Characterization and expression analysis of sox3 in medaka gonads
Quan Pu, Yuan Ma, Ying Zhong, Jing Guo, Lang Gui,**, Mingyou Li,*
a International Research Center for Marine Biosciences, Ministry of Science and Technology, Shanghai Ocean University, Shanghai, 201306, China
b Key Laboratory of Exploration and Utilization of Aquatic Genetic Resources, Ministry of Education, Shanghai Ocean University, Shanghai, 201306, China
Keywords:Medaka sox3 Gonad development Nervous system development
ABSTRACT Numerous studies have showed that sox3 is involved in neurogenesis and sex differentiation in vertebrates.However, the accurate expression pattern is still unclear in fish. In this study, medaka sox3 was isolated and its expression patterns were compared with germ cell gene vasa in adult gonads. By reverse transcriptionpolymerase chain reaction (RT-PCR) analysis, sox3 RNA expression was detected in the brain, eyes and gonads. By chromogenic and fluorescent in situ hybridization, sox3 was present in the oogonia and early stages of oocytes as well as granulosa cells and theca cells in the later stages; in the testis, sox3 was occurred in the Sertoli cells and sperm with a specific signal in the location of chromatid body of sperm; in the embryos, sox3 was expressed in the central nervous. These results suggest that medaka sox3 gene was occurred in somatic and germ cells in adult gonads of both sexes, involved in the process of spermatogenesis, as well as the development of the nervous system. This study provides a precise expression pattern of sox3 and suggests that sox3 may be involved in gonadal and nervous system development of fish.
1. Introduction
Thesox(Sry-related HMG box) gene family is related to the mammalian male-determining factorsrygene (Sinclair et al., 1990),described as the transcription factors that homological to the DNA-binding domain named high-mobility group (HMG-box), which can bind to the sequence (A/T) AACAAT.Soxgene is isolated from the mouse (Gubbay et al., 1990; Li, Hong, Gui, & Hong, 2012) and human(Sinclair et al., 1990) at the same time. Until now, more than 40 members of thesoxfamily are identified from a variety of species and perform different functions in organism (Guth & Wegner, 2008; Han et al., 2010).
Many studies have reported thatsoxfamily genes are involved in a wide range of development process, especially in sex determination(Gao et al., 2015; Lin, He, Gui, & Mei, 2020; She & Yang, 2015; Watanabe et al., 2016). Compared with othersoxgenes,sox3is a single-exon gene locates on the X chromosome which is expressed by progenitor cells(Foster & Graves, 1994), during the early development of central nervous system in vertebrates,sox3has a widely expression and plays important roles in embryonic and adult neurogenesis (Cheah & Thomas,2015; Hughes & Page, 2015; McAninch & Thomas, 2014; Okuda et al.,2006; Rogers et al., 2013; Takehana et al., 2014). In mouse,sox3is required for the formation of glial system of cerebellum (Cheah &Thomas, 2015) and hypothalamo-pituitary axis (Rizzoti et al., 2004). In chicken (Bylund, Andersson, Novitch, & Muhr, 2003) and zebrafish (Dee et al., 2008; Gou, Guo, Maulding, & Riley, 2018; Gou, Vemaraju, Sweet,Kwon, & Riley, 2018),sox3is involved in the neurogenesis and neural tube. Furthermore, in frog and Chinese loach,sox3has positive effects in the development of nervous system (Rogers, Archer, Cunningham,Grammer, & Casey, 2008; Xia et al., 2018).
Meanwhile,sox3plays an indispensable role in sex differentiation and gonadal development (Cheah & Thomas, 2015; Hughes & Page,2015; McAninch & Thomas, 2014; Okuda et al., 2006; Rogers et al.,2013; Takehana et al., 2014). In transgenic mouse, the mutation ofsox3results in male sex reversal in the uncommitted XX gonad (Sutton et al.,2011). The process of early spermatogenesis is blocked on account of a loss-function mutation ofsox3in the mouse testis (Raverot, Weiss, Park,Hurley, & Jameson, 2005). In fish, various expression profiles ofsox3reveal its distinct roles, mainly in testicular or ovarian development,such as black porgy (Shin, An, Park, Jeong, & Choi, 2009), red-spotted grouper (Yao, Zhou, Wang, Xia, & Gui, 2007) and catfish (Rajakumar& Senthilkumaran, 2014).Sox3Yloss-of-function causes XY sex reversal inOryzias dancena, but the expression pattern ofsox3has some contradictions as there is no detectable expression ofsox3in the gonads(Takehana et al., 2014). In zebrafish, knocking-out thesox3causes the development retardation of follicle and reduces fecundity (Hong et al.,2019). In brief, a unified and explicit expression pattern ofsox3is absented, hence much more studies on fish are needed.
