转运RNA衍生的小RNA功能及其研究方法
马剑峰,甘麦邻,朱砺,沈林園
转运RNA衍生的小RNA功能及其研究方法
马剑峰1,2,甘麦邻1,2,朱砺1,2,沈林園1,2
1. 四川农业大学动物科技学院,成都 611130 2. 四川农业大学,畜禽遗传资源发掘与创新利用四川省重点实验室,成都 611130
转运RNA衍生的小RNA (tRNA-derived small RNAs, tsRNA)是近年来发现的一类由成熟tRNA或tRNA前体通过特殊的作用机制加工产生的非编码RNA。根据其产生方式的不同,主要分为tRNA半分子(tRNA halves)和tRNA衍生片段(tRNA-derived RNA fragments, tRFs)两种类型。tsRNA广泛存在于各种生物体中,具有高度保守、结构稳定和组织表达特异性等特点。大量研究表明,多种细胞应激反应中tsRNA的表达显著上升,其参与应激反应的调控是一种保守的生物现象。tsRNA可通过调控转录本稳定性、调节翻译和作为表观遗传调控因子等方式在多种生物学过程中发挥重要作用,此外,tsRNA具有作为疾病生物标志物和治疗靶点的潜力,逐渐成为生物医学领域中的研究热点。本文从tsRNA的生物发生、功能和研究方法3个方面,对其研究现状进行了综述,以期为后续研究提供参考。
非编码RNA;转运RNA衍生片段;tsRNA;tRFs
人类基因组中编码蛋白质的基因仅占基因组序列的2%左右,而绝大多数转录本不编码蛋白质,研究表明至少有80%的序列被转录为非编码RNA (non-coding RNA, ncRNA)[1]。ncRNA包括核糖体RNA (ribosomal RNA, rRNA)、转运RNA (transfer RNA, tRNA)、microRNA、小干扰RNA (small interfering RNA, siRNA)、长链非编码RNA (long non-coding RNA, lncRNA)等[2],自20世纪50年代rRNA和tRNA被发现以来,ncRNA的功能开始被重视。rRNA和tRNA是中心法则的主要参与者,在蛋白质合成中发挥识别和转运的重要作用。随着高通量测序技术的快速发展,在动植物、细菌和真菌中都鉴定出一种长度大约为15~50 nt的ncRNA,因为来源于tRNA,所以将其称为tRNA-derived small RNAs (tsRNA);tsRNA结构稳定,保守性强,具有丰富的RNA修饰,在各种生物体中广泛表达[3~5]。
1977年,tsRNA最初由Borek等[6]在癌症患者的尿液中检测到,并且研究者们将tRNA分解产生的带有修饰的核苷酸作为癌症标志物[7],但当时tsRNA被认为是tRNA随机降解的产物。1992年,Saxena等[8]将血管生成素(Angiogenin, ANG)注射到非洲爪蛙()卵母细胞中,发现细胞中tRNA被降解,而rRNA和mRNA并没有被降解,证明了ANG是一种特异性水解tRNA的核酸酶。Thompson等[9]在氧化应激的酵母菌()、拟南芥()和HeLa细胞中检测到tsRNA的表达丰度增加,表明tRNA裂解是真核细胞应对氧化应激的一种保守的现象。直到2009年,tsRNA的产生机制才被初步揭示,Fu等[10]在哺乳动物细胞系(HepG2、HeLa、HEK293等)中发现,多种应激条件可以诱导血管生成素裂解tRNA的反密码子环产生tsRNA。
随着tsRNA产生机制的发现,tsRNA逐渐被证明参与多种调控过程,例如调节mRNA稳定性、翻译、rRNA合成、RNA逆转录等,进而调节细胞增殖、凋亡、迁移、周期等,在动物正常发育和疾病发生过程中发挥生物学功能[3]。tsRNA的表达具有组织和时空特异性,其异常表达可作为判断是否发生疾病的标志。但是,目前关于tsRNA的研究还处于起步阶段,其产生机制和具体生物学功能还没有被充分解析,其在正常发育和疾病发生过程中的作用还有待进一步的深入挖掘。本文总结了tsRNA的形成机制、生物学功能以及相关研究方法,以期为后续相关研究提供参考和借鉴。
1 tsRNA来源及命名原则
在早期的研究中,tsRNA被定义为RNA噪音,因此在测序数据的加工处理过程中被直接剔除掉。然而,随着对ncRNA的进一步理解和认识,同时生物信息学分析手段的不断提高,研究者们发现这些曾经被认为是垃圾序列的tsRNA片段其实是tRNA的降解产物,并不随机产生,可能发挥着特定的生物学功能。成熟tRNA以及前体tRNA在相关核酸酶的作用下通过断裂的方式形成短链的tsRNA。在真核生物中,tRNA基因由RNA聚合酶III转录生成tRNA前体(pre-tRNA)[11,12],成熟过程中tRNA前体的5ʹ前导序列和3ʹ尾巴序列分别被核糖核酸酶P (RNase P)和核糖核酸酶Z (RNase Z)切割,随后在tRNA核苷酸转移酶的作用下,在3ʹ端加上“CCA”序列[13],经过进一步碱基修饰和折叠等加工过程形成成熟的tRNA。当细胞在饥饿、缺氧、高温、氧化损伤等应激条件下[14],特定的核酸酶能对tRNA特定位点进行剪切形成具有功能性的tsRNA分子(表1)。对于tsRNA的命名目前还没有规范的标准,主要根据其生物学来源以及断裂位点命名,通常可以分为两种类型:tRNA半分子(tRNA-halves)和tRNA衍生片段(tRNA-derived fragments, tRFs)[14,24~26]。
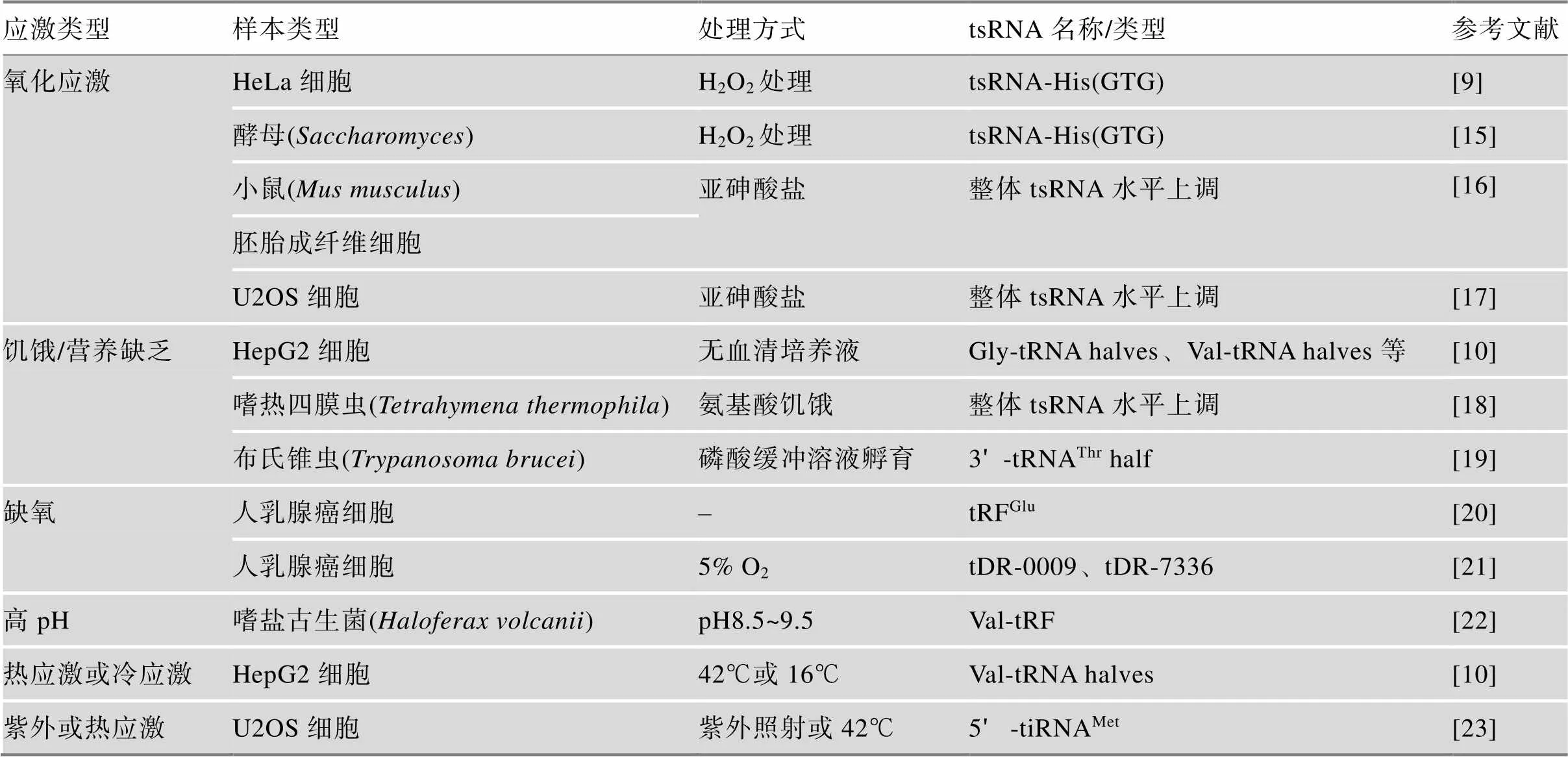
表1 应激诱导的相关tsRNA形成
