Beneficial effects of AOS-iron supplementation on intestinal structure and microbiota in IDA rats
Hong He,Hui Teng,Qun Hung,Dn He,Fengping An,Lei Chen,∗,Hongbo Song,∗College of Food Science,Fujin Agriculture nd Forestry University,Fuzhou 350002,Chin
bFujian Provincial Key Laboratory of Quality Science and Processing Technology in Special Starch,Fuzhou 350002,China
Keywords:
AOS-iron
Iron deficiency anemia
Rats
Intestinal tissue pathology
Gut microbiota
ABSTRACT
The objective of this study was to investigate the effects of agar oligosaccharide-iron(AOS-iron)on intestinal tissue pathology and microbiota in IDA rats induced by a low-iron diet,further to find the relationship between intestinal microbiota and iron metabolic disorders.After 4 weeks of AOS-iron supplementation,the fecal iron content of IDA rats markedly increased in a dose-dependent manner,only the damaged cecum and colon tissues in medium-dose(MD)and high-dose(HD)groups were repaired to the baseline,while the diversity of gut microbiota was improved even at low dose(LD).Furthermore,the supplementation of AOS-iron altered the composition of gut microbiota.At the genus level,the beneficial microbiota was enriched in AOS-iron groups,but the relative abundance of potential opportunistic pathogens obviously reduced compared to that in the anemia model(AM)group.Spearman’s correlation analysis revealed that biochemical parameters,including blood metabolic parameters,iron contents,body weight,GSH-PX and T-AOC activity,were positively correlated with SMB53,Anaerotruncus,Anaerostipes and Coprobacillus but negatively correlated with Morganella,Fusobacterium and Serratia.These findings indicated that AOS-iron effectively repaired the damaged intestinal tissue and ameliorated iron metabolic disorders by regulating gut microbiota desirably,which could provide references for the treatment of IDA.
1.Introduction
As an essential micronutrient for basic metabolic processes in almost all organisms,iron participates in the transport,exchange,and tissue respiration of oxygen,maintains the normal hematopoietic function[1].Insufficient iron intake,iron utilization disorders or massive iron loss may result in iron deficiency anemia(IDA)[2].IDA increases the risk of premature birth and infant death,and may impair children’s mental health and intellectual development[3].Recent studies have reported that iron deficiency or supplementation can change the composition of gut microbiota[4].
Iron supplement or iron fortified food is the most effective way to improve IDA.In fact,the research on iron supplement has gradually evolved from the traditional ferrous salt to the third-generation novel iron chelate.The first-generation iron supplement is traditional inorganic ferrous salt,such as ferrous sulfate,which are chemically unstable and easily combines with other substances in food,resulted in a low bioavailability of iron.In addition,the inorganic ferrous salt is absorbed in the form of ferrous ions,which easily generate free radicals,leading to lipid peroxidation and gastrointestinal side effects[5].The second-generation iron supplement(iron salts of organic acids),such as ferrous lactate,absorbed in the form of iron ion and molecular chelate,has light gastrointestinal irritation[6],but accompanied by adverse reactions[7].The third-generation novel iron supplement,mainly including sugariron chelate,peptide-iron chelate and amino acid-iron chelate,etc.,is absorbed in molecular form and has a high iron absorption rate[8].Our previous study[9]exhibited that the iron supplementation effect of agar oligosaccharide-iron complex(AOS-iron)was better than the traditional iron supplement.However,according to dietary bioavailability,5%–20% of the fortified iron was absorbed,and the remainder enters the colon and can be available for the gut microbiota[10,11].So far,there are few data on the effect of iron supplementation on the gut microbiota of IDA rats.
