Antioxidative, cytotoxic, and anti-metastatic potentials of Laurencia obtusa and Ulva lactuca seaweeds
Reem Al Monla, Yahya Salma, Achraf Kouzayha, Hala Gali-Muhtasib, Zeina Dassouki✉, Hiba Mawlawi✉
1Laboratory of Applied Biotechnology (LBA3B), AZM Center for Research in Biotechnology and its Applications, Doctoral School for Sciences and Technology, Lebanese University, Tripoli, Lebanon
2Department of Biology, American University of Beirut, Lebanon
3Center for Drug Discovery, American University of Beirut, Riad El Solh 1107 2020, Beirut, Lebanon
ABSTRACT
KEYWORDS: Macroalgae; Laurencia obtusa; Ulva lactuca;Reactive oxygen species; Apoptosis
1. Introduction
Cancer is a global burden that emerges as the most dreadful disease impacting all socio-economic levels[1]. At the national level,Lebanon has witnessed an increase in the rates of several cancer subtypes. For instance, over the last decade, incidence of colorectal cancer has been increasing significantly in this area and was ranked as one of the most common cancers in Lebanon[2]. Additionally,uterine cervical carcinoma is the second leading cause of tumors in women, surpassed only by breast cancer[3]. Due to poor prognosis,cervical cancer in Lebanon causes high mortality[4].
Chemotherapies are the first-line of choice adopted for cancer treatments. Unfortunately, their severe side effects including drug tolerance and toxicity could be detrimental[5]. Thus, there is a need for non-toxic natural compounds that act as chemotherapeutic drug sensitizers to potentiate the tumoricidal effects. Recently researchers renewed their interest in marine-based compounds as candidates for pharmaceutical use, specifically those derived from macroalgae[6].Thousands of bioactive marine-derived molecules have been reported to exhibit significant biological activities, including antimicrobial,anti-inflammatory, and anticancer effects[7].
Seaweeds contain high levels of essential proteins, vitamins,and polysaccharides with diverse biological activities, including immunomodulatory and anticancer effects[8]. Various proteins from red algae have gained significant attention for their cytotoxic and apoptotic potentials against a wide range of cancers[9,10].
The Lebanese coast is an interesting area in the Mediterranean region for studying diverse algal species[11]. Most published studies on the anticancer properties of Lebanese algae depend on colorimetric methods, without studying the mechanism of action of the obtained crude extracts or polysaccharides[12-14].
Ulva lactuca (U. lactuca) is a green algae, belonging to the division Chlorophyta, and it has been valorized to optimize its pharmaceutical, cosmeceutical, and nutraceutical potentials[15].In addition, the other studied algae Laurencia obtusa (L. obtusa),belonging to the genus Laurencia is mainly characterized by the presence of sesquiterpenes, di- and triterpenes, sterols, and alkaloids[16]. Their cytotoxic activities, specifically against cervical cancer and colon cancer, are poorly studied in the Mediterranean area.
In this paper, we investigated the apoptotic and anti-metastatic potential of the protein extracts and determined their effects on reactive oxygen species (ROS) generation in HCT-116 cells. Then we assessed cell cycle effects and amino acid content of the most potent extract from L. obtusa.
2. Materials and methods
2.1. Reagents
The following reagents were used in this study: Chloroform(Supelco), hexane (Sigma-Aldrich), methanol (Sigma-Aldrich),2,2-diphenyl-1-picrylhydrazyl (Sigma-Aldrich), 4′,6-diamidine-2′-phenylindole dihydrochloride (Roche, CAS 28718-90-3), fetal bovine serum (Sigma-Aldrich, CAS F4135), Dulbecco’s Modified Eagle’s Medium (Sigma-Aldrich, CAS D5796), zinc sulfate and barium hydroxide (Laboratory Chemicals, Lebanon), 2’,7’-dichlorofluorescein chloromethyl derivative (Thermo fisher, CAT C6827), phosphate buffer saline (Sigma-Aldrich, CAS 806552),adamantanamine hydrochloride (MERCK, CAS 665-66-7).
2.2. Marine algal materials
Red algae L. obtusa and green algae U. lactuca were collected from the Lebanese north coast of the Mediterranean. Fresh seaweeds were rinsed with tap water and associated epiphytes, salt, sand,microorganisms were removed. Algae were washed with ultrapure water, and then air dried in a shady place at room temperature (25-27 ℃) on absorbent paper, and finally ground to a fine powder. The algal species was identified by the macroalgae specialist professor Hiba Mawlawi based on the macroalgae taxonomy book[17]. The herbarium vouchers of L. obtusa (AZM-1102) and U. lactuca (AZM-1104) were kept in preservation at the Lebanese University, Doctoral School of Science and Technology.
2.3. Marine algal extraction
2.3.1. Lipids extraction
Dried and milled seaweeds (10 g) were extracted with 200 mL of (a)chloroform: methanol (CM 2:1) and (b) chloroform: hexane (CH 2:1),separately. After stirring, the mixture was centrifuged at 4 000 g for 20 min at 4 ℃. The collected supernatant containing lipids was evaporated under reduced pressure, by rotavap and residues “a” and “b” were used for protein extraction. The obtained extract was solubilized with their consequent solvent and then filtered. Finally, the lipid extract was rotavapped.