Medaka is an excellent model for studying the development of gonads, since it’s the first fish with male determining gene (Matsuda et al.,2002), it also has a lot of strains with transgenes specifically expressed in gonads (Li et al., 2009; Li, Hong, Gui, & Hong, 2012). In this study,sox3was isolated and its RNA expression was analyzed by RT-PCR in embryogenesis and adult tissues. In addition,in situhybridization was performed by compared with the germ cell gene markervasaon gonads sections. In general,sox3occurred in both somatic and germ cells of gonadal and was closely related to the process of spermiogenesis.
2. Materials and methods
2.1. Fish and embryos
Medaka was maintained at a constant temperature and photoperiod,which was 26◦C in 14 h light to 10 h dark cycle. The developmental stages of embryos were described by the study previously (Iwamatsu,2004). All experiments carried out with medaka in this study were conformed with guidance of the Laboratory Animal of Shanghai Ocean University Committee.
2.2. Molecular cloning and analysis of medaka sox3
The cDNA ofsox3was obtained by searching the database (http://www.ensembl.org/index.html) and the results showed thatsox3was a single-exon gene. A pair of primer was designed by primer 6.0 in order to further confirm the cDNA sequence ofsox3(Table 1). Nucleotide and protein sequences ofsox3were performed and analyzed by National Centre for Biotechnology Information (NCBI) and DNAMAN 8.0. Vector NTI suite 11.0 was used for multiple alignment of Sox3 protein sequences. Phylogenetic tree was constructed by the Mega 7.0 package with the method of nighbor-joining(NJ).
2.3. RT-PCR
Total RNA was extracted from adult tissues and disparate stages of embryos from medaka by using TRIzol reagent (Invitrogen, Carlsbad,CA), and cDNA was synthesized with an oligo(dT)18primer by using MMLV reverse transcriptase (TaKaRa, Shiga, Japan) from 1 μg RNA sample. The expression ofsox3was detected by reverse transcriptionpolymerase chain reaction (RT-PCR) withβ-actinserved as internal control. The primers used forsox3(ATGTATAACATGATGGAAACC and CTACCTCTCACTCACATCT GA) andβ-actin(TTCAACAGCCCTGCCATGTA and CCTCCAATCCAGA CAGTAT) were listed at Table 1. The detailed PCR amplification procedure is described in our previous study(Zhu, Gui, Zhu, Li, & Li, 2018).
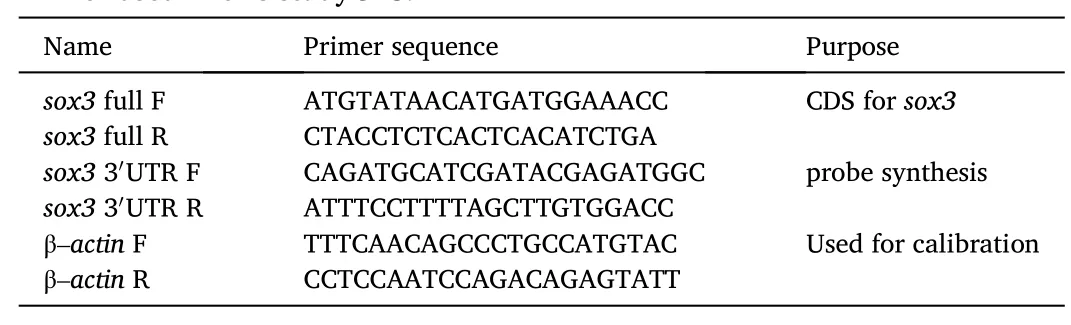
Table 1 Primer used in this studyCDS.