The gut microbiota,a complex micro-ecological system,which participates in the nutrient and energy metabolism processes,and is also important for the integrity of the mucosal barrier function[12].In recent years,studies have reported that the differences in microbial structure between individuals, which have been linked to a variety of conditions such as obesity and mood[13].Some diseases are often accompanied with the change in gut microbiota,the regulation of gut microbiota can be thought of as an effective approach for alleviating the diseases.Iron is an essential trace element for most intestinal bacteria,many of which have an active iron transport system and other mechanisms to scavenge iron[14,15].To receive iron from the micro-environment at its low concentrations,the microbiota has evolved sophisticated mechanisms such as the siderophore system[16].Deschemin et al.[17]have demonstrated that little iron could be stored in intestinal cells and transported to the organs in the absence of gut microbiota, and the reason might be that the gut microbiota influences iron uptake,transport and storage by modulating epithelial iron transporters.In this line,Hoppe et al.[18]suggested that adding probiotics to food may be a way to increase iron absorption.
Up-to date,only a few studies reported the effects of iron deficiency and iron supplementation on gut microbiota.Supplementation of iron source reduced theEnterobacteriaceaepopulation in the intestinal microbiota of iron deficient rats[19,20],while increasing the number ofEnterobacteriaceaein weaned piglets[21].Zimmermann et al.[11]reported that iron fortification interferes with the ratio ofEnterobacterfaecalistoBifidobacteriaandLactobacillusin African children. Alexandra et al.[4]found that irondeficient rats had significantly greater numbers ofLactobacilliandEnterobacteriaceaewhile a large significant decrease numbers ofRoseburiacompared to iron-sufficient rats,and the iron repletion partially restored bacterial populations.The experimentin vitrocolonic fermentation model inoculated with immobilized gut microbiota from the children showed that iron deficiency reduced the number ofRoseburia,Clostridium ClusterandBacteroidesand increased the number ofLactobacillusandEnterobacteriaceae,but had no impact on gut microbiota composition and activity under high-iron following normal iron conditions[22].So,the relationship between gut microbiota and iron metabolism should be further investigated.This research firstly studied the therapeutic effect of AOS-iron on cecum and colon tissues damage and gut microbiota disorder caused by IDA,tried to illustrate the relationship between gut microbiota and IDA biochemical parameters,including blood metabolic parameters,iron contents,GSH-PX,T-AOC and MDA levels and body weights.
2.Materials and methods
2.1.Materials
AOS-iron was synthesized by a chelation reaction of AOS and FeCl3according to our method described previously[23]and stored in desiccant before use. The iron ions were chelated to the AOS backbone of-OH and-COOH group,and the iron content was 14.03%.AOS-iron was soluble and stable at physiological pH,which can be almost completely absorbed in the form of iron chelate without generating free iron ions.Normal diet(45mg Fe/kg diet)and lowiron diet(12mg Fe/kg diet)were purchased from Trophic Animal Feed High-tech Co.,Ltd(Nantong,China)according to the American AIN93 standard.
2.2.Animals and experimental design
Sixty male SPF SD rats weighing(55±5)g were purchased from Shanghai SLAC Laboratory Animal Co.,Ltd(Shanghai,China).Rats were maintained in a hygienic and comfortable environment(temperature(23±2)◦C,humidity(50±10)%,and 12h/12h light/dark cycles).All rat operating procedures were authorized by the Laboratory Animal Ethics Committee of the College of Food Science,Fujian Agriculture and Forestry University.After 5 days of feeding with the basal diet,the rats were randomly divided into normal control group(NC,n=12)and a model group(n=48).During the experimental period, the rats were housed in cages(6 rats per cage)and allowed free access to food and deionized water.The NC group and the model group were fed with normal diet and low-iron diet,respectively.Blood was collected from the orbit of the rats in the model group.Food intake was recorded daily and the content of Hb was determined weekly.The IDA model was successfully established when the Hb content was less than 70g/L(4th weekend).
The IDA model rats were randomly divided into anemia model group(AM,n=12)and low dose,medium dose and high dose AOS-iron groups(LD,MD and HD,n=12).Rats in the LD,MD,HD groups were orally administered 1,2,4mg Fe/kg·bw AOS-iron solution,respectively.Rats in the NC and AM groups were orally administered with an equal volume of normal saline.Rats were administered at 9:00 am every day for 4 weeks.The AM and AOS-iron groups were always fed with a low-iron diet,while the NC group was always fed with the normal diet.
