact heavily among tilapia-producing countries and drastically reduce the worldwide tilapia production.
Furthermore, there are unexplored possibilities of host switching and adaptation of TiLV over time, and it may come up with a devastating result in the future. Future direction should emphasizing on finding out the host range or probable presence of any reservoir host in the system which helps in reccurance of the disease. Development of new and sensitive cell lines will also help in speedy recovery and propagation of virus for different basic studies. Since TiLV is a recently identified virus,continuous capacity building is needed to train and strengthen laboratory technicians and field aquatic health officers to detect and respond to TiLV outbreaks. The recent advances in diagnostics tools can be instrumental in diagnosing this particular virus however, hunt for more specific and sensitive method for diagnosis should go on. Also, extensive work may directed towards effectiveness of different drugs and chemicals against this particular virus. Studies on pathogenesis and prognosis of the diseases may also helps in development of adequate preventive measure. Farmers involve in tilapia farming are mainly lower income group; therefore it is necessary to develop low cost, precise diagnostic methods which do not require extensive laboratory facilities. Hence,more focused research needed for development of non-lethal sero diagogsis which will provide more accurate and instant diagnosis at farmers field. Although many questions related to the virus taxonomy and epidemiology remains unanswered, but the reports on its resistance to repeat infection opens a window for the possibilities of development of vaccine and resistant strains against TiLV. We all know the importance of vaccine for controlling viral or any other diseases. There must be extensive involvement of scientific work for development of effective vaccine against this deadly virus. Development of SPF tilapia’s or selectively bred resistant tilapia may be the ultimate answer to this deadly disease and extensive work may directed in this line.
Acknowledgements
The authors would like to thank the Dean, College of Fisheries, CAU(Imphal), Lembucherra, Tripura, 799210, India and Director ICAR-CIFA,Bhubaneswar, 751002, India for providing the necessary facilities. The contribution of Dr. Dibyendu Kamilya, College of Fisheries, Tripura for English editing is highly acknowledged.
 Aquaculture and Fisheries2022年1期
Aquaculture and Fisheries2022年1期
- Aquaculture and Fisheries的其它文章
- A review of gynogenesis manipulation in aquatic animals
- Classification and morphology of circulating haemocytes in the razor clam Sinonovacula constricta
- Potential probiotic and health fostering effect of host gut-derived Enterococcus faecalis on freshwater prawn, Macrobrachium rosenbergii
- Utility of gillnets for selectively targeting penaeids off Iran
- Refining tickler chains for penaeid trawls
- The deformation characteristics and flow field around knotless polyethylene netting based on fluid structure interaction (FSI)one-way coupling