2.3.2. Protein extraction
A total of 100 mL of ultrapure water (40 ℃) was added to the residues “a” and “b” obtained from extraction in the previous step.Protein “a” was obtained from the residue extracted by CM solvent,whereas protein “b” was obtained from the CH solvent lipid-free residue. The solution was stirred for 24 h at 40 ℃, filtered, and then 20 mL of ultrapure water (60 ℃) was added. To precipitate protein,1 mL of zinc sulfate (1 M) and 1 mL of barium hydroxide (1 M)were added under constant stirring for 3 min, and then centrifuged at 5 000 g for 10 min at 4 ℃. The resulting pellet containing the protein extracts was lyophilized. The supernatants “a” and “b” were used for polysaccharides extraction.
2.3.3. Polysaccharide extraction
To eliminate traces of proteins, the two supernatants obtained previously were filtered separately, and 50 mL of ultrapure water was added to the obtained filtrate, followed by centrifugation at 5 000 g for 10 min at 4 ℃ (twice). The collected supernatants “a” and “b” were then lyophilized to obtain polysaccharide a and b (PS a and b).
2.4. Physicochemical analysis
2.4.1. Protein quantification
Biuret method is a well-known method used for dosing colorimetric proteins. A total of 500 µL of the protein sample was added in the presence of 1 500 µL of biuret reagent. After vortexing, the samples were incubated for 15 min in the dark at 37 ℃. Bovine serum albumin was used as a standard, and the absorbance was measured at 340 nm.
2.4.2. Total carbohydrate extraction and quantification
Reducing sugar content was calculated using Dubois colorimetric phenol-sulphuric acid method. The samples were hydrolyzed by concentrated acid (11 M HSO) for 1 h at 37 ℃, then the acid strength was diluted to 1 M. Next, the reaction was boiled for 2 h.The absorbances were read at 492 nm in a spectrophotometer and concentrations were determined by glucose standards[18].
2.4.3. Lipid content
The lipid content of each algae extract was determined based on the mass of pellets obtained from lipid extraction. Yield was calculated using the formula:
% of lipids = (mass of lipid extract/mass of sample) × 100
2.5. 2,2-Diphenyl-1-picrylhydrazyl (DPPH) free radical scavenging assay
The scavenging effects of samples against DPPH radicals were monitored according to the method of the previous study[19].Seaweed extracts were aliquoted into a series of concentrations (100,200, 300 and 500 µg/mL). One mL of freshly prepared 0.16 mM methanol DPPH solution was added and incubated in the dark for 30 min. The absorbance was measured at 517 nm. All the tests were performed in triplicates. The percentage of free radical scavenging was calculated using the formula below:
Free radical scavenging (%) = [(control OD−sample OD)/control OD] × 100
Where control OD is optical density of the control and sample OD optical density of the tested sample.
2.6. Cell line and cell culture
Human cervical cancer cells (HeLa), and colorectal cancer cells(HCT-116) were purchased from the American Type Culture Collection. Cells were cultured in Dulbecco’s Modified Eagle Medium at 37 ℃ in a humidified atmosphere of 5% COand 95% air. Media were supplemented with 1% penicillin-streptomycin (100 U/mL) and 10% heat-inactivated fetal bovine serum.
2.7. Cell viability assay
Cells were seeded in a 96 well plate at a seeding density of 10and left to adhere overnight. At 80% confluency, cells were treated with various concentrations of extracts (5, 10, 100 and 500 µg/mL). After 24 and 48 h treatments with different extracts, cells were incubated with 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide(MTT), for 3 h at 37 ℃ in the dark. Untreated cells were used as control of viability (100%). The mean absorbance values of three experiments were expressed as percentage of viability relative to the control. The cell growth assay is an MTT-based method that measures the ability of metabolically active cells to convert tetrazolium salt into a blue formazan product, the absorbance of which is recorded at 550 nm.
2.8. Wound-healing migration assay
HCT-116 cells were seeded in a 24 well plate and cultured in a growing medium for 24 h. At 90% confluency, a scratch wound was applied with a sterile 200 µL tip. Cell debris was washed twice with 1× phosphate buffer saline (PBS). Then cells were incubated with algal extract at two different concentrations (10 and 100 µg/mL). Images were captured using a camera coupled to an optical microscope. Images of the wounds were taken directly after scratch at 0 h, then after 12 and 24 h of extract incubation. The surface area was analyzed by Image J analysis program.
2.9. DAPI staining
Morphological changes of treated cells were investigated under a fluorescent microscope using 4′,6-diamidino-2-phenylindole(DAPI) staining. Briefly, the cells were treated with L. obtusa and U.lactuca protein b extracts at two different concentrations (100 and 500 µg/mL). After 24 h, cells were washed with PBS, fixed, and stored overnight at −20 ℃. Cells were then stained with DAPI in dark condition. A fluorescent microscope (Zeiss Axio) was used for visualization.