2.4. In situ hybridization
Gonadal sections and embryos chemicalin situhybridization (SISH)and fluorescentin situhybridization (FISH) were performed to detect the expression pattern of medakasox3, SISH was stained with BCIP/NBT and FISH was visualized by using the (TSA™) Plus Fluorescence Systems according to the manufactures’ instruction (Life Technologies, Carlsbad,CA) as described by our previous studies (Li et al., 2011; Yuan, Chen,Zhu, Yuan, & Li, 2018). Briefly, the pGEM-T vector contained 760 bp partial regions ofsox3ORF with 3′UTR was linearized for the synthesis of anti-sense and sense RNA probes, the T7 or SP6 promoter was selected according to the direction of transcription by using the digoxigenin(DIG) or FITC RNA Labelling Kit (Roche, Basel, Switzerland) forsox3andvasa. Nucleus was stained with DAPI, the Gold anti-fade reagent(Invitrogen, Carlsbad, CA) was used for sealing slides.
2.5. Microscopy
Leica TCS SP8 Laser Scanning Confocal Microscope (Leica, Germany)and Nikon Ds-Ri2 camera (Nikon, Tokyo, Japan) were used for observation and photography as described (Sun, Gui, Liu, Hong, & Li, 2020).
3. Results
3.1. Isolation of medaka sox3
The medakasox3ORF was 912 nt, and encoded 303 amino acids,which was 94%, 92%, 92%, 82%, 72% and 71% identical to that of zebrafish,Japanese eel,gibel carp,catfish,mouse and human (Fig. 1A and B). Medaka Sox3 had a highly conserved peptide of 99-aa called HMG-box that was almost identical in different species (Fig. 1B). The phylogenetic tree elucidated that the Sox3 formed two major branches in the evolution of vertebrates, with the closest relationship to black porgy (Acanthopagrus schlegelii) in the fish sub-clade (Fig. 2A). As thesox3-bearing regions on the chromosomes of different organisms contained the same genes, which meant thatsox3exhibits a conserved synteny from fish to human (Fig. 2B).
3.2. RT-PCR analysis of sox3
The expression ofsox3in adult tissues and embryo was detected by RT-PCR. In embryos,sox3was persistently expressed from 2-cell stage to fry and peaked in the gastrula stage during embryogenesis that meantsox3was maternally provided (Fig. 3A). In adult tissues,sox3had high expression in central nervous system such as brain and eyes, the signal was also detected in the gonads (Fig. 3B).
3.3. Embryonic expression of sox3 RNA by chromogenic ISH
Then, the expression ofsox3in the embryos was further explored by chromogenic ISH,sox3was widely expressed in the blastula and neurula embryos (Fig. 3C and D). Until the 3-day embryo, the signal showed a tendency toward centralization of the nervous system (Fig. 3E). In the 7-day embryos, the signal became more specific that enriched in the forebrain, midbrain, posterior brain and spine (Fig. 3F).
3.4. Gonadal expression of sox3 RNA by chromogenic ISH

Fig. 2. Phylogenetic tree and chromosome synteny of sox3. (A) Phylogenetic tree of sox3. Bootstrap values are given. Accession numbers were followed the organism.(B) Chromosome synteny of sox3. Chr, chromosome. Grp, linkage group. Numerals in parentheses are chromosomal positions (http://www.ensembl.org). Scale bars: 0.02.
SISH was performed on gonadal sections in order to further explore the subcellular distribution ofsox3expression. A mature ovary of medaka contains a few of oogonia and a mass of oocytes at different stages was selected to detectsox3expression. In the ovary, oocytes are surrounded by two kinds of somatic cells, one is the inner single layer granulosa cells and another is the outer layer theca cells. Granulosa cells can produce the estrogen, meanwhile, the steroidal precursors of estrogen are supplied by theca cells during the development. These hormones play essential roles in vitellogenesis, maturation of oocytes and secondary sex characteristics. By chromogenic ISH,sox3RNA was strongly expressed in oocytes at the early stages (I-II), but the signal in oogonia was hard to distinguished. In the later stages of oocytes (III–V),sox3signal was mainly detected in the somatic cells such as granulosa cells and theca cells (Fig. 3G and H). The signal ofsox3was not detected in the negative control that used the sense probe on the sections (data not shown).