在哺乳动物中tRNA-halves是在应激反应下由血管生成素特异性地剪切成熟体tRNA反密码子环形成的28~36 nt的小RNA[10,27],这一过程受到tRNA修饰的调控因此只能切割特定位点产生tRNA- halves[28],它也被称为tRNA-drived stress-induced small RNA (tiRNA),根据其对应的成熟tRNA序列可以分为5ʹ-tiRNA和3ʹ-tiRNA。此外,在氧化应激过程中,酵母中的Rny1p酶能促进tRNA的裂解[15],大肠杆菌中PrrC、大肠杆菌素D和E5也能特异性的切割tRNA的反密码子环[29]。
tRFs的长度为15~32 nt,根据其断裂位点可以分为5种类型[30,31]。如图1所示,tRF-1是前体tRNA在成熟过程中由RNase Z或ELAC2剪切3ʹ端形成的片段,因其含poly-U序列又称为3ʹU-tRF。tRF-5来自成熟tRNA的5ʹ端,其切割位点在成熟tRNA的D环或D环与反密码子环之间,可以进一步分为3种亚型(tRF-5a、tRF-5b、tRF-5c)。tRF-3由成熟tRNA裂解TΨC环所产生,其3ʹ末端带有成熟tRNA的CCA序列,根据长度不同可分为tRF-3a和tRF-3b。tRF-2则包含反密码子环及其两端的茎序列,i-tRF (internal tRNA fragments)对应的是成熟tRNA内部序列的片段,目前对这两种tRFs的研究还很少,其确切的产生机制还不完全清楚[32~34]。
2 tsRNA生物学功能
近年来研究表明,tsRNA广泛存在于细菌、真菌、动植物多物种中,并且在进化上具有保守性[35]。tsRNA是具有调节功能的小非编码RNA,通过多种机制发挥生物学作用,包括调节mRNA稳定性、调控翻译过程和作为表观遗传调控因子等。目前,已在不同类型的细胞系、组织或体液中发现了大量的tsRNA,相关研究进一步揭示了tsRNA可能参与疾病发生过程(表2)。
2.1 调节mRNA稳定性
tsRNA在结构和大小上与microRNA相似。研究早期,部分tsRNA被认为是microRNA[62],miR-1280通过抑制靶基因ROCK1表达来抑制直肠癌的转移,但后来发现miR-1208来源于tRNALeu[49]。tsRNA具有microRNA类似的作用,通过靶向mRNA调节其稳定性。Zhang等[39]在胃癌细胞中证实tRF-3019a来源于tRNAAlaAGC,通过靶向FBXO47 mRNA的3ʹUTR区降低其表达。Li等[63]在基因敲除的小鼠胚胎成纤维细胞中鉴定出tRNAHisGTG和tRNALeuCAG的片段,这些tsRNA不依赖于产生,随后的研究表明它们可以引导AGO2蛋白裂解靶mRNA。有研究报道在HEK293细胞中,5ʹ- tsRNA和3ʹ-tsRNA优先与AGO1、AGO3和AGO4结合,而不与AGO2结合[64]。Goodarzi等[20]发现了来自tRNAGlu、tRNAAsp、tRNAGly和tRNATyr的tsRNA,它们通过与RNA结合蛋白YBX1竞争性结合来抑制乳腺癌细胞中多种原癌基因转录本的稳定性。5ʹ- tRFGln能够抑制翻译,并且不需要与mRNA互补的靶序列,其作用机制依赖于保守残基“GG”[65]。因此tsRNA能发挥microRNA类似的调控功能,但其具体调控机制与microRNA并不完全相同。

图1 tsRNA分类与作用机制
2.2 调控蛋白质的翻译
tsRNA能够通过多种机制调控蛋白质的翻译过程。除了上述调节mRNA稳定性外,tsRNA还能通过影响核糖体的生物发生、结合核糖体、影响翻译起始过程等调控翻译过程。
2.2.1 影响核糖体的生物发生
核糖体是一个以rRNA为功能核心的复合物,核糖体生物发生是一个复杂而精确调控的细胞过程,不同的非编码RNA能参与调控其rRNA生成[66]。Couvillion等[67]研究表明,tsRNA作为原生动物四膜虫() rRNA剪接复合体的组成部分参与rRNA合成过程,剪接复合体中包含3ʹ-tsRNA、Twi12蛋白、Tan1蛋白和外切酶Xrn2,结合后Twi12稳定并激活Xrn2的外切酶活性,进而切割前体rRNA。Kim等[68]研究表明,HeLa细胞中3ʹ-tsRNALeuCAG可以结合核糖体蛋白RPS28的mRNA增强其翻译,RPS28翻译的减少会阻断18S核糖体RNA成熟并诱导细胞凋亡。随后在小鼠和人类的研究中也进一步证实了tsRNALeuCAG通过与编码区靶位点相互作用调节RPS28的翻译,表明其功能在不同物种中具有保守性[69]。
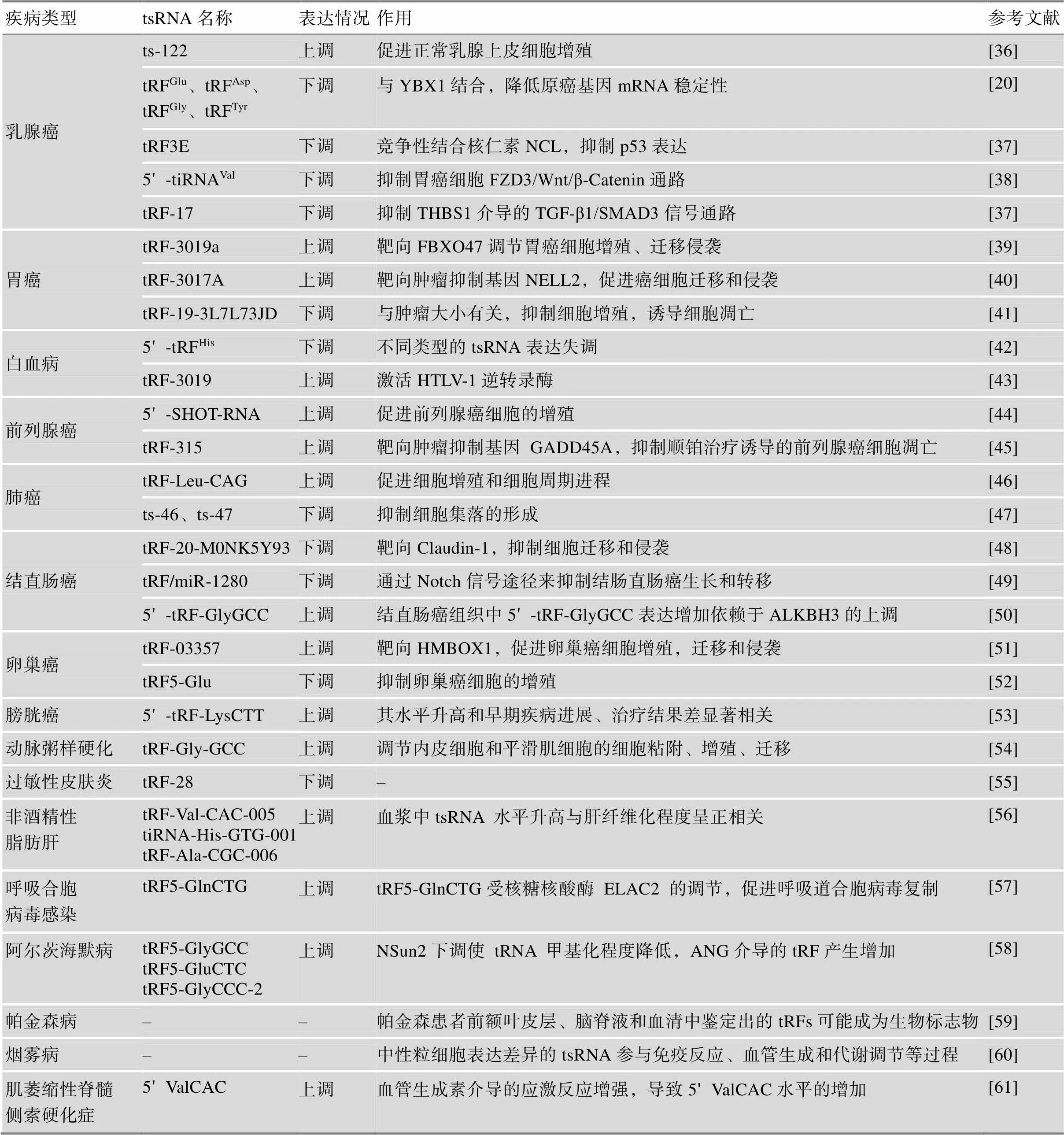
表2 与疾病相关的tsRNA
2.2.2 tsRNA与核糖体结合
tsRNA能与核糖体成分结合调控翻译过程。Gebetsberger等[22]对古细菌中与核糖体共同纯化的小RNA进行测序,发现在高pH值条件下生长的细胞中5ʹ-tRFVal表达丰度高,该片段与小核糖体亚基结合抑制整体翻译水平。最近的研究发现[70],Val-tRF与小核糖体亚基结合干扰翻译起始复合物的形成,将其导入酵母菌和大肠杆菌中也能够抑制蛋白质的合成。布鲁氏锥虫()中的3ʹ-tiRNAThr在古细菌和酵母菌中通过类似的作用来介导刺激翻译[19],这些研究表明,tsRNA与核糖体结合可能是一种跨物种的保守的调控机制。
2.2.3 调控翻译起始过程
翻译调控主要发生在翻译起始水平,因为这是蛋白质生物合成中最关键和限速的步骤。细胞受到应激后,真核起始因子eIF2α磷酸化导致应激颗粒的形成,可抑制整体蛋白质的合成[71],Yamasaki等[23]研究表明,亚砷酸盐、热休克或紫外线照射促进了血管生成素切割tRNA,细胞中tiRNA积累导致不依赖于eIF2α磷酸化的应激颗粒形成。Anderson研究小组[72,73]发现5ʹ-tiRNAAla和5ʹ-tiRNACys在其5ʹ末端带有4~5个鸟嘌呤残基的序列,能够形成分子间RNA G四聚体(RG4)来代替翻译起始因子eIF4复合物,进一步研究发现RG4与eIF4G结合,破坏40S核糖体对mRNA的识别,激活应激反应中的细胞保护作用[71]。此外,YB-1是细胞质中应激颗粒的组成部分,RG4结构能与YB-1结合,抑制翻译起始复合物形成[74]。
