Identification of maize leaf diseases using improved convolutional neural network
Bao Wenxia, Huang Xuefeng, Hu Gensheng, Liang Dong
Identification of maize leaf diseases using improved convolutional neural network
Bao Wenxia1, Huang Xuefeng1, Hu Gensheng1※, Liang Dong2
(1.,,230601,; 2.,,230039,)
Maize leaf disease is a serious problem in the process of agricultural production. Controlling maize leaf disease is of great significance to improving maize yield and quality, maintaining food security, and promoting agricultural development. In this study, traditional machine learning methods often needed spot segmentation and feature extraction to identify maize leaf disease, but due to the subjective and exploratory nature of artificial feature extraction, the result of feature extraction seriously affected the accuracy of disease recognition. The Common convolutional Neural Network (CNN) had many parameters, which made it difficult to converge and had low generalization ability. Aiming at the problems such as low accuracy of traditional methods for leaf disease identification and weak model generalization ability of maize, this study presented an improved CNN, which included seven convolutional layers, four maximum pooling layers, one Inception module, one ResNet module, two Global Average Pooling (GAP) layers, and one SoftMax classification layer. The CNN improved the traditional CNN structure. The backbone network of the model was composed of a convolutional layer stack with a size of 3×3 and a feature fusion network composed of the Inception module and ResNet module. The 3×3 convolutional layer stack was used to increase the area size of the feature map, and the Inception module and ResNet module were combined to extract the distinguishable features of the maize leaf disease. At the same time, the improved CNN used the GAP instead of the full connection layer to optimize the training time and improve the training accuracy. This study randomly scaled, flipped, and rotated the original image of the data set to obtain the enhanced image, and took 80% of the image as the training data set, and the rest as the test data set. The size of the image was uniformly modified to 256×256 pixels for training. The improved CNN in this study randomly abandoned some neurons and their connections during the training process, reducing the number of intermediate layer features. Selecting appropriate Dropout effectively solved the problem of model overfitting. At the same time, the study showed that the learning rate controlled the speed of adjusting the weights of the neural network based on the loss gradient. In order to find the best model parameters, this study optimized and selected the batch size, learning rate, and Dropout parameters, and determined that the validation accuracy of the model was the highest when the batch size was 64, the learning rate was set to 0.001, and the dropout parameter was set to 30%, thus further improving the efficiency of the model. Based on 3 852 maize data sets in PlantVillage and 110 maize leaf blight data captured in the field, this study compared the test accuracy of the traditional model and the improved CNN. The experimental results showed that compared with classical Machine learning models such as K-Nearest Neighbor (KNN), Support Vector Machine (SVM), Back Propagation Neural Networks (BPNN) and deep learning models such as AlexNet, VGG16, ResNet, and Inception-v3, the accuracy of improved CNN in this study reached over 98%. The classical machine learning model had a maximum recognition rate of 77%. In order to show the performance of the improved CNN, the recall rate, average precision, and F1-score of different models were compared. The results showed that the precision of the improved CNN was 95.74%, the recall rate was 99.41% and F1-score was 97.36%, which further improved the stability of the model and provided a reference for further research on corn disease detection and recognition.
disease; maize; convolutional neural network; feature extraction; parameter selection
0 Introduction
Maize was an important food, feed, and cash crop in most parts of the world. At present, China’s maize planting area was about 25 million hm2, distributed in around 24 provinces, municipalities, and autonomous regions. Annual maize production accounted for 90% of coarse grain production and played an important role in agricultural production and the national economy. However, the incidence of maize leaf diseases, such as maize rust and leaf blight, has become widespread, and the degree of damage has increased. Maize leaf diseases have critically affected the yield and quality of maize. Therefore, the prevention and treatment of maize leaf diseases are essential in improving maize yield and quality, maintaining food security, and promoting agricultural development.
When maize leaf diseases occur, the diseased leaves show symptoms that differ from normal leaves. In the early stage of maize rust, the diseased spots are distributed on both sides of the leaves and scattered in pale yellow long to ovate brown small purulent scars. When the scars break, rust-colored powders are emitted. In the late stage of the disease, the spots appear black in a round or oblong shape. In severe cases, maize rust can infect the ears, bracts, and even male flowers. In the early stage of maize leaf blight, the leaves first exhibit water-stained blue-grey spots that spread along the veins to both ends. In the late stage of the disease, the spots show a central light brown color and a dark brown long-stem type on the outer edge. In severe cases, the merged spots cause the entire maize leaf to die. The maize leaf spot disease appears as pinpoints that fade green-yellow in the early stage and spread along the veins. Later spots on the leaf surface showing elliptical grey or light brown with no distinct edges. In severe cases, large areas of maize leaves are killed.