2.10. Quantitative ROS determination
The generation of intracellular ROS was monitored quantitatively using CM-HDCFDA. HCT-116 cells were seeded in 96-well culture plates and allowed to adhere overnight in a COincubator at 37 ℃.Cells were treated with two different concentrations of protein extracts(100 and 500 µg/mL) from both algae and incubated for 4 h at 37 ℃.At the end of the treatment period, cells were incubated with a 20 µM working solution of CM-HDCFDA dye in PBS at 37 ℃ for 40 min.Fluorescence intensity was detected using a microplate fluorometer at an excitation wavelength of 485 nm and an emission wavelength of 528 nm.
2.11. Cell cycle analysis
After 24 h of treatment with protein extracts (500 µg/mL) of L. obtusa, cells were collected and centrifuged. The pellets were washed with ice cold PBS, fixed, and stored at −20 ℃ overnight.Subsequently, cells were washed twice with PBS and incubated with 200 µg/mL RNAse A for 40 min at 37 ℃, then stained with 0.625µg/mL of propidium iodide for 30 min. The fluorescence intensity was measured by flow cytometry using a Fluorescence Activated Cell Sorter and analyzed using Cell Quest.
2.12. Amino acids profile of L. obtusa protein b extract by high performance liquid chromatography (HPLC)
2.12.1. Hydrolysis
Sample was added to 6 M hydrochloric acid and 1% of phenol in a sealed tube. The hydrolysis was carried out at 110 ℃ for 16 h. After hydrolysis, the contents were neutralized and dried with nitrogen gas.
2.12.2. Derivatization procedure
Amino acids were derivatized using a precolumn procedure. An aliquot of 600 µL of sample or standards was added with 1 200 µL of a 200 mM borate buffer (pH 10). Then, 1 200 µL of 15 mM FMOCCl was added and derivatization occurred for 15 min. The reaction was stopped by the addition of 1 200 µL of 300 mM adamantanamine hydrochloride, and the reaction was left for 1 min[20]. Then, the samples and standards were filtered and analyzed by HPLC.
2.12.3. HPLC analysis
An LC-1500 HPLC system was equipped with a diode-array detector set at 263 nm. Samples were then injected with a 20 µL loop using a C-18 column. The column was operated at ambient temperature with a flow rate of 1 mL/min. Buffer A (50 mM acetate buffer set at pH 4.2), and buffer B (acetonitrile) were added. Amino acids were separated with the following gradient elution conditions (min/A%):0/72, 3/72, 27/55, 32/0, 37/0, 39/72, and 47/72.
2.13. Statistical analysis
All statistical analyses (t-test and one-way ANOVA) were performed using GraphPad Prism 7 (version 7.0, GraphPad Software Inc., La Jolla, CA, USA). P < 0.05 was considered significant.Quantitative data are expressed as mean ± standard deviation from the indicated set of experiments.
3. Results
3.1. U. lactuca and L. obtusa antioxidant activity
As shown in Figure 1, all the extracts of both studied algae demonstrated dose-dependent inhibition of DPPH free radicals at 100-500 µg/mL. Significant antioxidant activity was noted in both protein extracts of U. lactuca, specifically, the protein b extract with(40.33±3.28)% inhibitory effect observed at 500 µg/mL (Figure 1A). The protein b extract had higher scavenging activity than other extracts from U. lactuca. Besides, L. obtusa extracts showed stronger antioxidant activity than green algae extracts. The protein b extract of L. obtusa inhibited 60.81% of the DPPH free radicals at 500 µg/mL. A relatively lower percentage (30.00±2.36)% was noted in the protein a extract of the red algae (Figure 1B). Lipid extracts from both algae had minimal antioxidant activity in comparison to other extracts (Figure 1A and B).