2.3 作为表观遗传调控因子
表观遗传学指在DNA序列不改变的情况下,基因功能发生可遗传变化。表观遗传学主要包括DNA甲基化、乙酰化、组蛋白修饰和ncRNA等,这些表观遗传信息和DNA序列信息共同参与调控基因的表达[75,76]。大量研究表明,tsRNA可以影响不同的表观遗传过程来调节基因的表达。Argonaute蛋白分为AGO和PIWI两个家族,其中AGO蛋白广泛表达而PIWI主要在生殖细胞中表达,一类长度为24~32 nt的piRNA能与PIWI蛋白形成复合物,通过表观遗传参与基因的调控[77,78]。tsRNA和piRNA的长度相似,也可以和PIWI蛋白相互作用来参与表观遗传的调控。Couvillion等[79]证实了tsRNA与四膜虫PIWI蛋白Twi12相关,两者表达丰富呈高度正相关,在之后研究中发现Twi12-tsRNA复合物能够调控核内RNA的加工[67]。Zhang等[80]发现,tsRNAs可以调节人单核细胞的组蛋白甲基化,这些单核细胞在IL-4刺激下分化为树突状细胞,在分化的细胞中鉴定出表达下调的piRNA,并与tRNAGlu的5'端序列相匹配,5ʹ-tsRNAGlu与PIWIL4相互作用促进H3K9甲基化从而抑制的转录。动植物基因组中含有大量的转座因子,它是一种对基因组有潜在危害的可移动基因组DNA,其转录活性通常被表观遗传标记抑制,如DNA甲基化和组蛋白修饰[81]。在着床前的胚胎发育过程中,大多数表观遗传标记消失,此过程中需要其他的机制来保护基因组[82]。tsRNA可以通过靶向转座子引物结合位点,抑制转座子活性。Schorn等[83]发现3ʹ-tsRNACCA在(SET domain bifurcated histone lysine methyltransferase 1)敲除的胚胎干细胞中高表达,而在(DNA methyltransferase 1)敲除的细胞中没有。Dnmt1是哺乳动物细胞中主要DNA甲基转移酶,介导DNA甲基化导致非LTR逆转录转座子的抑制[84],而Setdb1介导H3K9三甲基化(H3K9me3)在内源性和外源性逆转录转座子沉默中起着关键作用[85]。Schorn等[83]研究表明tsRNA可能作为新的表观遗传调控因子,调节LTR逆转录转座子的表达。
3 tsRNA研究方法
随着在更多物种中tsRNA被发现,深入研究其生物学功能以及参与的调控机制成为新的研究热点。目前,tsRNA的研究思路主要是通过高通量测序筛选出组间差异表达的tsRNA,然后验证确定目标分子,围绕目标分子进行体内外的功能验证和调控网络的构建。
3.1 tsRNA相关数据库
目前国内外学者对tsRNA的研究主要集中在癌症等疾病方面,tsRNA常作为与microRNA相似的小RNA进行研究,从测序数据中鉴别出哪些小RNA属于tsRNA尤为重要,为此研究者们建立了多个用于tsRNA研究的数据库(表3)。Kumar等[86]建立了第一个tsRNA的数据库tRFdb,包括了人和小鼠等8个物种的tsRNA序列,通过序列或ID可以检索到其相应的类型、tRNA基因位置和有关实验。tRF2Cancer[87]是一个用于癌症研究的数据库,支持用户上传测序数据和CLIP-Seq数据,以鉴定tsRNA、预测靶标和分析生物学功能等。Schuster等[88]开发了专门用于多物种精子RNA分析的数据库SpermBase,可用于精子tsRNA分析。PtRFdb是由Gupta等[89]开发的植物tsRNA数据库。Li等[30]开发的tRFtarget能有效地预测tsRNA靶基因的结合位点、结合区域以及稳定性,应用于tsRNA的功能研究。tsRBase[90]是目前最全面、最系统的tsRNA数据库,集成了多个物种的tsRNA表达模式,该数据库收集了来自不同组织/细胞系,或不同处理和遗传背景下的样本,有助于描述不同条件下tsRNA的特定表达模式。这些数据库能帮助研究者们对tsRNA进行深入研究,但目前仍缺乏对这些数据库的准确性和灵敏性的综合评估。
3.2 tsRNA鉴别和定量方法
微阵列和RNA-seq是高通量检测tsRNA表达情况的有效工具[33]。Farina等[36]利用微阵列分析了乳腺上皮细胞系中的tsRNA表达变化,检测到对肿瘤抑制因子RUNX1敏感的tsRNA。Wang等[91]对乳腺癌组织中tsRNA表达谱进行了研究,鉴定了其中30个显著差异的tsRNA。Xu等[92]对多发性骨髓瘤进行了RNA-seq,鉴定出33个上调和22个下调的tsRNA并对其功能进行了分析,发现tsRNA可能参与骨髓瘤的发生。目前使用广泛的小RNA测序建库流程存在一定缺陷,会导致测序结果产生系统性偏差[93]。与microRNA相比,tsRNA带有大量的RNA修饰会妨碍cDNA建库过程中5ʹ和3ʹ接头的连接和逆转录酶的作用[94],导致含有特殊修饰的tsRNA无法被检测,因此需要将总RNA进行预处理以提高测序文库构建的效率。最近,Shi等[95]开发了PANDORA-seq来克服这一问题。PANDORA-seq主要利用两种酶(AlkB和T4PNK)处理,将RNA末端的3ʹ-P和2ʹ3ʹ-CP转换为3ʹ-OH,5ʹ-OH转换为5ʹ-P,去掉了部分RNA甲基化,如m1A、m3C、m1G、m22G。其测序结果揭示了小鼠组织中与之前研究完全不同的小RNA表达图谱,并且能够通过Northern blot实验验证。