Professional plant pathologists identified diseases from the different symptoms of a plant, but manual identification of the disease was costly and inefficient. Compared with professional plant pathologists, farmers who grew crops did not have the relevant expertise and experience to determine the type of crop disease[1]. In recent years, machine learning methods had been widely used for plant disease identification. These methods usually required spot segmentation and feature extraction. The extracted features were used to train machine learning models and ultimately achieve plant disease identification[2-3]. Commonly used plant disease features were texture features[4-5], color features[6], visual features[7], and vegetation index features[8]. Classical machine learning methods for classification and identification of plant diseases were support vector machines[3-5], K-means clustering[9], simple linear iterative clustering[10], distance metrics[11], comprehensive discriminant analysis[12], random forests[13], probabilistic neural networks[5], and BP neural networks[14]. These methods required manual extraction and selection of plant disease features. Given the subjectivity and exploratory nature of manual feature extraction, the abovementioned machine learning methods had difficulty in obtaining accurate recognition results.
Deep learning methods, especially the Convolutional Neural Network (CNN) methods, had made breakthroughs in identifying plant diseases. Different from classical machine learning methods that required manual extraction of specific features, the CNN learned the advanced robust features of the target directly from the original image to improve identification accuracy[15-19]. In the field of plant diseases recognition, the CNN had better performance than traditional machine learning methods[20-23]. Liu et al.[23]designed a double convolution neural network structure and applied it to maize leaf disease identification, and the accuracy of the improved model reached more than 90%. The model structure of this double network required a large amount of computation, and the model architecture was more complex. For the transplant performance of the algorithm, a lightweight network model should be further selected. Zhang et al.[24]used two improved deep convolution neural network models GoogLeNet and Cifar10 to improve the fitting ability of the model. Lv et al.[25]designed a neural network based on backbone AlexNet architecture, named DMS-Robust AlexNet. In the DMS-Robust AlexNet, dilated convolution and multi-scale convolution were combined to improve the capability of feature extraction. However, existing deep learning models usually used extremely deep structures to extract the features of the target. The number of parameters needed to be calculated for the fully connected layer of the model was very large. In addition, training a deep learning model required many training samples. Collecting or producing large numbers of training samples was difficult and expensive. In the absence of sufficient training samples, the above deep learning methods showed difficulty in obtaining high identification accuracy.
To avoid the subjectivity and uncertainty of manual feature extraction and improve the accuracy of the identification of maize leaf diseases, and the improved CNN was used in this study. Combining the advantages of VGG16, ResNet, and Inception-v3, the improved CNN extracted the distinguishable features of maize leaf diseases with a simple structure and used a global average pooling layer to replace the fully connected layer to reduce the number of parameters of the model. In addition, by augmenting the training samples and selecting the parameters of the model, the identification performance of the model for maize leaf diseases was improved.
1 Materials and methods
1.1 Image acquisition
Some of the maize images used in this study were from the PlantVillage dataset (https://github.com/spMohanty/ PlantVillage-Dataset), which was open to all users for performing the experiments. There were 3 852 images of the maize crops. The images were divided into four classes out of which three were diseased class and one was a healthy class, including images of healthy maize leaves (1 162 images), grey leaf spot (513 images), common rust (1 192 images), and maize leaf blight (985 images). Some of the maize leaf blight images (110 images) were captured with a handheld digital camera (EOS 80D SLR, Canon, Japan) in the field near Changjiang West Road, Hefei City, Anhui Province, China (31°51'53'' N, 117°14'33'' E), all the collected images were shown in Fig.1. The samples were labeled according to the disease category, and the label name was the category of the disease. The image size was uniformly modified to 256×256 pixels.
1.2 Image augmentation
Training a CNN required many training samples. If the number of training samples was insufficient, then the CNN might encounter overfitting. Deep learning models usually performed better with large data sets, so to achieve this, this study extended the data set by creating an augmented sample from an existing sample. The technique of generating a manual sample from the original sample by making some changes to the original sample was called data augmentation. Data scaling allowed for a much greater diversity of data and ultimately increases the size of the data available to train the model. CNN was known to process changes in images and classify objects, even if they were in different locations. This was the basic premise of data expansion. In order to train CNN, a large amount of data was needed so that it learned and extracted more features. The process of collecting data sets was related to money, manpower, computing resources, and time consumption. Therefore, this study to make some small changes to the image, such as scaling, rotation, translation, etc., to expand the existing data. The benefit of generating more training examples was that it made the model more robust and generic, thereby increasing its accuracy. Some images in the data set needed to go through several steps of data expansion. In this study, image transformation was used to process the training sample data. The image was randomly rotation by 45 degrees, and the rotation and scaling ratios were adjusted to 0.2. Some augmented images were shown in Fig.2. Finally, in order to meet the data balance of each disease category, the augmented data set included grey leaf spot (1 000 images) and Leaf blight (1 000 images). The healthy maize and the common rust did not strengthen and the original samples remained unchanged. Different leaf disease datasets with augmented or non-augmented were shown in Table 1, 80% of the augmented image were used in the train samples.