2.3.Sample collections
Three rats were randomly selected from each group to collect their fresh fecal samples(1.00g;wet weight)on week 4,6,and 8 into a 2.5mL sterile centrifuge tube and stored at-80◦C for high throughput sequencing.After 4 weeks of iron supplementation,all rats were fasted for 12h,and sacrificed under chloral hydrate anesthesia and then dissected.The cecum and colon tissues were removed quickly,washed with normal saline,and immersed in formaldehyde solution.There were no signs of accidental or significant toxicity throughout the experiment and all rats survived.
2.4.Fecal iron content
The fecal iron content was measured by an atomic absorption spectrophotometer(AA-6300C,Shimadzu,Japan)according to the modified method of Sadeghi et al.[24]
2.5.Histological analyses of cecum and colon tissue samples
Cecum and colon tissues were fixed in 10% neutral formalin,embedded in paraffin and cut into 5μm slices.After hematoxylin and eosin(HE)staining,the morphology of cecum and colon tissues was observed at a magnification of 100 times using a BA210T microscope equipped with a camera(Motic,Xiamen,China).
2.6.Gut microbiota analysis
2.6.1.DNA extraction and PCR amplification
Total genomic DNA samples were extracted from rat feces using the Fast DNA SPIN extraction kit(MP Biomedicals,Santa Ana,CA,USA)according to the manufacturer’s instructions.The V3-V4 hypervariable regions of the 16S rDNA gene from fecal microbiota were amplified using specific primers(forward primer 338F 5'-ACTCCTACGGGAGGCAGCA-3'and reverse primer 806R 5'-GGACTACHVGGGTWTCTAAT-3').PCR was performed according to the previous description of Peat et al.[25],and reaction conditions were initial denaturation at 98◦C for 2min,followed by 28 cycles of 98◦C denaturation for 15s,55◦C annealing for 30s,72◦C extension for 30s,and finally 72◦C extension for 5min,store at 10◦C.PCR amplification products were excised from a 2% agarose gel,and then purified using an Axyprep DNA Gel Extraction Kit(Axygen Biosciences,Union City,CA,US)and fluorescence quantification usingthe Quant-iT PicoGreen dsDNA Assay Kit(Invitrogen,Carlsbad,CA,USA)according to the manufacturer’s instructions.

Table 1Effects of AOS-iron administration on the food intake and fecal iron content in rats fed on an iron deficiency diet for 4,6 and 8 weeks.
2.6.2.Sequencing and bioinformatic analysis

Fig.1.The colon and cecum tissue sections of different rats at the 8 weeks:(A)Cecum of NC group;(B)Colon of NC group;(C)Cecum of AM group;(D)Colon of AM group;(E)Cecum of LD group;(F)Colon of LD group;(G)Cecum of MD group;(H)Colon of MD group;(I)Cecum of HD group;(J)Colon of HD group.

Table 2The abundance and diversity statistical analysis of gut microbiota in rats.
2×300 bp paired-end(PE)sequencing was performed using a high throughput Illumina Miseq platform.The high-quality sequence was assigned to samples according to the barcode information,and the sequence was processed and analyzed using Quantitative Insights into Microbial Ecology(QIIME,v1.8.0).The 16S rDNA gene sequence was assigned to the operational taxonomic unit(OTU)with 97% sequence similarity by UCLUST(Edgar,2010),and then classified using the Greengenes database(Release 13.8,http://greengenes.secondgenome.com/).Alpha diversity estimates were calculated with the Shannon index.PCA and PCoA analyses were used to investigate the similarity of community structure between different samples. The heatmap was constructed based on the relative abundance of different gene level using a random forest algorithm with R package “randomForest”,and the biochemical metabolic network on the gut microbiota correlation was visualized using Cytoscape(http://www.cytoscape.org/).