实时荧光定量逆转录–聚合酶链式反应(real time quantitative reverse transcription-polymerase chain reaction, qRT-PCR)和Northern 印迹(Northern blot)是用来定量tsRNA的最成熟的技术,常用于验证RNA-seq的结果。Wang等[96]利用qRT-PCR验证了差异表达tsRNA在早期乳腺癌患者血浆、组织中的表达丰度,确定了6个tsRNA可以作为早期乳腺癌的潜在诊断标志物。Shi等[97]去除了总RNA的修饰后,再用qRT-PCR检测肾小球足细胞中部分tsRNA的表达。此外,Lee等[98]将总RNA进行凝胶纯化后,把其中17~26 nt的小RNA用于合成cDNA再进行PCR,其实验结果与总RNA进行qRT-PCR结果一致。与qRT-PCR相比,Northern blot检测灵敏度较低,Su等[99]通过Northern blot检测到了胎盘中表达丰度较高tsRNA,验证了RNA-seq结果的真实性。McArdle等[100]开发了一种基于电催化铂纳米颗粒的电化学直接检测方法,能够特异性的检测血浆中3种tsRNA,并且与qPCR获得的结果相同,为快速检测tsRNA提供了新的研究思路。
3.3 tsRNA过表达和干扰技术
过表达或RNA干扰技术常被用来进行功能获得性或缺失性研究。通过导入人工合成的带有修饰的核苷酸来过表达或抑制目标tsRNA,观察细胞、动物表型的变化或靶基因、靶蛋白的变化,是目前研究tsRNA功能的常用方法。与microRNA研究方法类似,tsRNA mimics和inhibitors是最常用的技术手段[101]:mimics是运用化学方法合成的双链RNA,能模拟内源性tsRNA进而增强内源性tsRNA的功能,其进入细胞后与Argonaute蛋白结合形成沉默复合物,进一步与靶基因mRNA结合调控其表达;inhibitor是化学合成的单链RNA,是专门针对细胞中特异的靶tsRNA的抑制剂,转染至细胞后与tsRNA互补结合,削弱内源性tsRNA的效应。Shen等[102]对小鼠3T3-L1前脂肪细胞瞬时转染tRFGluTTCmimics或inhibitors,发现tRFGluTTC具有促进脂肪细胞增殖的功能。锁核酸(locked-nucleic acids, LNAs)是一种具有双环结构的寡核酸衍生物,其2ʹ-O位和4ʹ-C位通过缩水作用形成氧亚甲基桥,由于结构的特殊性其具有与天然核酸的高亲和力、碱基错配识别能力和抗核酸酶解特点[103]。使用LNAs合成的tsRNA抑制剂,在tsRNA的功能研究中取得不错的效果。Goodarzi等[20]使用反义LNAs抑制肿瘤细胞中的tRFGlu、tRFAsp和tRFGly,导致RNA结合蛋白YBX1与原癌基因mRNA的结合更加稳定,揭示了tsRNA的肿瘤抑制作用。Kim等[68]用LNA/ASO混合物抑制3ʹ-tsRNALeuCAG,显著降低了HCT-116细胞活性,并且证明了LNA并不影响成熟tRNA的功能。
有研究报道,一些tRNA基因的差异表达导致tRNA衍生片段丰度的变化,而不是成熟tRNA丰度的变化[104]。此外,通过过表达亲本tRNA实现了上调tsRNA的表达。例如,Green等[105]用tRNA过表达质粒转染软骨细胞,使tRF-3003a的水平显著升高,并且其抑制靶基因的效果和转染mimics相似。Kuscu等[106]的研究中也实现了过表达亲本tRNA使tRF-3水平升高。与化学合成的tsRNA类似物相比,亲本tRNA产生的内源性tsRNA更有利于功能性研究,但过表达一种tRNA可能会产生不同类型的tsRNA,对其功能研究是否会有影响还尚未见报道。
3.4 其他研究方法
此外,研究人员还利用RNA pulldown、RIP(RNA immunoprecipitation)、CLIP (crosslinking immunoprecipitation)、PAR-CLIP (photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation)、CLASH (cross-linking ligation and sequencing of hybrids)等分子生物学实验技术来分析tsRNA与核酸以及蛋白质的相互作用(表4),随着对tsRNA的深入研究,创新更多高效的技术手段,将有利于帮助理解tsRNA的功能。
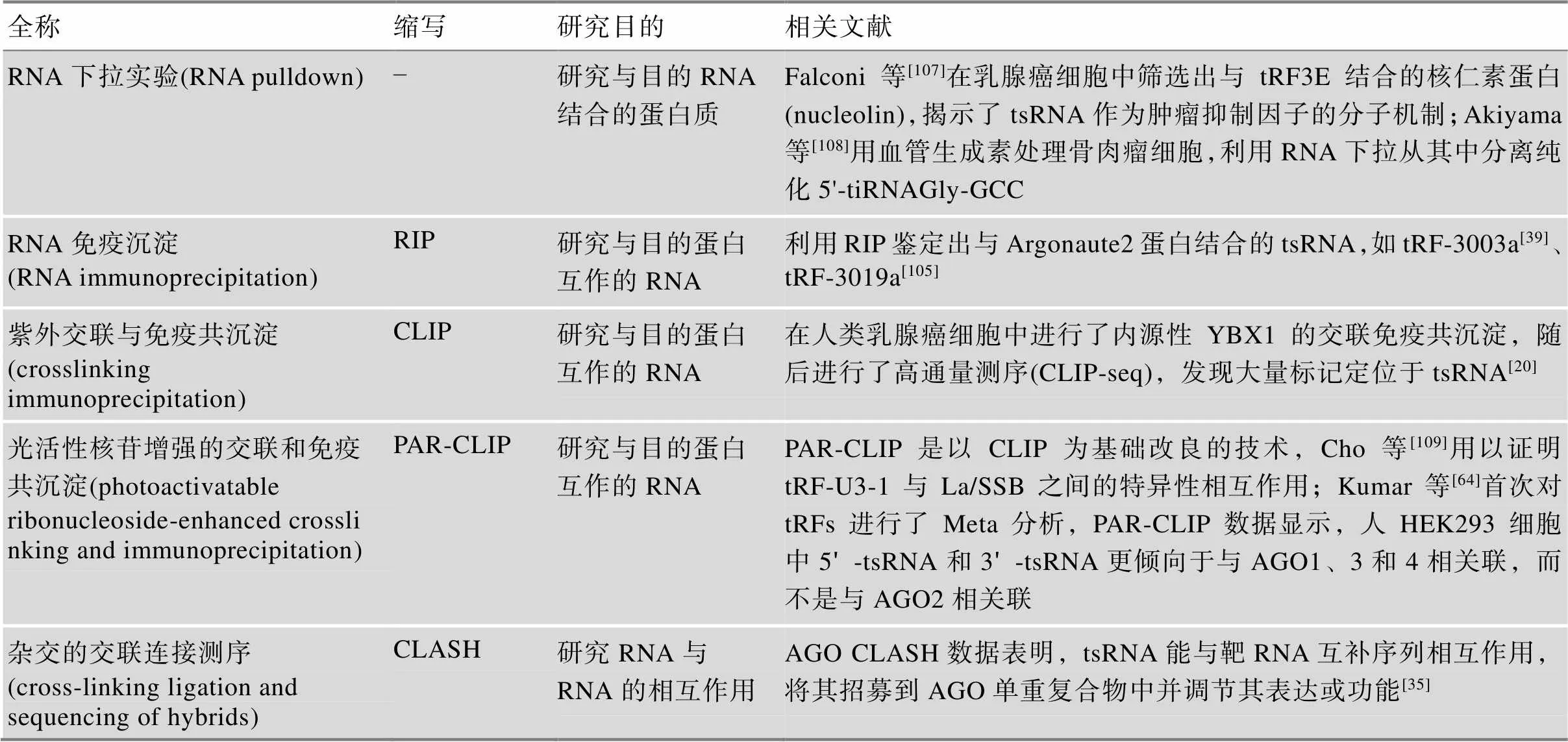
表4 tsRNA与蛋白质/RNA相互作用的研究方法
4 结语与展望
早在20世纪70年代tsRNA就已经被检测出来,但长期以来一直认为其是tRNA随机降解产物,直到最近10年,部分tsRNA的产生机制和生物学功能才逐渐被发现。tsRNA通过调控转录本稳定、调节基因表达等途径在正常生物学过程和疾病发生中发挥重要作用,目前tsRNA的研究还面临着一些问题:(1) tsRNA命名规则还没有统一;(2) tsRNA产生机制还未研究透彻,例如i-tRF、tRF-2产生方式还不清楚,不同物种中tsRNA产生机制是否保守也有待进行深入研究;(3)目前对其功能的研究几乎都是利用化学合成的RNA序列,而内源性的tsRNA具有多种RNA修饰,因此很可能忽略了RNA修饰对tsRNA功能的影响。
尽管目前对tsRNA的认识还不完全,但它作为一种新的功能性小非编码RNA,在生物体中有着极为重要的作用,相信在未来的研究中将揭示其更多的生物学意义。