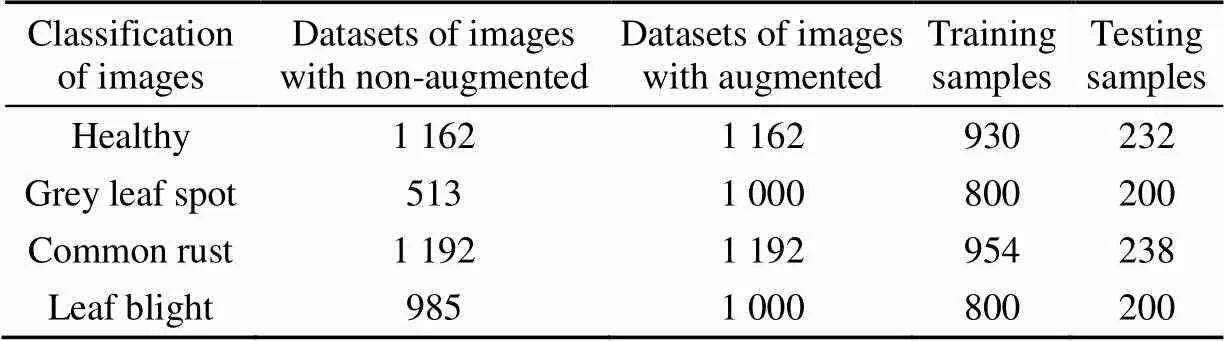
Table 1 Different leaf disease datasets with augmented or non-augmented
1.3 Model architectures
The common CNN had a large number of parameters, which made it difficult to converge and had low generalization ability. In view of the traditional methods for maize leaf disease identification accuracy was low and the model generalization ability was weak.Therefore, Combining the advantages of VGG16, ResNet, and Inception-v3, this study presented an improved CNN, which included seven convolutional layers, four maximum pooling layers, one Inception module, one ResNet module, two global average pooling layers, and one SoftMax classification layer. The structure of the improved CNN was shown in Fig.3.

Note: Conv represents the convolution operation; the product of 3 integers, including, the size of the convolution kernels as the product of the first number and the second number, the number of convolution kernels as the third number; 2×2 represents the size of the pooling area; S represents the stride; GAP is the Global Average Pooling.
The CNN improved the traditional CNN structure. The backbone network of the model was composed of a convolutional layer stack with a size of 3×3 and a feature fusion network composed of the Inception module and ResNet module.The stack of a 3×3 convolutional layer was used to increase the area size of the feature map.A 7×7 or 5×5 convolutional layer had many parameters, so the improved CNN model used a smaller 3×3 convolutional layer. The stacking of 3×3 convolutional layers achieved the same receptive field as 5×5 or 7×7 with few parameters. In addition, The Inception module and ResNet module were combined to extract the distinguishing features of a maize leaf disease. In this study, there were many ways to improve the performance of the network, such as hardware upgrades, larger data sets, and so on. But in general, the most direct way to improve network performance was to increase the depth and width of the network. Where, the depth of the network was only the number of layers of the network, and the width referred to the number of channels in each layer. However, this method brought two disadvantages. One was that when the depth and width increased continuously, the parameters that needed to be learned also increase continuously, and huge parameters were prone to overfitting. The other was to increase the size of the network uniformly, which would lead to an increase in computation. A good way to address both issues was to use the Inception module. Therefore, this study chose the Inception module from the improved CNN. However, due to the deepening of the network, features would be more and more abundant, and the model might lead to greater errors. Therefore, the improved CNN used the ResNet module and Inception module combined area to extract features to solve the problem of gradient explosion and gradient disappearance. The Inception module increased the ability of network feature extraction, especially in the front end of the network, which increased the diversity of features. The feature indicated that the size should be reduced gently, from the input to the output. The dimension of the feature representation was only represented as a superficial representation of information, which lost some important factors such as the correlation structure. High-dimensional information was more suitable for local processing in the network. In a convolutional network, increasing the nonlinear activation response step by step decoupled more features, and the network would be trained faster. Spatial aggregation could be embedded in low dimensions without decreasing network representation. Balance the depth and width of the network. The optimal performance of the network could be achieved by balancing the number of filters per layer and the number of layers of the network. Increasing the width and depth of the network improved the performance of the network, but the performance improvement obtained by increasing both in parallel was the largest. Therefore, computing resources should be reasonably allocated to the width and depth of the network.The CNN used the convolutive layer as a feature extractor to extract advanced feature mapping and input these feature mapping into the complete connection layer to stretch a long feature vector, which was then sent to the Softmax classifier. Its disadvantage was that the full connection layer had too many parameters, which was easy to reduce the training speed and lead to over-fitting. In the improved CNN, the global pooling layer replaced all the full connection layers on CNN. In the global pooling layer, each layer extracted the feature map of the last convolutional layer to generate a feature point constructs all feature points into vectors, and then the vectors were directly fed back into the Softmax layer for the corresponding category of each classification task. One advantage was that the global pooling layer on the full connection layer was the category corresponding to the extracted feature graph classification. Another advantage in the global pooling layer was that there was no need to optimize the parameters, which naturally avoided over-fitting. In addition, the global pooling layer combined spatial information, so the constructed feature vectors were more robust to spatial transformation. For the feature map after the middle layer, the GAP achieved dimensionality reduction of parameters and enhanced the ability of the generalization effect.