2.7.Statistical analysis
All data were analyzed by one-way ANOVA and Duncan’s multiple range tests.Statistical differences were considered significant atP<0.05.All results were expressed as mean ± standard deviation(SD).SPSS software version 21.0(IBM Corp.,Armonk,NY)was used for all analyses.
3.Results
3.1.Changes in food intake and fecal iron content
As summarized in Table 1,the changes in food intake and fecal iron content were detected at the weekend of 4th,6th,and 8th of the experiment. After 4 weeks of iron deficiency, the IDA model was successfully established,i.e.,the food intake and fecal iron content in the AM and AOS-iron groups were significantly lower(P<0.05)than that in the NC group,and there was no significant difference in food intake and fecal iron content between the AM and AOS-iron groups(P>0.05).During iron supplementation,the food intake and fecal iron content of all rats increased with feeding time.At the weekend of 6th and 8th,the food intake of the AM group was still significantly lower than that in the NC group,while significantly increased in LD,MD and HD groups and recovered to baseline(P<0.05).Moreover,the fecal iron content of AOS-iron groups was significantly increased in a dose-dependent manner compared to the AM group(P<0.05).
3.2.Histomorphological changes in the cecum and colon
Microscopic analysis of rat cecum and colon sections was shown in Fig.1.At the end of the 8th week,compared to the NC group,the tissues of the cecum and colon(Fig.1C,1D)in the AM group were seriously destroyed,including intestinal mucosal erosion and fold atrophy,and the narrowed thickness of submucosa. In addition,the epithelial cells and lamina propria cells of the cecum almost disappeared,and obvious hyperemia was observed in the submucosal layer of the colon.After 4 weeks of iron supplementation(week 8),cecum and colon lesions caused by a low-iron diet were improved with increasing dose of AOS-iron.In the LD group,the degree of intestinal mucosal erosion in the cecum and colon was improved,but the size of mucosal fold and the thickness of submucosa were not significantly different from those in the AM group.On the other hand,the morphological structure of cecum and colon in the MD and HD groups was similar to the NC group.
3.3.The abundance and diversity of gut microbiota
The operational taxonomic unit (OTU) count and Shannon index of the gut microbiota were decreased after feeding with the lowiron diet for 4 weeks(Table 2),especially the Shannon index in the AM and AOS-iron groups were significantly lower(P<0.05)than that in the NC group,indicating that IDA obviously reduced the relative abundance and diversity of gut microbiota.However,after 2 weeks(week 6)of iron supplementation,the OTU count and Shannon index in the AOS-iron groups were significantly higher than that of the AM group(P<0.05).Especially,continued iron supplementation for 4 weeks(week 8),the OTU count and the Shannon index were restored to normal levels in the LD,MD and HD groups,and there was no significant difference among the three AOS-iron groups(P>0.05),indicating that even the low dose of AOS-iron can effectively improve the abundance and diversity of gut microbiota.
Additionally,as shown in Fig.2,principal component analysis(PCA)and principal coordinates analysis(PCoA)were conducted to analyze the overall structural changes in the gut microbiota.PCA scores indicated that fecal microbiota in the AM group exhibited a significant structural shift along the negative direction of the first principal component(PC1)compared to the NC group(Fig.2A).In contrast,the administration of AOS-iron significantly restored the variation induced by iron deficiency along the positive direction of PC1.Collectively, the structure of fecal microbiota altered by the low-iron diet was partially reversed by AOS-iron,especially in the MD and HD groups.Compared with the NC group,the overall microbial community structure of the rats in AM group changed significantly,while that in the LD,MD and HD groups was similar with that in NC group,indicating that there was an obvious difference in the microbial community structure between IDA rats and all AOS-iron rats(Fig.2B).Furthermore,the structures of intestinal microbiota in the MD and HD groups were closer to that of normal rats.So,PCoA further validated the results of PCA.