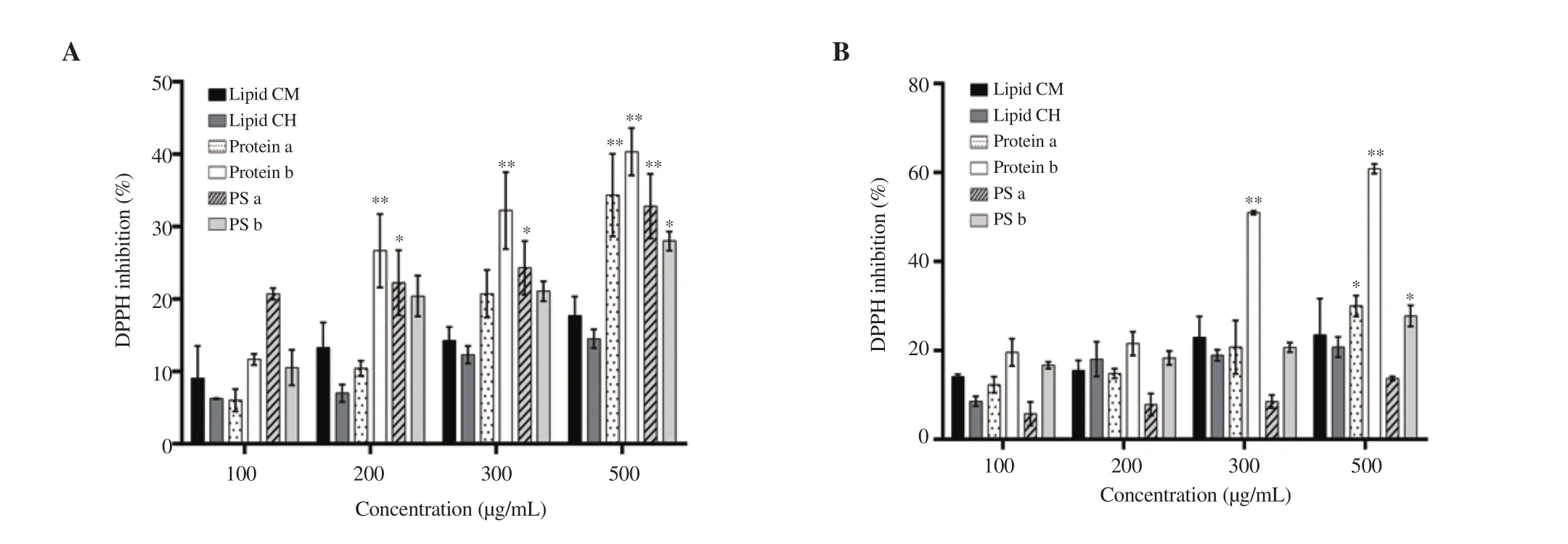
Figure 1. DPPH scavenging activity of different extracts from (A) Ulva lactuca (U. lactuca) and (B) Laurencia obtusa (L. obtusa). Data are expressed as mean ± standard deviation. Significant differences are indicated as *P < 0.05 and **P < 0.01 compared with the control (n=3). PS: polysaccharide, CM:chloroform:methanol, CH: chloroform:hexane.
Moreover, the results showed that L. obtusa had a higher protein content than that of U. lactuca, however the red algae had a lower lipid concentration. Additionally, both algae had low polysaccharide yield (Supplementary Table 1).
3.2. Cytotoxic effect of the U. lactuca and L. obtusa protein extracts on cell viability
Isolated protein extracts of U. lactuca and L. obtusa showed the highest antioxidant activity in comparison to all other extracts.Hence, their cytotoxicity against two cancer cell lines was next assessed in order to confirm the correlation between antioxidant and cytotoxic potentials of the proteic algal extracts. Upon treatment for 24 or 48 h, protein extracts of both algae induced a significant decrease in cell viability in a dose- and time-dependent manner(Figures 2 and 3). Indeed, the protein a and b extracts of U. lactuca exerted strong cytotoxic activity against HCT-116 cell line at 24 and 48 h (Figure 2A and B). HeLa cells were less sensitive to protein extracts, with (42.90±4.88)% and (40.27±2.00)% of viable cells after 48-h treatment of protein a and b extracts at 500 µg/mL, respectively(Figure 2C and D).
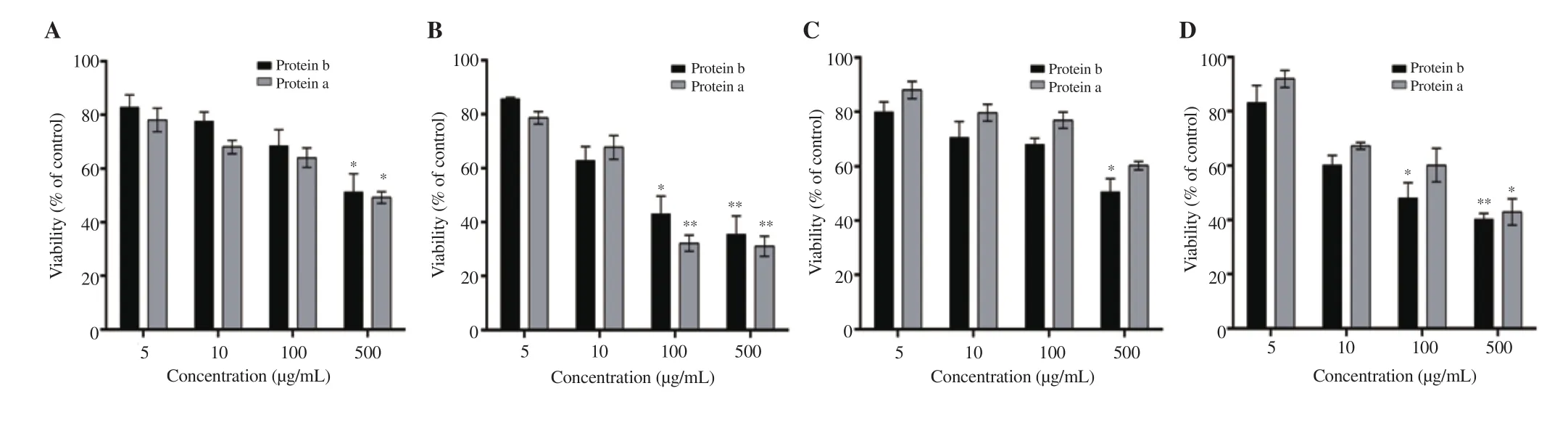
Figure 2. Effect of the protein extracts a and b of the green algae U. lactuca against HCT-116 (A: 24 h and B: 48 h) and HeLa cells (C: 24 h and D: 48 h).Significant differences are indicated as *P < 0.05 and **P < 0.01 compared with the control (n=3).