1.4 Model evaluation criteria
To evaluate the performance of the model, this study selected the following indicators used to evaluate the performance of the classifier: Recall (%), Precision (%), F1-score (%), and Accuracy (%) as the following expression (1), (2), (3), and (4) respectively.
where TP is the True Positive, FN is the False Negative, FP is the False Positive.
1.5 Model parameter optimization
In this study, if the parameters of the model were too many and the training samples were too few, the trained model was easy to produce overfitting phenomenon. When training neural networks, the problem of over fitting was often encountered. The specific performance of overfitting was that the loss function of the model on the training data was small, and the accuracy was high. But in the testing data, the loss function was relatively large and the accuracy was low. The depth of the CNN model increases, there was a problem of overfitting. During the training process, some neurons and their connections were randomly discarded, which reduced the number of features in the middle layer. Therefore, choosing the appropriate dropout effectively solved the overfitting problem of the model. The learning rate controlled the speed of adjusting neural network weights based on the loss gradient. If the learning rate was small, the gradient descent would be slow. If the learning rate was large, the gradient descent step crossed the optimal value. In this study, choosing a better learning rate was more conducive to the model fitting. This study chose the different batch size, dropout value, the learning rate was used to optimize the model. The results were shown in Table 2.
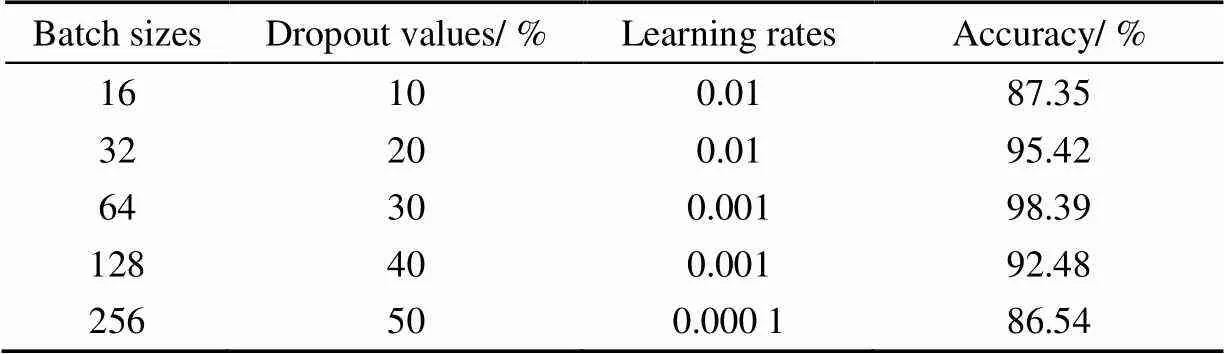
Table2 Optimization results of different parameters
It could be seen that different model parameters affected the accuracy of the verification set. Therefore, it was necessary to select appropriate parameters to train the model. This study founded that when batch size was equal to 64, dropout When the value was set to 30% and the learning rate was adjusted to 0.001, the training accuracy of the model was 98.39%. Therefore, the best parameters were used to train the model, and the research showed that the value of model parameters affected the performance of the model. The accuracy of maize leaf disease was improved by selecting suitable model parameters.
2 Results and analysis
2.1 Analysis of experimental results based on machine learning
In this study, the improved CNN was compared with classical machine learning models. The selected classical machine learning models included SVM[26], BP neural networks[14], and KNN[27]. All algorithms used maize images from the testing set in Table 1 to test the model's indicators, including images of healthy maize leaves (232 images), grey leaf spot (200 images), common rust (238 images), and maize leaf blight (200 images), a total of 870 images of maize. The average values of TP, FP, True Negative (TN), and FN of the classical machine learning models and the improved CNN for the testing samples of all classes were shown in Table 3.

Table 3 Calculation results of various indexes of different algorithms
In order to evaluate the comprehensive performance of the model, the average value of all evaluation indices was selected as the basis. As shown in Table 3, The TP value of the improved CNN model was 758. The TP value of SVM was only 532. This result was caused by the improved model's improved ability to predict positive samples. The maximum value of FN of traditional machine learning was 76, and then the value of FN of the improved model in this study was 3, which showed the value of the improved model. According to these values, the precision, recall, and F1-score of the model were calculated, as shown in Table 4.