In the medaka testis, there are a number of efferent cysts and each cyst contains different stages of germ cells. In addition, there are two types of somatic cells playing important roles in the development of testis, one is the Sertoli cells which also known as nurse cells, located in the spermatogenic cysts, the other is Leydig cells located between the spermatogenic cysts and produced the androgen hormones during the reproduction. Results showed thatsox3RNA was weakly expressed in Sertoli cells, but highly expressed in sperm, and a considerable amount of the signal was concentrated in the location of the chromatid body(Fig. 3I and J).
3.5. Ovarian expression of sox3 and vasa by FISH

Fig. 3. Expression of sox3 RNA. (A and B) RT-PCR analysis of medaka sox3 in embryos and adult tissues. (A) In the embryos, sox3 was expressed as early as two cells stage and peaked in the stages of gastrula, neurula and 3 day. (B) In the tissues, sox3 was high expressed in brain, eyes and gonads, the expression in the ovary was higher than in the testis. (C– F) In situ hybridization in embryos, sox3 had non-specific expression in the stages of blastula and neurula with a tendency toward centralization of the nervous system until third day. (G and H) Adult gonad sections were hybridized to antisense probe, sox3 was expressed in the early stages of oocysts (I−II), the signal of granulosa and theca cells in the later stages (III–V) could also be detected. (I and J) Sox3 occurred in sperm, a considerable amount of the signal was concentrated in the CB of sperm. I–V, stages of oocytes; gc, granulosa cells; tc, theca cells; sg, spermatogonia; sc, spermatocytes; st, spermatids; sm, sperm;se, Sertoli cells. di, diencephalon; ey, eye; mb, midbrain; hb, hindbrain; sc, spinal cord. Scale bars, 100 μm.
The chromogenic SISH had verified the specificity of thesox3expression in gonads, to precisely identify thesox3expression patterns,the co-localization ofsox3withvasa, which is a well-studied germ cell maker in multitudinous species including medaka, was further performed by using FISH (Li et al., 2009; Li, Zhao, Wei, Zhang, & Hong,2015; Yuan, Li, & Hong, 2014). In the ovary,vasasignal was expressed throughout oogenesis with the strongest signal in oogonia and enriched in the primary oocytes, its expression was continuously decreased with the development of oocytes (Fig. 4B).Sox3was co-localized withvasaat the early stages (I−II) of oocytes (Fig. 4A and C), besides, thesox3positive signal could also be detected in oogonia compared withvasa(Fig. 4C), andsox3was also occurred in granulosa cells and theca cells in the later stages (III–V) of oocytes, which was different fromvasa(Fig. 4D–F).
3.6. Testicular expression of sox3 and vasa by FISH
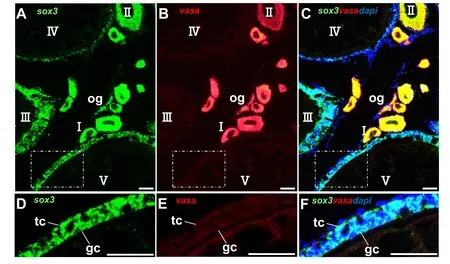
Fig. 4. Expression of sox3 and vasa RNA in the ovary. (A–F) Adult ovarian cryosections were hybridized to antisense probe. The signals were stained for the sox3(green) and vasa (red) by FISH. Nuclei were stained with DAPI (blue). (A) The signal of sox3 was detected in the germ cells in the early stages (I−II) and granulosa cells, theca cells in the later stages (III–V) of oocytes. (B) Vasa signal was expressed throughout oogenesis, with the strongest signal in oogonia and primary oocytes,the expression of vasa was continuously decreased with the development of oocytes. (C) The signal of sox3 and vasa was co-localized in the early stages (I−II) with the signal of oogonia can also be detected. (D–F) Enlargement of (A–C), sox3 was expressed in the granulosa cells and theca cells in the later stages of oocytes different from vasa. I–V, stages of oocytes. og, oogonia gc, granulosa cells; tc, theca cells. Scale bars, 100 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Meanwhile, the expression pattern ofsox3withvasawas compared on testicular sections by using FISH. In the testis, thevasasignal was abundant in spermatogonia, then reduced from spermatocytes, spermatids to sperm with the development of testis (Fig. 5B). In contrast, thesox3RNA was hardly detected in the spermatogonia and spermatocytes,but highly expressed in the sperm as well as weakly expressed in Sertoli cells, (Fig. 5A). In addition, the signal ofsox3showed a polarity phenomenon in the stage of round spermatid and became more concentration when developed into the elongated spermatid, expressed stronger in the location of chromatoid body (CB) of sperm (Fig. 5D–G).