Figure 3 shows that protein b extract of L. obtusa exhibited the most potent effect against all tested cancer cell lines. HCT-116 cell line was found to be more sensitive to these extracts, specifically protein b extract of L. obtusa at 48 h [(21.41±3.77)%]. Protein b extract of L.obtusa exhibited the lowest ICat 48 h [(18.63±1.27) µg/mL].
Figure 3. Effect of the protein extracts a and b of the red algae L. obtusa against HCT-116 (A: 24 h and B: 48 h) and HeLa cells (C: 24 h and D: 48 h). Data are expressed as mean ± SD. Significant differences are indicated as *P < 0.05 and **P < 0.01 compared with the control (n=3).
3.3. Anti-metastatic effect of protein b extracts of both algae in HCT-116 cells
Figure 4 shows that protein b extracts of L. obtusa and U. lactuca significantly decreased the rate of gap closure in a dose- and time-dependent manner. All treatments revealed a significantly lower rate in gap closure in comparison with the control at 24 h. The wound healing effects of L. obtusa (26.70±8.24)% and U. lactuca(36.79±7.00)% at 100 µg/mL were significantly lower than the control (76.23±7.05)% (Figure 4A). The difference in gap closure rate between control and treated cells could also be noted visually by the optical microscope images (Figure 4B). This result indicates the potential of these extracts, especially L. obtusa protein b extract to inhibit HCT-116 cancer cell migration and invasion in vitro.
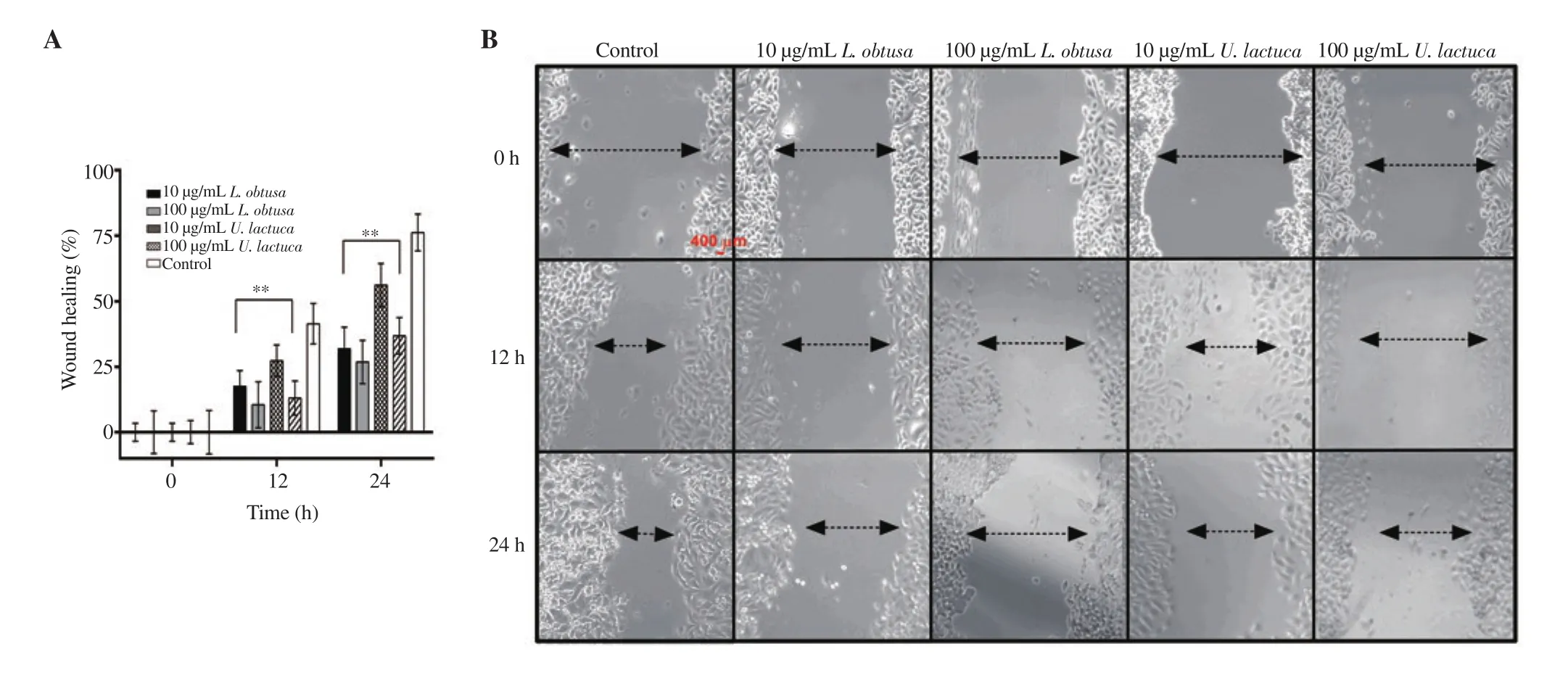
Figure 4. In vitro anti-metastatic potential of protein b extracts of U. lactuca and L. obtusa on HCT-116 cell migration by wound healing assay. Wound was created by a straight line scratch across the cancer cell monolayer. A: Percentage of cell-covered area at 12 and 24 h compared with 0 h. B: Representative images of two independent experiments done in triplicate, using a 10 × magnification and scale bar= 400 µm. Data are expressed as mean ± SD. Significant differences are indicated as *P < 0.05 and **P < 0.01 compared with the control.