In this study, the precision of classical machine learning models was 77.23% because of the subjectivity and uncertainty of the classical machine learning models to manually extract the image features of maize leaf diseases. This study used the improved CNN to automatically extract the image features of maize leaf diseases, and the precision was 95.74%. In Table 4, the values of precision, recall, and F1-score of the improved CNN were better than those of the classical machine learning models. Traditional machine learning needed to extract features manually. Features were easy to be damaged in the process of extraction, which led to the reduction of model fitting ability. The improved CNN learned features better and improved the stability of the model. At the same time, the index of accuracy was selected to evaluate the identification performance of different models on varying maize leaf diseases. Accuracy referred to the number of correctly identified maize leaf diseases of a certain type divided by the total number of such diseases in the testing set. The identification results of the classical machine learning models and the improved CNN on different maize leaf diseases were shown in Table 5.

Table 4 Classification results of different algorithms

Table 5 Accuracy of different algorithms for healthy leaves and three kinds of diseased leaves%
As shown in Table 5, The classical machine learning model KNN obtain an identification accuracy of 77%. However, the improved CNN correctly identified all healthy maize leaves and maize rust leaves in the testing set. This was because the color characteristics of healthy maize leaves and rust maize leaves were obviously different. For maize leaf blight and grey leaf spot, which were not clearly distinguishable from color and texture features. The SVM algorithm was a classification task based on small samples, so it was difficult to implement calculations for large-scale training samples. As for the data set in this study, the accuracy of SVM for grey leaf spot only reached 54%, while that of the improved CNN reached 98%. In this study, through the accuracy of different algorithms on healthy leaves and three kinds of disease leaves, it was concluded that, compared with the improved CNN, the classical machine learning model had a much worse recognition effect on leaf disease. Compared with the SVM algorithm, improved CNN achieved better results. Therefore, it was a better choice to use a convolutional neural network to extract features automatically.
2.2 Analysis of experimental results based on CNN models
In this study, the improved CNN was compared with the commonly used CNN models. The selected CNN models were AlexNet, ResNet, VGG16, Inception-v3, and GoogLeNet[22]. A total of 870 maize images in the testing set in Table 2 were used to test model indicators for all models. The values of TP, FP, TN, and FN of different CNN models were shown in Table 6.
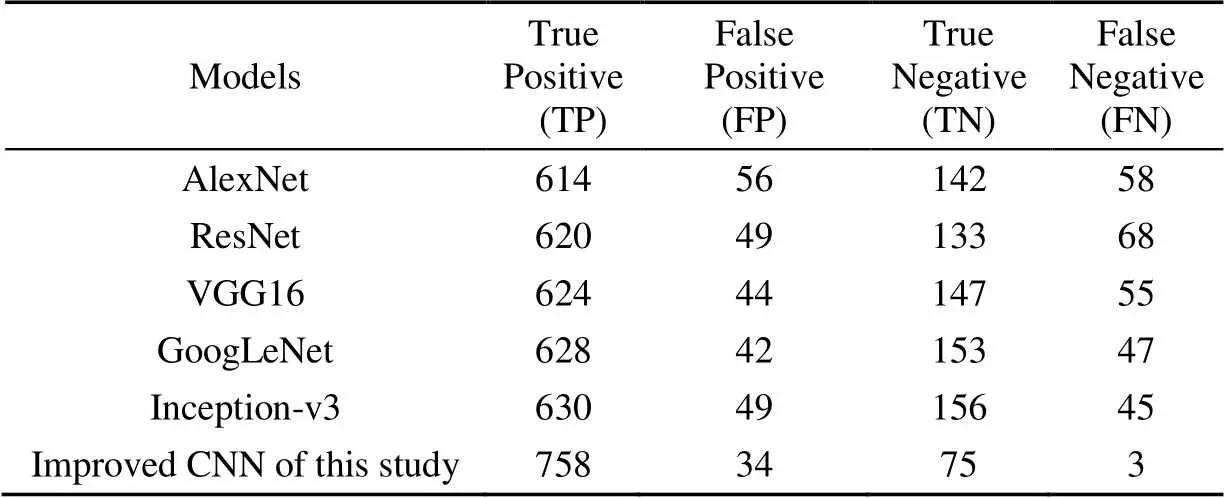
Table 6 Calculation results of various indices of different Convolutional Neural Network (CNN) models
As shown in Table 6, The TP value of the classical deep learning model AlexnNet was 614, while the improved model reached 758, which indicated that the improved model had an increased ability to recognize positive samples. Inception-v3 used multi-scale convolution to extract features, but the TN value was only 156. The improved model in this study not only integrated multi-scale information but also increased the model's ability to identify negative samples, so the TN value of the improved model was 75. The values of precision, recall, and F1 score of different CNN models as shown in Table 7.