4. Discussion
A lot of studies have demonstrated thatsox3gene plays essential roles in embryogenesis, neurogenesis and gonadal development in vertebrates (Cheah & Thomas, 2015; Okuda et al., 2006; Rogers et al.,2013). Although, the function ofsox3has been reported, the accurate expression pattern is still unclear. In this study, medakasox3was cloned and RNA expression in developing embryos and adult tissues were analyzed. The current results showed that the medakasox3contained thesoxBsubfamily sequence that encoded the HMG-box and theSoxpmotif which was in accordance with several other studies (Bergstrom,Young, Albrecht, & Eicher, 2000; Gao et al., 2015; Hughes & Page,2015) and highly conserved from fish to human (Focareta & Cole, 2016).
The early stages of the embryogenesis especially the neurogenesis is critical for fish. It is report that SoxB1 group genes are involved in these phases (Avilion et al., 2003; McAninch & Thomas, 2014; Nitta et al.,2006). In zebrafish, the central nervous system is destroyed and subsequently inhibits some aspects of neurogenesis when thesox3gene is knocked-down (Dee et al., 2007). In our present study,sox3RNA was detected in the tissues belonged to nervous system such as brain and eyes which suggested thatsox3may be correlated with the nervous system, same results are also obtained in the studies of frog (Archer, Jin,& Casey, 2011) and mouse (Cheah & Thomas, 2015). In addition,medakasox3was highly expressed in nervous system during embryogenesis with a few studies have demonstrated the same results in fish(Okuda et al., 2006; Wagner & Podrabsky, 2015). By the way, numerous studies support that the development of gonad is regulated primarily by the hypothalamic pituitary gonadal (HPG) axis of the neuroendocrine system in fish (Chen, Hu, & Zhu, 2013). HPG can affect the maturation of gonad by regulating gene expression, Piwil1 protein which is essential for germline maintenance and gonadal development is confirmed in turbot (Scophthalmus maximus) that regulated by HPG both in testis and ovary (Wang, Wang, & Liu, 2018). The results of tissues and embryos suggested that medakasox3gene may play a crucial role in the individual development and the development process associated with the HPG axis. However, more studies are needed for illuminating the exactly role and mechanism ofsox3in fish, especially the complex regulatory mechanisms in development processes associated with the HPG axis.
The present study revealed thatsox3RNA also expressed in the gonads, hints its roles on gonadal development in medaka, which was in consistent with that in teleost such asOryzias dancena(Takehana et al.,2014) and red-spotted grouper (Yao et al., 2007). In ovary, the ovonic communication between somatic cells and oocytes is important for oogenesis. During the follicular development, somatic cells are involved in regulation of maintaining cell homeostasis (Yuan et al., 2015), while apoptosis of granulosa cells is pivotal for follicular development through atresia of many early follicles to ensure growth and maturation of some dominant follicles (Matsuda, Inoue, Manabe, & Ohkura, 2012). According to the current study, thesox3positive signal occurred in oogonia and early stages of oocytes. More strikingly,sox3was expressed in the granulosa and theca cells. The presence ofsox3in young oocytes,oogonia and somatic cells in the later oocytes may be relevant to the development of ovary, as is the case in red-spotted grouper (Yao et al.,2007) and Japanese eel (Jeng et al., 2018), a high expression ofsox3during the ovary development, suggest thatsox3may play more important role in oogenesis.