[1] ENCODE Project Consortium, Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermüller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaöz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Löytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, NISC Comparative Sequencing Program, Baylor College of Medicine Human Genome Sequencing Center, Washington University Genome Sequencing Center, Broad Institute, Children's Hospital Oakland Research Institute, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Shahab A, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Zhang X, Xu M, Haidar JN, Yu Y, Ruan Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrímsdóttir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project., 2007, 447(7146): 799–816.
[2] Goodall GJ, Wickramasinghe VO. RNA in cancer., 2021, 21(1): 22–36.
[3] Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing., 2010, 16(4): 673–695.
[4] Andersen KL, Collins K. Several RNase T2 enzymes function in induced tRNA and rRNA turnover in the ciliate., 2012, 23(1): 36–44.
[5] Rosace D, López J, Blanco S. Emerging roles of novel small non-coding regulatory RNAs in immunity and cancer., 2020, 17(8): 1196–1213.
[6] Borek E, Baliga BS, Gehrke CW, Kuo CW, Belman S, Troll W, Waalkes TP. High turnover rate of transfer RNA in tumor tissue., 1977, 37(9): 3362– 3366.
[7] Speerm J, Gehrkep CW, Kuo KC, Waalkes TP, Borek E. tRNA breakdown products as markers for cancer., 1979, 44(6): 2120–2123.
[8] Saxena SK, Rybak SM, Davey RT Jr, Youle RJ, Ackerman EJ. Angiogenin is a cytotoxic, tRNA-specific ribonuclease in the RNase A superfamily., 1992, 267(30): 21982–21986.
[9] Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes., 2008, 14(10): 2095–2103.
[10] Fu HJ, Feng JJ, Liu Q, Sun F, Tie Y, Zhu J, Xing RY, Sun XZ, Zheng XF. Stress induces tRNA cleavage by angiogenin in mammalian cells., 2009, 583(2): 437–442.
[11] Liapi E, van Bilsen M, Verjans R, Schroen B. tRNAs and tRNA fragments as modulators of cardiac and skeletal muscle function., 2020, 1867(3): 118465.
[12] Raina M, Ibba M. tRNAs as regulators of biological processes., 2014, 5: 171.
[13] Maraia RJ, Lamichhane TN. 3′ processing of eukaryotic precursor tRNAs., 2011, 2(3): 362–375.
[14] Zong TY, Yang YY, Zhao H, Li L, Liu MX, Fu XX, Tang GZ, Zhou H, Aung LHH, Li PF, Wang JX, Wang ZB, Yu T. tsRNAs: novel small molecules from cell function and regulatory mechanism to therapeutic targets., 2021, 54(3): e12977.
[15] Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in., 2009, 185(1): 43–50.
[16] Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, Anderson P, Pan T, Hatzoglou M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress., 2012, 287(51): 42708–42725.