3.4. Morphological alterations in colon cancer cells treated with U. lactuca and L. obtusa extracts
Normal morphology of the control untreated cancer cells was observed (Figure 5A). Protein b extracts of both algae promoted a dose-dependent increase in apoptotic bodies in HCT-116 cells.Changes in nuclear morphology, chromatin condensation, and DNA fragmentation were noticeable at 100 and 500 µg/mL (Figure 5B-E).A higher extent of cell death was visualized in L. obtusa extract in comparison with U. lactuca (Figure 5E).
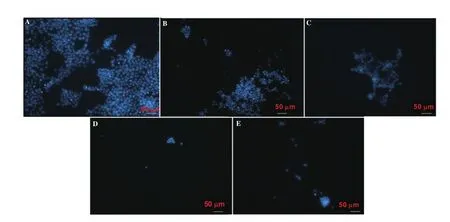
Figure 5. Immunofluorescent images of HCT-116 cells treated with L. obtusa and U. lactuca protein b extracts. A: HCT-116 cells in the absence of U. lactuca and L. obtusa extracts; B: cells treated with 100 µg/mL U. lactuca extract; C: cells treated with 100 µg/mL L. obtusa; D: cells treated with 500 µg/mL U. lactuca; E:cells treated with 500 µg/mL L. obtusa. Nuclei were counterstained with DAPI (blue) at × 40 magnification, and scale bar = 50 µm.
3.5. Effect of protein extracts on ROS generation in HCT-116 cells
As shown in Figure 6, upon treatment with L. obtusa extracts, the intracellular ROS levels were enhanced significantly in a dosedependent manner. In contrast, U. lactuca extracts did not induce a significant increase in ROS levels at both tested concentrations (only 1.37 folds at 500 µg/mL), suggesting the lack of involvement of ROS generation in its apoptotic cell death pathway.
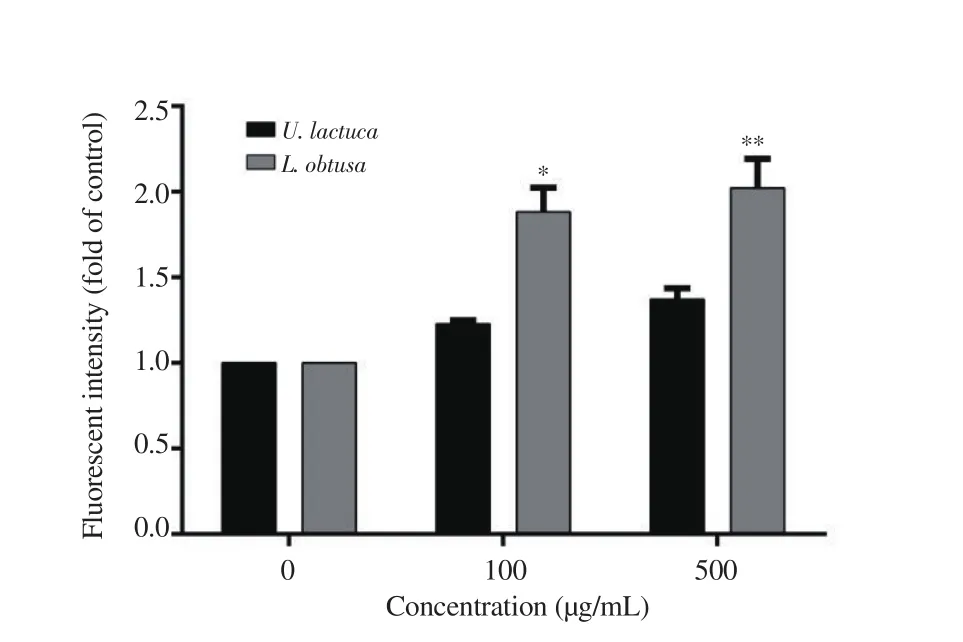
Figure 6. Quantitative analysis of DCFDA intensity (ROS generation) upon treatment with increasing concentrations of protein b extracts from U. lactuca and L. obtusa. Data are expressed as mean ± SD. Significant differences are indicated as *P < 0.05 and **P < 0.01 compared with the control (n=3).