Fig. 5. Expression of sox3 and vasa RNA in the testis. (A–G) Adult testicular cryosections were hybridized to antisense probe. The signals were stained for the sox3 RNA (green) and vasa (red) by FISH. Nuclei were stained with DAPI (blue). (A) The sox3 signal was hardly detected in the spermatogonia, spermatocytes and spermatids, peaked in sperm, weakly expressed in Sertoli cells. (B) The vasa signal was peaked in spermatogonia and persists at reduced levels from spermatocytes to sperm. (D–G) Enlargement of C, the sox3 signal was colocalized with vasa and dispersed in the cytoplasm at the stage of round spermatid, showed a polarity phenomenon in the stage of elongating spermatid, condensed to the final form corresponding to the mature CB at later stages. se, Sertoli cells; sc, spermatocytes; sg,spermatogonia; st, spermatid; sm, sperm. Scale bars: 25 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Sox3is reported to be an analog of SRY in testis and bounds up with testicular development (Bergstrom et al., 2000; Fernandino, Guilgur, &Somoza, 2006; Takehana et al., 2014). In the testis, medakasox3RNA was found in somatic cells, such as Sertoli cells, as well as highly expressed in sperm with a considerable amount of the signal concentrated in the location of CB of sperm like the case ofvasain medaka (Li et al., 2009; Li et al., 2015; Yuan et al., 2014). The schematic illustration expression patterns ofsox3RNA in medaka testis was showed in Fig. 6.In mammalian, CB is considered as the center for mRNA storage and germ cell processing (Ginter-Matuszewska et al., 2011). Vasaand germ cell nuclear factor are all reported to accumulate in CB in early spermatids (Nguyen Chi et al., 2009). Previous study shows that the CB of sperm is related to the nucleolus during spermatogenesis in the tilapia(Peruquetti, Taboga, & De Azeredo-Oliveira, 2010). Moreover, a study reports thatsox3indirectly stimulates the proliferation of somatic cells and spermatogonia induced by 11-ketotestosterone (11-KT) in adult catfish (Rajakumar & Senthilkumaran, 2016). Based on these data,sox3might play an important role in spermatogenesis of fish. However, more information aboutsox3in testis is needed.
In conclusion, we demonstrate the accurate expression pattern ofsox3in medaka and the expression in the CB of sperm firstly. The present results suggest thatsox3was present in the oogonia and early stages of oocytes as well as granulosa cells and theca cells in the later stages;sox3was occurred in the Sertoli cells and sperm with a specific signal in the location of chromatid body of sperm in the testis. Thus,sox3occurs in both the somatic and germ cells.
CRediT authorship contribution statement
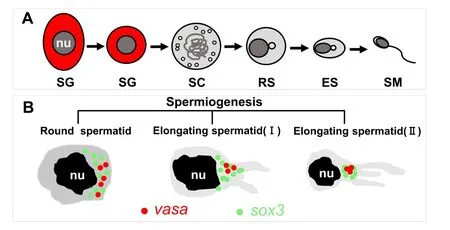
Fig. 6. Schematic illustration expression patterns of sox3 RNA in medaka testis.(A) Sperm formation. sg, spermatogonium; sc, primary spermatocytes; rs, round spermatid; es, elongated spermatid; sm, mature sperm; nu, nucleus. (B) In round spermatids, sox3 was dispersed in the cytoplasm colocalized with vasa and tended to concentrated in the CB. In elongating spermatids, CB start condensing, condensed to the final form corresponding to the mature CB.
M Li and L Gui conceive and design the experiments. Q Pu and Y Ma performed the experiments. Y Zhong, L Gui and M Li analyzed the data. J Guo contributed reagents and materials. Q Pu, L Gui and M Li wrote and corrected the manuscript.L. G and M. Y. L conceive and design the experiments. Y. L. Z. and Y. Z. performed the experiments. Y. L. Z., Y. Z., L.G and M. Y. L analyzed the data. J. G. and M. Y. L contributed reagents and materials. Y. L. Z., Y. Z., L. G and M. Y. L wrote and corrected the manuscript.
Acknowledgement
This work was supported by the National Key R & D Program of China (2018YFD0901205) and National Natural Science Foundation of China (31672700).
 Aquaculture and Fisheries2022年1期
Aquaculture and Fisheries2022年1期
- Aquaculture and Fisheries的其它文章
- A review of gynogenesis manipulation in aquatic animals
- Tilapia Lake Virus (TiLV) disease: Current status of understanding
- Classification and morphology of circulating haemocytes in the razor clam Sinonovacula constricta
- Potential probiotic and health fostering effect of host gut-derived Enterococcus faecalis on freshwater prawn, Macrobrachium rosenbergii
- Utility of gillnets for selectively targeting penaeids off Iran
- Refining tickler chains for penaeid trawls