Table 7 Classification results of different Convolutional Neural Network (CNN) models
In this study, given that CNN models automatically extract the disease features of maize leaves, the values of precision, recall, F1-score, and accuracy in Table 7 and Table 8 were almost above 90%. The improved CNN distinguished different maize leaf diseases by combining the Inception module and the ResNet module to extract detailed features of the maize leaf disease images. The precision of the improved model was 95.74%.Different convolutional neural networks had different feature extraction capabilities, so different results were obtained from Table 8. As for the recognition of maize disease, GoogLeNet achieved about 94% accuracy of healthy maize leaves and common rust, mainly because GoogLeNet adopted multi-scale convolution to extract features of maize leaf disease, thus increasing the capability of nonlinear mapping of features. However, compared with the improved CNN, the accuracy of leaf blight and grey leaf spot was 6% lower. This was because leaf blight and grey leaf spot had complex backgrounds and only relying on multi-scale features cannot achieve good results. Therefore, this study combined the ResNet module to further add features.

Table 8 Accuracy of different Convolutional Neural Network (CNN) models for healthy leaves and three kinds of diseased leaves%
2.3 Analysis of experimental results based on the latest identification model of maize leaf disease
In this study, the improved CNN was compared with the latest identification models of maize leaf disease. The testing data came from the PlantVillage (870 images) and field images of maize leaf blight (110 images). The experimental results were shown in Table 9. In this study, the improved CNN achieved good experimental results in the recognition of maize disease with a simple background, with the accuracy reaching about 98.87%. However, for maize leaf blight in the field, the improved CNN in this study reached 98.73%. The experimental results showed that the proposed identification model had better identification accuracy for maize leaf diseases than the latest identification models. The improved CNN further improved the identification accuracy of maize leaf disease images taken in the field by fusing the Inception module and ResNet module.

Table 9 Accuracy of maize leaf disease under different datasets
2.4 The ablation study used field images of maize leaf blight
In order to verify the rationality of the improved CNN structure, an ablation study was carried out in this experiment. The improved CNN was called the baseline network in this experiment after removing the Inception and ResNet modules. The baseline network, baseline network + Inception, baseline network + ResNet, and improved CNN were compared and analyzed. The testing data used maize leaf blight images taken from the field. The results were shown in Table 10. The experimental results showed that only using the baseline network without adding Inception and ResNet modules, it was difficult to extract the essential characteristics of the targets, and the identification accuracy of the model was sufficient. If only the Inception module or ResNet module was added, the identification accuracy of the model was improved, but it was not as good as the fusion structure of the Inception module and ResNet module. The improved CNN fully extracted the effective features of the diseased area and improved accuracy.

Table 10 Accuracy of the ablation study used field images of maize leaf blight
3 Conclusions
Deep learning models were used to identify different plant leaf diseases. This study proposed an improved Convolutional Neural Network (CNN) for the identification of maize leaf diseases. The conclusion was as follows:
1) The improved CNN combined the Inception module and ResNet module to extract detailed features of maize leaf disease images. Compared with traditional machine learning algorithms, CNN improved the accuracy of the model. The accuracy of improved CNN in this study was 98.73%.
2) This study found that when batch size was equal to 64, the dropout value was set to 30%, and the learning rate was adjusted to 0.001, the accuracy of model training was the highest. The study showed that the value of model parameters affected the performance of the model. The identification accuracy of maize leaf disease was improved by selecting suitable model parameters.
3) The average recall rate, average precision, and F1-score of different models were compared. The results showed that the precision of the improved CNN was 95.74%, the recall rate was 99.41% and F1-score was 97.36%, which further improves the stability of the model. At the same time, the method provides a reference for the further research of maize disease detection and identification.
The current models were limited to the images of maize leaves with a simple background and a single disease. In practical application, it may be necessary to identify the disease directly in the field, and the same leaf had multiple symptom characteristics of disease types. In the future, this study should further enrich the image data set of maize disease, make full use of the multi-scale characteristics of disease symptoms, and establish an end-to-end disease recognition model, to further improve the accuracy and practicability of the model.
[1]Miller S A, Beed F D, Harmon C L. Plant disease diagnostic capabilities and networks[J]. Annual Review of Phytopathology, 2009, 47(1): 15-38.
[2]Zhao Y X, Wang K R, Bai Z Y, et al. Bayesian classifier method on maize leaf disease identifying based images[J]. Computer Engineering & Applications, 2007, 43(5): 193-195.