[17] Su ZL, Kuscu C, Malik A, Shibata E, Dutta A. Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3–mediated gene silencing., 2019, 294(45): 16930–16941.
[18] Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in., 2005, 280(52): 42744–42749.
[19] Fricker R, Brogli R, Luidalepp H, Wyss L, Fasnacht M, Joss O, Zywicki M, Helm M, Schneider A, Cristodero M, Polacek N. A tRNA half modulates translation as stress response in., 2019, 10(1): 118.
[20] Goodarzi H, Liu XH, Nguyen HCB, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement., 2015, 161(4): 790–802.
[21] Cui YY, Huang Y, Wu XW, Zheng MJ, Xia YQ, Fu ZY, Ge H, Wang S, Xie H. Hypoxia-induced tRNA-derived fragments, novel regulatory factor for doxorubicin resistance in triple-negative breast cancer., 2019, 234(6): 8740–8751.
[22] Gebetsberger J, Zywicki M, Künzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in., 2012, 2012: 260909.
[23] Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression., 2009, 185(1): 35–42.
[24] Oberbauer V, Schaefer MR. tRNA-derived small RNAs: biogenesis, modification, function and potential impact on human disease development., 2018, 9(12): 607.
[25] Guzzi N, Bellodi C. Novel insights into the emerging roles of tRNA-derived fragments in mammalian development., 2020, 17(8): 1214–1222.
[26] Ma ZL, Zhou JB, Shao Y, Jafari FA, Qi PF, Li YL. Biochemical properties and progress in cancers of tRNA-derived fragments., 2020, 121(3): 2058–2063.
[27] Tao EW, Cheng WY, Li WL, Yu J, Gao QY. tiRNAs: A novel class of small noncoding RNAs that helps cells respond to stressors and plays roles in cancer progression., 2020, 235(2): 683–690.
[28] Prehn JHM, Jirström E. Angiogenin and tRNA fragments in Parkinson’s disease and neurodegeneration., 2020, 41(4): 442–446.
[29] Thompson DM, Parker R. Stressing out over tRNA cleavage., 2009, 138(2): 215–219.
[30] Li NS, Shan NY, Lu LG, Wang ZH. tRFtarget: a database for transfer RNA-derived fragment targets., 2021, 49(D1): D254–D260.
[31] Park J, Ahn SH, Shin MG, Kim HK, Chang S. tRNA-derived small RNAs: novel epigenetic regulators., 2020, 12(10): 2773.
[32] Martinez G. tRNA-derived small RNAs: new players in genome protection against retrotransposons., 2018, 15(2): 170–175.
[33] Xie YY, Yao LP, Yu XC, Ruan Y, Li Z, Guo JM. Action mechanisms and research methods of tRNA-derived small RNAs., 2020, 5(1): 109.
[34] Zhu LW, Ge JX, Li TW, Shen YJ, Guo JM. tRNA-derived fragments and tRNA halves: the new players in cancers., 2019, 452: 31–37.
[35] Kumar P, Kuscu C, Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs)., 2016, 41(8): 679–689.
[36] Farina NH, Scalia S, Adams CE, Hong DL, Fritz AJ, Messier TL, Balatti V, Veneziano D, Lian JB, Croce CM, Stein GS, Stein JL. Identification of tRNA-derived small RNA (tsRNA) responsive to the tumor suppressor, RUNX1, in breast cancer., 2020, 235(6): 5318–5327.
[37] Falconi M, Giangrossi M, Zabaleta ME, Wang JB, Gambini V, Tilio M, Bencardino D, Occhipinti S, Belletti B, Laudadio E, Galeazzi R, Marchini C, Amici A. A novel 3′-tRNAGlu-derived fragment acts as a tumor suppressor in breast cancer by targeting nucleolin., 2019, 33(12): 13228–13240.
[38] Mo DP, Jiang P, Yang YN, Mao XL, Tan XY, Tang X, Wei D, Li B, Wang XM, Tang L, Yan F. A tRNA fragment, 5′-tiRNAVal, suppresses the Wnt/β-catenin signaling pathway by targeting FZD3 in breast cancer., 2019, 457: 60–73.
[39] Zhang F, Shi JX, Wu ZH, Gao P, Zhang WX, Qu BC, Wang X, Song YX, Wang ZN. A 3′-tRNA-derived fragment enhances cell proliferation, migration and invasion in gastric cancer by targeting FBXO47., 2020, 690: 108467.
[40] Tong LH, Zhang WX, Qu BC, Zhang F, Wu ZH, Shi JX, Chen XW, Song YX, Wang ZN. The tRNA-derived fragment-3017A promotes metastasis by inhibiting NELL2 in human gastric cancer., 2021, 10: 570916.
[41] Shen YJ, Xie YY, Yu XC, Zhang SS, Wen QY, Ye GL, Guo JM. Clinical diagnostic values of transfer RNA-derived fragment tRF-19-3L7L73JD and its effects on the growth of gastric cancer cells., 2021, 12(11): 3230–3238.
[42] Veneziano D, Tomasello L, Balatti V, Palamarchuk A, Rassenti LZ, Kipps TJ, Pekarsky Y, Croce CM. Dysregulation of different classes of tRNA fragments in chronic lymphocytic leukemia., 2019, 116(48): 24252–24258.
[43] Ruggero K, Guffanti A, Corradin A, Sharma VK, De Bellis G, Corti G, Grassi A, Zanovello P, Bronte V, Ciminale V, D’Agostino DM. Small noncoding RNAs in cells transformed by human t-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase., 2014, 88(7): 3612–3622.
[44] Honda S, Loher P, Shigematsu M, Palazzo JP, Suzuki R, Imoto I, Rigoutsos I, Kirino Y. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers., 2015, 112(29): E3816–E3825.
[45] Yang CW, Lee M, Song G, Lim W. tRNALys-derived fragment alleviates cisplatin-induced apoptosis in prostate cancer cells., 2021, 13(1): 55.
[46] Shao Y, Sun QL, Liu XM, Wang P, Wu RQ, Ma ZL. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer., 2017, 90(5): 730–738.
[47] Balatti V, Nigita G, Veneziano D, Drusco A, Stein GS, Messier TL, Farina NH, Lian JB, Tomasello L, Liu CG, Palamarchuk A, Hart JR, Bell C, Carosi M, Pescarmona E, Perracchio L, Diodoro M, Russo A, Antenucci A, Visca P, Ciardi A, Harris CC, Vogt PK, Pekarsky Y, Croce CM. tsRNA signatures in cancer., 2017, 114(30): 8071–8076.
[48] Luan N, Chen YQ, Li QS, Mu YL, Zhou Q, Ye X, Deng Q, Ling LM, Wang J, Wang JW. TRF-20-M0NK5Y93 suppresses the metastasis of colon cancer cells by impairing the epithelial-to-mesenchymal transition through targeting Claudin-1., 2021, 13(1): 124–142.
[49] Huang BQ, Yang HP, Cheng XX, Wang D, Fu SY, Shen WC, Zhang Q, Zhang LJ, Xue ZY, Li Y, Da YR, Yang Q, Li ZS, Liu L, Qiao L, Kong Y, Yao Z, Zhao P, Li M, Zhang RX. tRF/miR-1280 suppresses stem cell–like cells and metastasis in colorectal cancer., 2017, 77(12): 3194–3206.
[50] Wu YM, Yang XL, Jiang GM, Zhang HS, Ge LC, Chen F, Li JX, Liu HL, Wang HS. 5′-tRF-GlyGCC: a tRNA- derived small RNA as a novel biomarker for colorectal cancer diagnosis., 2021, 13(1): 20.