3.4.Composition of intestinal microbiota
To assess the effects of AOS-iron on fecal microbiota composition,the V3-V4 regions of 16S rRNA genes from fecal samples were sequenced using the high-throughput sequencing technology based on the Illumina MiSeq platform.At the phylum level,the relative abundance of different groups of gut microbiota was shown in Fig.3.In general,the gut microbiota mainly consists of three phylum levels,of which the Firmicutes are dominant,followed by Bacteroidetes and Proteobacteria.After 4 weeks of iron deficiency,the relative abundance decrease of Firmicutes in the AM group and the increase of Bacteroidetes and Proteobacteria were obvious compared with that in the NC group.However,after 2 weeks of AOS-iron intervention(week 6),the increase in the relative abundance of Firmicutes and the decrease of Proteobacteria in the AOS-iron groups were significant compared to the AM group,especially the relative abundances of Firmicutes and Proteobacteria in the MD and HD groups were similar to that in the NC group.On the other hand,the abundances of Actinobacteria, Cyanobacteria, Tenericoutes and Fusobacteria did not change significantly.After AOS-iron intervention for 4 weeks(week 8),the gut microbiota feature in AOS-iron groups was similar to that in the rats fed 2 weeks of AOS-iron diet,and the relative abundance of Firmicutes and Proteobacteria was further improved.Thus,AOS-iron intervention 4 weeks with the low,medium and high dose recovered the relative abundance of Firmicutes and Proteobacteria to near normal level.

Fig.2.Diversity analysis of intestinal microbiota of different rats for 8 weeks:(A)PCA analysis;(B)PCoA analysis.

Fig.3.Relative abundance of the gut microbiota in rats at the phylum level at 4th,6th and 8th weeks.
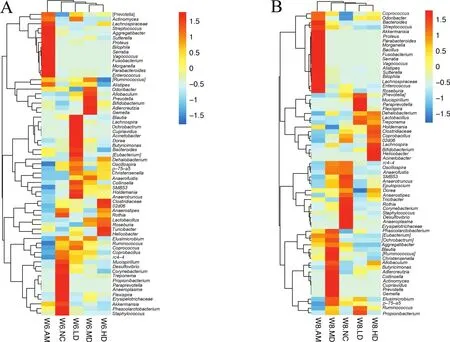
Fig.4.Heatmap of fecal microbiota at the genus level based on relative abundance.The microbiota names are shown on the right.(A):the fecal samples from rats reared for 6 weeks;(B):the fecal samples from rats reared for 8 weeks.
The metagenomic analysis at the genus level revealed that IDA induced by the low-iron diet changed the composition of gut microbiota,but AOS-iron intervention inhibited this adverse situation(Fig.4).At the weekend of 6th and 8th,65(Fig.4A)and 67(Fig.4B)bacteria at the genus level were identified,respectively,which were significantly affected by iron deficiency or AOS-iron supplementation.Compared with the group that supplementation of the AOS-iron for 2 weeks(week 6),two more weeks of feeding(week 8)significantly promoted the relative abundance ofLactobacillus,Coprobacillus,Bifidobacterium,Anaerotruncus,Anaerostipes,Butyricimonas,Ruminococcus,especially in the HD group(P<0.05);in contrast,Bacteroides,Streptococcus,Proteus,Parabacteroides,Morganella,Fusobacterium,Serratia,Sutterella,BilophilaandEnterococcusmarkedly reduced in the AOS-iron groups compared with that of the AM group(P<0.05)(Fig.4A and B).These results imply that AOS-iron showed a positive effect on regulating the dysbiosis of gut microbiota induced by the low-iron diet.
Spearman’s correlation analysis was performed to examine the connection between the gut microbiota composition and host biochemical parameters.The biochemical parameters of IDA have been reported by our previous study[9],including blood metabolic parameters(Serum ferritin,Hb,RBC,HCT and MCH),iron contents(Serum iron,Fecal iron and Liver iron),GSH-PX,T-AOC and MDA levels in serum and liver,as well as body weight.