[3]王燕妮, 贺莉. 基于多分类SVM的石榴叶片病害检测方法[J]. 计算机测量与控制, 2020, 28(9):191-195. Wang Yanni, He Li. Detection method of pomegranate leaf disease based on multi-classification SVM[J]. Computer Measurement & Control, 2020, 28(9): 191-195. (in Chinese with English abstract)
[4]Dhingra G, Kumar V, Joshi H D. A novel computer vision based neutrosophic approach for leaf disease identification and classification[J]. Measurement, 2018, 135(1): 782-794.
[5]Pantazi X E, Moshou D, Tamouridou A A. Automated leaf disease detection in different crop species through image features analysis and one class classifiers[J]. Computers and Electronics in Agriculture, 2019, 156(1): 96-104.
[6]Barbedo J G A, Koenigkan L V, Santos T T. Identifying multiple plant diseases using digital image processing[J]. Biosystems Engineering, 2016, 147(1): 104-116.
[7]Yigit E, Sabanci K, Toktas A, et al. A study on visual features of leaves in plant identification using artificial intelligence techniques[J]. Computers and Electronics in Agriculture, 2019, 156(1): 369-377.
[8]Patrick A, Pelham S, Culbreath A, et al. High throughput phenotyping of tomato spotted wilt disease in peanuts using unmanned aerial systems and multispectral imaging[J]. IEEE Instrumentation & Measurement Magazine, 2017, 20(3): 4-12.
[9]朱颢东, 申圳. 基于余弦定理和K-means的植物叶片识别方法[J]. 华中师范大学学报:自然科学版, 2014, 48(5):650-655. Zhu Haodong, Shen Zhen. Plant leaf identification method based on cosine theorem and K-means[J]. Journal of Central China Normal University: Natural Sciences Edition, 2014, 48(5): 650-655. (in Chinese with English abstract)
[10]Tetila E C, Machado B B, Belete N A D S, et al. Identification of soybean foliar diseases using unmanned aerial vehicle images[J]. IEEE Geoence and Remote Sensing Letters, 2017, 14(12): 2190-2194.
[11]Dhingra G, Kumar V, Joshi H D. Basil leaves disease classification and identification by incorporating survival of fittest approach[J]. Chemometrics and Intelligent Laboratory Systems, 2019, 186(1): 1-11.
[12]Wang F, Song L, Omasa K, et al. Automatically diagnosing leaf scorching and disease symptoms in trees/shrubs by using RGB image computation with a common statistical algorithm[J]. Ecological Informatics, 2017, 38(1): 110-114.
[13]JohannesA, Picon, A G, EchazarraJ, et al. Automatic plant disease diagnosis using mobile capture devices, applied on a wheat use case[J]. Computers and Electronics in Agriculture, 2017, 138(1): 200-209.
[14]Xiao M H, Ma Y, Feng Z X, et al. Rice blast recognition based on principal component analysis and neural network[J]. Computers and Electronics in Agriculture, 2018, 154(1): 482-490.
[15]Lenz I, Lee H, Saxena A. Deep learning for detecting robotic grasps[J]. International Journal of Robotics Research, 2015, 34(4): 705-724.
[16]李艳红, 樊同科. 基于深度残差的多特征多粒度农业病虫害识别研究[J]. 湖北农业科学, 2020, 59(16):153-157. Li Yanhong, Fan Tongke. Research on identification of agricultural insects based on depth residual network with multi-feature and multi-granularity[J]. Hubei Agricultural Sciences, 2020, 59(16): 153-157. (in Chinese with English abstract)
[17]Dyrmann M, Karstoft H, Midtiby H S, et al. Plant species classification using deep convolutional neural network[J]. Biosystems Engineering, 2016, 151(1): 72-80.
[18]孔建磊,金学波,陶治,等. 基于多流高斯概率融合网络的病虫害细粒度识别[J]. 农业工程学报,2020,36(13):148-157. Kong Jianlei, Jin Xuebo, Tao Zhi, et al. Fine-grained recognition of diseases and pests based on multi-stream Gaussian probability fusion network[J]. Transactions of the Chinese Society of Agricultural Engineering (Transactions of the CSAE), 2020, 36(13): 148-157. (in Chinese with English abstract)
[19]Quan L Z, Feng H Q, Lv Y J, et al. Maize seedling detection under different growth stages and complex field environments based on an improved Faster R–CNN[J]. Biosystems Engineering, 2019, 184(1): 1-23.
[20]Zhang S W, Zhang S B, Zhang C L, et al. Cucumber leaf disease identification with global pooling dilated convolutional neural network[J]. Computers and Electronics in Agriculture, 2019, 162(1): 422-430.
[21]Agarwal M, Singh A, Arjaria S, et al. ToLeD: Tomato leaf disease detection using convolution neural network[J]. Procedia Computer Science, 2020, 167(1): 293-301.