[51] Zhang MM, Li FF, Wang J, He WZ, Li Y, Li HY, Wei ZL, Cao YX. tRNA-derived fragment tRF-03357 promotes cell proliferation, migration and invasion in high-grade serous ovarian cancer., 2019, 12: 6371–6383.
[52] Zhou K, Diebel KW, Holy J, Skildum A, Odean E, Hicks DA, Schotl B, Abrahante JE, Spillman MA, Bemis LT. A tRNA fragment, tRF5-Glu, regulates BCAR3 expression and proliferation in ovarian cancer cells., 2017, 8(56): 95377–95391.
[53] Papadimitriou M-A, Avgeris M, Levis P, Papasotiriou ECh, Kotronopoulos G, Stravodimos K, Scorilas A. tRNA-derived fragments (tRFs) in bladder cancer: increased 5′-tRF-LysCTT results in disease early progression and patients’ poor treatment outcome., 2020, 12(12): 3661.
[54] He XQ, Yang YY, Wang Q, Wang JR, Li SF, Li CR, Zong TY, Li XL, Zhang Y, Zou YL, Yu T. Expression profiles and potential roles of transfer RNA-derived small RNAs in atherosclerosis., 2021, 25(14): 7052-7065.
[55] Meng L, Jiang L, Chen J, Ren HJ, Gao ZQ, Wu F, Wen YY, Yang LJ. Transfer RNA‑derived fragment tRF‑28‑QSZ34KRQ590K in plasma exosomes may be a potential biomarker for atopic dermatitis in pediatric patients., 2021, 21(5): 489.
[56] Huang P, Tu B, Liao HJ, Huang FZ, Li ZZ, Zhu KY, Dai F, Liu HZ, Zhang TY, Sun CZ. Elevation of plasma tRNA fragments as a promising biomarker for liver fibrosis in nonalcoholic fatty liver disease., 2021, 11(1): 5886.
[57] Choi E-J, Wu WZ, Zhang K, Lee I, Kim I-H, Lee YS, Bao XY. ELAC2, an enzyme for tRNA maturation, plays a role in the cleavage of a mature tRNA to produce a tRNA-derived RNA fragment during respiratory syncytial virus infection., 2021, 7: 609732.
[58] Wu WZ, Lee I, Spratt H, Fang X, Bao XY. tRNA- derived fragments in alzheimer’s disease: implications for new disease biomarkers and neuropathological mechanisms., 2021, 79(2): 793–806.
[59] Magee R, Londin E, Rigoutsos I. TRNA-derived fragments as sex-dependent circulating candidate biomarkers for Parkinson’s disease., 2019, 65: 203–209.
[60] Li LZ, Liu P, Wang RL, Huang YY, Luo JC, Jiao LQ, Tao Z, Zheng YM, Fan JF, Zhao HP, Han ZP, Luo YM. Pathophysiological significance of neutrophilic transfer RNA-derived small RNAs in asymptomatic moyamoya disease., 2021, 10(5): 1086.
[61] Hogg MC, Rayner M, Susdalzew S, Monsefi N, Crivello M, Woods I, Resler A, Blackbourn L, Fabbrizio P, Trolese MC, Nardo G, Bendotti C, van den Berg LH, van Es MA, Prehn JHM. 5′ValCAC tRNA fragment generated as part of a protective angiogenin response provides prognostic value in amyotrophic lateral sclerosis., 2020, 2(2): fcaa138.
[62] Venkatesh T, Suresh PS, Tsutsumi R. tRFs: miRNAs in disguise., 2016, 579(2): 133–138.
[63] Li ZH, Ender C, Meister G, Moore PS, Chang Y, John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs., 2012, 40(14): 6787–6799.
[64] Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets., 2014, 12: 78.
[65] Sobala A, Hutvagner G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells., 2013, 10(4): 553–563.
[66] Lafontaine DLJ. Noncoding RNAs in eukaryotic ribosome biogenesis and function., 2015, 22(1): 11–19.
[67] Couvillion MT, Bounova G, Purdom E, Speed TP, Collins K. Apiwi bound to mature tRNA 3′ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus., 2012, 48(4): 509–520.
[68] Kim HK, Fuchs G, Wang SC, Wei W, Zhang Y, Park H, Roy-Chaudhuri B, Li P, Xu JP, Chu K, Zhang FJ, Chua M-S, So S, Zhang QC, Sarnow P, Kay MA. A transfer-RNA-derived small RNA regulates ribosome biogenesis., 2017, 552(7683): 57–62.
[69] Kim HK, Xu JP, Chu K, Park H, Jang H, Li P, Valdmanis PN, Zhang QC, Kay MA. A tRNA-derived small RNA regulates ribosomal protein S28 protein levels after translation initiation in humans and mice., 2019, 29(12): 3816-3824.e4.
[70] Gebetsberger J, Wyss L, Mleczko AM, Reuther J, Polacek N. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress., 2017, 14(10): 1364–1373.
[71] Lyons SM, Kharel P, Akiyama Y, Ojha S, Dave D, Tsvetkov V, Merrick W, Ivanov P, Anderson P. eIF4G has intrinsic G-quadruplex binding activity that is required for tiRNA function., 2020, 48(11): 6223–6233.
[72] Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation., 2011, 43(4): 613–623.
[73] Lyons SM, Gudanis D, Coyne SM, Gdaniec Z, Ivanov P. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs., 2017, 8(1): 1127.
[74] Lyons SM, Achorn C, Kedersha NL, Anderson PJ, Ivanov P. YB-1 regulates tiRNA-induced stress granule formation but not translational repression., 2016, 44(14): 6949–6960.
[75] Yu MQ, Lu BJ, Zhang JS, Ding JW, Liu PY, Lu Y. tRNA-derived RNA fragments in cancer: current status and future perspectives., 2020, 13(1): 121.
[76] Ling C, Rönn T. Epigenetics in human obesity and type 2 diabetes., 2019, 29(5): 1028–1044.
[77] Iwasaki YW, Siomi MC, Siomi H. PIWI-interacting RNA: its biogenesis and functions., 2015, 84(1): 405–433.
[78] Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, Zamore PD. PIWI-interacting RNAs: small RNAs with big functions., 2019, 20(2): 89–108.
[79] Couvillion MT, Sachidanandam R, Collins K. A growth- essentialPiwi protein carries tRNA fragment cargo., 2010, 24(24): 2742–2747.
[80] Zhang X, He X, Liu C, Liu J, Hu QF, Pan T, Duan XB, Liu BF, Zhang YW, Chen JL, Ma XR, Zhang X, Luo HH, Zhang H. IL-4 inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA To upregulate CD1a molecules on monocytes/dendritic cells., 2016, 196(4): 1591–1603.
[81] Saze H. Epigenetic regulation of intragenic transposable elements: a two-edged sword., 2018, 164(5): 323–328.
[82] Kim HK, Yeom JH, Kay MA. Transfer RNA-derived small RNAs: another layer of gene regulation and novel targets for disease therapeutics., 2020, 28(11): 2340–2357.
[83] Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. LTR-Retrotransposon control by tRNA-Derived small RNAs., 2017, 170(1): 61–71.
[84] Min B, Park JS, Jeong YS, Jeon K, Kang YK. Dnmt1 binds and represses genomic retroelements via DNA methylation in mouse early embryos., 2020, 48(15): 8431–8444.
[85] Fukuda K, Shinkai Y. SETDB1-mediated silencing of retroelements., 2020, 12(6): 596.
[86] Kumar P, Mudunuri SB, Anaya J, Dutta A. tRFdb: a database for transfer RNA fragments., 2015, 43(D1): D141–D145.