3.6. Effect of protein b extract from L. obtusa on cell phase distribution in HCT-116 cells
Protein extract of L. obtusa was the only extract to induce ROS generation in HCT-116, and exhibited the highest antioxidant,cytotoxic, and anti-migratory effect. Thus, we decided to focus on the effect of this protein b extract from red algae on cellular deregulation. Figure 7 shows statistically significant changes in colon cancer cell cycle progression, upon algal extract treatment. A significant increase in the sub-Gpopulation was noted in HCT-116 cells treated with 500 µg/mL concentration of L. obtusa protein b extract [(60.38±3.21)%] compared with the control [(28.47±1.65)%],which was associated with a significant decrease in the percentages of cells in G, S and G/M phases (Figure 7C).
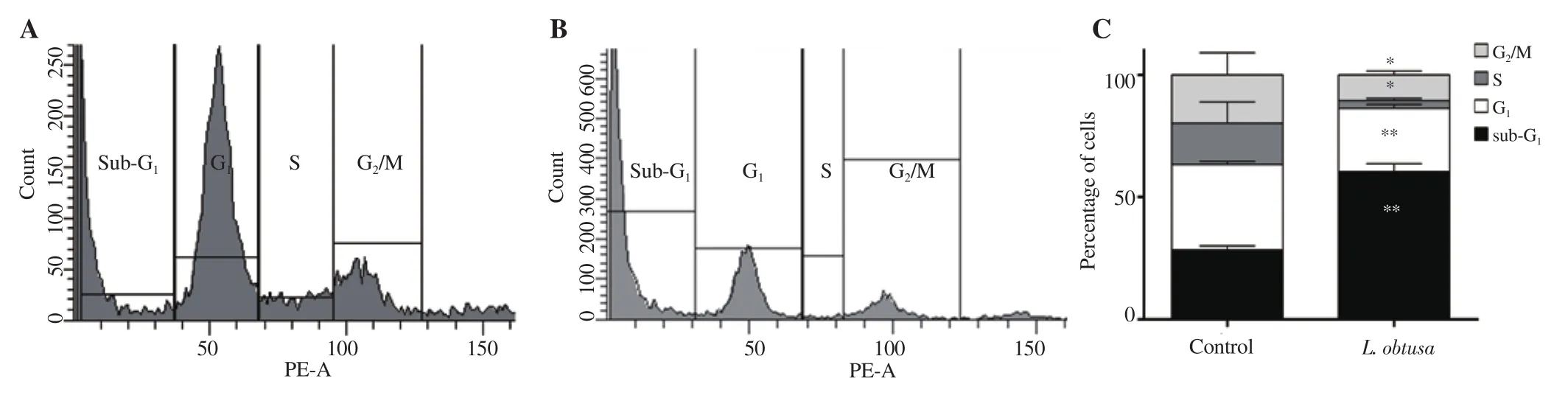
Figure 7. Cell cycle profile of HCT-116 colon cancer cells after treatment with L. obtusa protein b extracts. A: Distribution of cells in sub-G1, G1, S, and G2/M phases in the control. B: HCT-116 treated with L. obtusa at 500 µg/mL for 24 h. C: Percentage of cells in each cycle in control and treated cells. Data are expressed as mean ± SD. Significant differences are indicated as *P < 0.05 and **P < 0.01 compared with the control (n=3).
3.7. HPLC amino acid analysis of protein b extract from L.obtusa
HPLC profiles of a mixture of Fmoc-derived standards and protein b hydrolyzed samples were obtained and detected with HPLC coupled UV detector (Supplementary Figure 1A and B). The results revealed that glycine, arginine, and threonine, followed by aspartic and glutamic acid are the most abundant types of amino acids(Supplementary Table 2).
4. Discussion
Natural biocompounds from marine organisms, specifically those derived from seaweeds, have regained researchers’ attention worldwide. Their high biodiversity allows them to act as a rich reservoir for unique secondary metabolites characterized by multifarious pharmacological activities against chronic diseases,including cancer[21]. Seaweeds are one of the most promising candidates for potent anticancer agents. Some of the compounds from these algal species are in clinical trials, but more research is needed to validate their anticancer potential[22]. In this sense, the present study investigated the antioxidant activity of L. obtusa and U. lactuca, by preparing lipidic, proteinaceous, and polysaccharide extracts. Then, we determined the apoptotic potential of the protein extracts against HCT-116 cells, and assessed cell cycle alterations and amino acid content of the most potent extract from L. obtusa.
Seaweeds exhibit antioxidant systems to resist environmental stress by producing bioactive products and enzymes[23]. In this regard,we focused primarily on the antioxidant properties of the obtained algal extracts, as they have not been examined previously in the area. The results of DPPH scavenging assay revealed significant antioxidant activity in protein extracts of U. lactuca, and even a higher activity was recorded in L. obtusa. The antioxidant activity of U. lactuca and L. obtusa was comparable to or even better than previously reported extracts of the same species collected from other geographical regions[24-26]. The studied green algae U. lactuca showed a higher lipid content than L. obtusa, but the latter showed a higher concentration of proteins (Supplementary Table 1), which is in agreement with the literature since red algae are known to be richer in proteins than other macroalgal species[27].