[22]孙俊,谭文军,毛罕平,等. 基于改进卷积神经网络的多种植物叶片病害识别[J]. 农业工程学报,2017,33(19):209-215. Sun Jun, Tan Wenjun, Mao Hanping, et al. Recognition of multiple plant leaf diseases based on improved convolutional neural network[J]. Transactions of the Chinese Society of Agricultural Engineering (Transactions of the CSAE), 2017, 33(19): 209-215. (in Chinese with English abstract)
[23]刘永波, 雷波, 曹艳, 等. 基于深度卷积神经网络的玉米病害识别[J]. 中国农学通报, 2018, 34(36):159-164. Liu Yongbo, Lei Bo, Cao Yan, et al. Maize diseases identification based on deep convolutional neural network[J]. Chinese Agricultural Science Bulletin, 2018, 34(36): 159-164. (in Chinese with English abstract)
[24]Zhang X H, Qiao Y, Meng F F, et al. Identification of maize leaf diseases using improved deep convolutional neural networks[J]. IEEE Access, 2018, 6: 30370-30377.
[25]Lv M J, Zhou G X, He M F, et al. Maize leaf disease identification based on feature enhancement and DMS-Robust AlexNet[J]. IEEE Access, 2020, 8: 57952-57966.
[26]Griffel L M, Delparte D, Edwards J. Using support vector machines classification to differentiate spectral signatures of potato plants infected with potato virus Y[J]. Computers and Electronics in Agriculture, 2018, 153(1): 318-324.
[27]Vijay S, Anshul S, Shanu S. An automatic leaf recognition system for plant identification using machine vision technology[J]. International Journal of Engineering Science and Technology, 2013, 5(4): 874-879.
[28]Syarief M, Setiawan W. Convolutional neural network for maize leaf disease image classification[J]. Telecommunication Computing Electronics and Control, 2020, 18(3): 1376-1381.
[29]Priyadharshini R A, Arivazhagan S, Arun M, et al. Maize leaf disease classification using deep convolutional neural networks[J]. Neural Computing and Applications, 2019, 31(12): 8887: 8895.
[30]Lin Z Q, Mu S M, Shi A J, et al. A novel method of maize leaf disease image identification based on a multichannel convolutional neural network[J]. Transactions of the ASABE, 2018, 61(5): 1461-1474.
基于改进卷积神经网络模型的玉米叶部病害识别
鲍文霞1,黄雪峰1,胡根生1※,梁 栋2
(1. 安徽大学电子信息工程学院,合肥 230601;2. 安徽大学互联网学院,合肥 230039)
准确识别玉米病害有助于对病害进行及时有效的防治。针对传统方法对于玉米叶片病害识别精度低和模型泛化能力弱等问题,该研究提出了一种基于改进卷积神经网络模型的玉米叶片病害识别方法。改进后的模型由大小为3×3的卷积层堆栈和Inception模块与ResNet 模块组成的特征融合网络两部分组成,其中3×3卷积层的堆栈用于增加特征映射的区域大小,Inception模块和ResNet 模块的结合用于提取出玉米叶片病害的可区分特征。同时模型通过对批处理大小、学习率和 dropout参数进行优化选择,确定了试验的最佳参数值。试验结果表明,与经典机器学习模型如最近邻节点算法(K- Nearest Neighbor,KNN)、支持向量机(Support Vector Machine,SVM)和反向传播神经网络(Back Propagation Neural Networks,BPNN)以及深度学习模型如AlexNet、VGG16、ResNet 和Inception-v3相比,经典机器学习模型的识别率最高为77%,该研究中改进后的卷积神经网络模型的识别率为98.73%,进一步提高了模型的稳定性,为玉米病害检测与识别的进一步研究提供了参考。
病害;玉米;卷积神经网络;特征提取;参数选择
Bao Wenxia, Huang Xuefeng, Hu Gensheng. et al. Identification of maize leaf diseases using improved convolutional neural network[J]. Transactions of the Chinese Society of Agricultural Engineering (Transactions of the CSAE), 2021, 37(6): 160-167. (in English with Chinese abstract)doi:10.11975/j.issn.1002-6819.2021.06.020 http://www.tcsae.org
鲍文霞,黄雪峰,胡根生,等. 基于改进卷积神经网络模型的玉米叶部病害识别[J]. 农业工程学报,2021,37(6):160-167. doi:10.11975/j.issn.1002-6819.2021.06.020 http://www.tcsae.org
2020-08-11
2020-11-08
Natural Science Foundation of China (61672032, 31971789); The Open Research Fund of the National Engineering Research Center for Agro-Ecological Big Data Analysis & Application Anhui University (AE2018009); CERNET Innovation Project (NGII20190617)
Bao Wenxia, Associate professor, research interests: image processing and pattern recognition. Email:bwxia@ahu.edu.cn
Hu Gensheng, Professor, research interests: image processing. Email:1414451346@qq.com
10.11975/j.issn.1002-6819.2021.06.020
TP391.4
A
1002-6819(2021)-06-0160-08