[87] Zheng LL, Xu WL, Liu S, Sun WJ, Li JH, Wu J, Yang JH, Qu LH. tRF2Cancer: a web server to detect tRNA-derived small RNA fragments (tRFs) and their expression in multiple cancers., 2016, 44(W1): W185–W193.
[88] Schuster A, Tang C, Xie YM, Ortogero N, Yuan SQ, Yan W. SpermBase: a database for sperm-borne RNA contents., 2016, 95(5): 99.
[89] Gupta N, Singh A, Zahra S, Kumar S. PtRFdb: a database for plant transfer RNA-derived fragments., 2018, 2018: bay063.
[90] Zuo YL, Zhu L, Guo ZX, Liu WR, Zhang JT, Zeng Z, Wu QB, Cheng J, Fu X, Jin Y, Zhao Y, Peng Y. tsRBase: a comprehensive database for expression and function of tsRNAs in multiple species., 2021, 49(D1): D1038–D1045.
[91] Wang XM, Yang YN, Tan XY, Mao XL, Wei D, Yao YF, Jiang P, Mo DP, Wang T, Yan F. Identification of tRNA-derived fragments expression profile in breast cancer tissues., 2019, 20(3): 199–213.
[92] Xu C, Fu YF. Expression profiles of tRNA-derived fragments and their potential roles in multiple myeloma., 2021, 14: 2805–2814.
[93] Benesova S, Kubista M, Valihrach L. Small RNA- sequencing: approaches and considerations for miRNA analysis., 2021, 11(6): 964.
[94] Drino A, Oberbauer V, Troger C, Janisiw E, Anrather D, Hartl M, Kaiser S, Kellner S, Schaefer MR. Production and purification of endogenously modified tRNA- derived small RNAs., 2020, 17(8): 1104– 1115.
[95] Shi JC, Zhang YF, Tan DM, Zhang XD, Yan MH, Zhang Y, Franklin R, Shahbazi M, Mackinlay K, Liu SC, Kuhle B, James ER, Zhang LW, Qu YC, Zhai QW, Zhao WX, Zhao LL, Zhou CC, Gu WF, Murn J, Guo JT, Carrell DT, Wang YS, Chen XM, Cairns BR, Yang XL, Schimmel P, Zernicka-Goetz M, Cheloufi S, Zhang Y, Zhou T, Chen Q. PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications., 2021, 23(4): 424–436.
[96] Wang JY, Ma G, Li MH, Han X, Xu J, Liang MD, Mao XR, Chen X, Xia TS, Liu XA, Wang S. Plasma tRNA fragments derived from 5′ ends as novel diagnostic biomarkers for early-stage breast cancer., 2020, 21: 954–964.
[97] Shi HM, Yu MY, Wu Y, Cao YP, Li SW, Qu GT, Gong J, Gan WH, Zhang AQ. tRNA-derived fragments (tRFs) contribute to podocyte differentiation., 2020, 521(1): 1–8.
[98] Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs)., 2009, 23(22): 2639–2649.
[99] Su ZL, Frost EL, Lammert CR, Przanowska RK, Lukens JR, Dutta A. tRNA-derived fragments and microRNAs in the maternal-fetal interface of a mouse maternal- immune-activation autism model., 2020, 17(8): 1183–1195.
[100] McArdle H, Hogg MC, Bauer S, Rosenow F, Prehn JHM, Adamson K, Henshall DC, Spain E. Quantification of tRNA fragments by electrochemical direct detection in small volume biofluid samples., 2020, 10(1): 7516.
[101] Lu TX, Rothenberg ME. MicroRNA., 2018, 141(4): 1202–1207.
[102] Shen LY, Tan ZD, Gan ML, Li Q, Chen L, Niu L, Jiang DM, Zhao Y, Wang JY, Li XW, Zhang SH, Zhu L. tRNA-derived small non-coding RNAs as novel epigenetic molecules regulating adipogenesis., 2019, 9(7): 274.
[103] Veedu RN, Wengel J. Locked nucleic acids: promising nucleic acid analogs for therapeutic applications., 2010, 7(3): 536–542.
[104] Torres AG, Reina O, Stephan-Otto Attolini C, Ribas de Pouplana L. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments., 2019, 116(17): 8451–8456.
[105] Green JA, Ansari MY, Ball HC, Haqqi TM. tRNA- derived fragments (tRFs) regulate post-transcriptional gene expression via AGO-dependent mechanism in IL-1β stimulated chondrocytes., 2020, 28(8): 1102–1110.
[106] Kuscu C, Kumar P, Kiran M, Su Z, Malik A, Dutta A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner., 2018, 24(8): 1093–1105.
[107] Falconi M, Giangrossi M, Zabaleta ME, Wang JB, Gambini V, Tilio M, Bencardino D, Occhipinti S, Belletti B, Laudadio E, Galeazzi R, Marchini C, Amici A. A novel 3′-tRNAGlu-derived fragment acts as a tumor suppressor in breast cancer by targeting nucleolin., 2019, 33(12): 13228–13240.
[108] Akiyama Y, Kharel P, Abe T, Anderson P, Ivanov P. Isolation and initial structure-functional characterization of endogenous tRNA-derived stress-induced RNAs., 2020, 17(8): 1116–1124.
[109] Cho H, Lee W, Kim G-W, Lee S-H, Moon J-S, Kim M, Kim HS, Oh J-W. Regulation of La/SSB-dependent viral gene expression by pre-tRNA 3′ trailer-derived tRNA fragments., 2019, 47(18): 9888–9901.
The function and research methods of tRNA-derived small RNAs (tsRNA)
Jianfeng Ma1,2, Mailin Gan1,2, Li Zhu1,2, Linyuan Shen1,2
tRNA-derived small RNA (tsRNA), a class of small non-coding RNAs processed from the precursor tRNA or mature tRNA, broadly exist in organisms and have features including high conservation, structural stability and tissue specificity. According to the different paths of biogenesis, tsRNA is classified into two major types, tRNA halves and tRNA-derived RNA fragments (tRFs). Many studies have revealed that the expression of tsRNA is significantly increased under a variety of cellular stress responses, and its involvement in the regulation of stress responses is conserved across different species. tsRNA plays an important role in various biological processes by regulating transcript stability, protein translation and epigenetic processes.Recent researches have shown that tsRNAhas the potential as disease biomarkers and therapeutic targets, making it the focus of biomedical research. In this review, we summarize the research on tsRNA from aspects of biogenesis, function and research methods in order to provide a reference for relevant research.
non-coding RNA;tRNA-derived fragments; tsRNA; tRFs
2021-07-16;
2021-09-17
国家自然科学基金项目(编号:31902135,31972524)和四川省科技厅基础研究项目(编号:2021YJ0265,2021YFYZ0007,2020YFN0147)资助[Supported by the National Natural Science Foundation of China (Nos. 31902135, 31972524) and the Sichuan Science and Technology Support Program (Nos. 2021YJ0265, 2021YFYZ0007, 2020YFN0147)]
马剑峰,在读硕士研究生,专业方向:动物遗传育种。E-mail: 1098991954@qq.com
朱砺,博士,教授,研究方向:猪遗传育种与繁殖。E-mail: zhuli7508@163.com
沈林園,博士,讲师,研究方向:猪遗传育种与繁殖。E-mail: shenlinyuan0815@163.com
10.16288/j.yczz.21-256
2021/11/24 15:44:21
URI: https://kns.cnki.net/kcms/detail/11.1913.R.20211123.1547.002.html
(责任编委: 宋旭)