Many investigations confirm the positive correlation between antioxidant and the cytotoxic potential of molecules[28], and this relation was further validated in our study. The antioxidative protein extracts also showed strong inhibition of cell viability in cancer cell lines studied (HCT-116 and HeLa) in a time and dose-dependent manner. This data is consistent with other studies, which showed a potent cytotoxic activity, against colon cancer, of L. obtusa and U.lactuca collected from various regions[29-31].
Activation of metastasis is a key hallmark in cancer pathogenesis and is associated with cell migration from their original sites to other regions[32]. Interestingly, low concentrations (10 µg/mL) of protein extracts that exhibited weak antiproliferative capacities, were able to significantly reduce cell migration at 12 and 24 h treatments. In this regard, our data indicate that protein extract of L. obtusa possesses the strongest antimetastatic activity, followed by comparable antimigratory potential of U. lactuca, and Colpomenia sinuosa phenolrich extract obtained from our previous study[33]. Following the confirmation of the antiproliferative and anti-metastatic potential of the algal protein extracts, we aimed to elucidate whether these extracts can induce intracellular ROS by aggravating mitochondrial metabolism. High basal oxidative stress in cancer cells makes them sensitive to treatments through ROS induction and disruption of the cellular homeostasis, and this is proposed as an anti-cancer drug strategy in several studies[34]. Accordingly, our results on L.obtusa protein b treatment validated this strategy, as the levels of intracellular ROS were elevated upon treatment of HCT-116 cells.These data correlate with other findings, where a marked increase in ROS levels was also detected in cancer cells treated with various red algae extracts collected from different origins[35,36]. Therefore,we can consider L. obtusa protein extract as a double-edged sword,which acts as a strong radical scavenging activity, according to our DPPH results, and as a pro-oxidant in cancer cells to induce cytostatic activity over HCT-116 cells.
Bioactive agents that could activate apoptosis in cancer cells have the potential to be used in the pharmaceutical industry[37]. Moreover,DAPI staining analysis demonstrated that the exposure of HCT-116 cells to protein extracts of both algae induced apoptosis dosedependently. Indeed, HCT-116 cells lost its integrated regular shape,with non-defined boundaries, in addition to nuclear blebbing and chromatin condensation noted at high concentrations.
Apoptosis and control of cancer cells cycle progression is an effective strategy for inhibiting tumor growth[38]. Our data indicated that treatment of HCT-116 cells with 500 µg/mL protein extract of L. obtusa resulted in a significant increase in the sub-Gpopulation, implying that this extract may act to repress cancer cell proliferation via inhibition of cell cycle progression. The recorded sub-Gincrease suggests the presence of apoptotic cell death, which further confirmed the morphological changes and apoptotic features observed by DAPI. This pattern in cell cycle alteration was also noted in different Laurencia extracts from different regions, and in multiple types of cancer[35,39].
Proteins derived from seaweeds exhibit strong antioxidant,anticancer, and anti-inflammatory activities. To identify the chemical composition and the concentration of amino acid residues present in L. obtusa protein b extract, HPLC analysis was performed and confirmed that glycine, arginine, and threonine, followed by aspartic and glutamic acid are the most abundant types of amino acids present in this extract. Consistent with our results, other studies reported that alanine, glycine, and asparagine had the highest concentrations in different macroalgae[40,41].
Despite the great cytotoxic potential of the studied algal proteic extracts, Western blot analysis must be performed to unravel the pathway of apoptosis. Moreover, brown algal extracts from Lebanon were proved to be safe on normal colon cells[33], similarly extracts from Ulva and Laurencia must be further tested.
In conclusion, the seaweeds U. lactuca and L. obtusa collected from the Lebanese coast and their protein extracts are a valuable source of antioxidant and cytostatic compounds that target colon and cervical cancer cell lines. One of the main findings of this study was to recognize the apoptotic cell death induced by protein b extract of L. obtusa, which occurred via a ROS-mediated pathway and caused significant cell cycle alterations and morphological changes in cells.Thus, the protein extracts of L. obtusa could be used as functional foods owing to their antioxidant and cytotoxic properties. However,more studies are needed to test the in vivo anti-cancer activity of the extracts and further unravel the key pathways and molecules involved in cell death.
Conflict of interest statement
The authors declare no conflict of interest.
Funding
This work was funded by the Lebanese University grant.
Acknowledgments
The authors would like to express gratitude for the Doctoral School for Sciences and Technology for providing many reagents and complete support (Lebanese University, Azm center, Tripoli,Lebanon).
Authors’ contributions
RAM and YS contributed equally as they performed the experiments mostly together. HM, HGM, ZD, and RAM conceived and designed the research study. Intellectual content and literature research was done by RAM, HM, YS, and HGM. Moreover, RAM,HM, and YS analyzed the data. Preparation and drafting of the manuscript was achieved by RAM, ZD, and HM. HM, YS, AK, and HGM edited and revised the manuscript. Statistical analysis was calculated by RAM, YS, and HGM. All authors have read and agreed to the published version of the manuscript.