Spearman correlation analysis demonstrated that the abundance of several specific gut bacteria was strongly correlated with the IDA-related parameters.Ruminococcu,SMB53,Coprobacillus,Anaerotruncus,Treponema,AnaerostipesandBlautiawere positively correlated with abnormal parameters RBC,Hb,MCH,HCT,serum ferritin,iron contents,GSH-PX and T-AOC levels,whileFusobacterium,Serratia,Morganella,Bacteroides,Enterococcus,Parabacteroides,ProteusandVagococcusdisplayed a negative relationship.Moreover,the MDA content in serum and liver displayed an opposite trend(Fig.5A).The network further showed that the blood metabolic parameters,iron contents,GSH-PX and TAOC levels and body weight were positively correlated withSMB53,Anaerotruncus,CoprobacillusandAnaerostipesbut negatively correlated withFusobacterium,SerratiaandMorganella.Furthermore,it was clear that MDA content positively correlated withFusobacterium,SerratiaandMorganella(Fig.5B).
4.Discussion

Fig.5.Spearman’s correlations analysis between the microbiota and biochemical parameters at the genus level.(A):Heatmap shows the correlation between gut microbiota of significant differences and biochemical parameters.The degree of change of color correspond the extent of association between gut microbiota and biochemical parameters.(B):Visualization of the correlation network in line with partial correlation between the gut microbiota of significant differences and the parameters associated with IDA metabolism disorder.Each node corresponds to the gut microbiota at the genus level and biochemical indexes.The solid red line and blue line corresponds the positive and negative correlation,respectively.Only the significant edges are drawn in the network using the Spearman correlation test(|r|>0.6,FDR adjusted P < 0.01).
We found that the food intake(Table 1)in the AM group significantly decreased,indicating that IDA affects the appetite of the rats leading to growth retardation[26].Surprisingly,at the weekend of 8th,the fecal iron content(Table 1)of rats in the AM group was significantly lower than that in the NC and AOS-iron groups,which was consistent with the results of Dostal et al.[27].On the other hand,the intestinal mucosa is not only an important site to absorb nutrients,but also a protective barrier against the invasion of harmful substances and pathogenic microorganisms[28].This study has proven that IDA could cause the damage of intestinal mucosa,which was in consistence with previous studies[29].Indeed,intestinal tissue lesions are associated with oxidative stress[30,31].Oxidative stress is an imbalance between oxidation and anti-oxidation[32],and excessive free radical production leads to lipid peroxidation[33],resulting in abnormal biofilm structure and cell damage.This study confirmed that cecum and colon lesions(Fig.1)of rats caused by a low-iron diet were repaired by feeding AOS-iron.Moreover,the intestinal tract in MD and HD groups also returned to normal.So,AOS-iron supplementation can effectively treat intestinal tissue damage in IDA rats.
It is well known that the intestinal microbial community is essential for host physiology and metabolism.Iron as part of the symbiosis between the microbiota and host may be critical to maintaining the composition and metabolism of gut microbiota[34].The low dose of AOS-iron was sufficient to positively affect the relative abundance and diversity of gut microbiota in rats in the 8th weeks(Table 2).Moreover,at the phylum level,our study suggested that supplementation of AOS-iron increases the abundance of Firmicutes,while reduces the abundance of Proteobacteria.Andoh et al.[35]suggested that the abundance of phyla Firmicutes in lean people was significantly lower than that in obese people.Although the bacterial communities of rats in all groups were dominated by Firmicutes, IDA reduced the relative abundance of Firmicutes.It was observed that the body weight[9]and the abundance of Firmicutes(Fig.3)in IDA rats increased after the AOS-iron supplementation,indicating that the abundance of Firmicutes was improved due to the intervention of AOS-iron,which resulted in body weight gain.Proteobacteria is a microbial signature of gut microbiota imbalance.The abundance of Proteobacteria increase during metabolic disorders induced by environmental or host factors,and return to normal during normal physiological metabolism[36].Previous results displayed that IDA caused disorder of iron metabolism in rats[9],which was positively correlated to the change of the relative abundance of Proteobacteria.Therefore,the increase in the abundance of Firmicutes and the reduction in that of Proteobacteria effectively improved the symptoms of IDA.
To further understand the intrinsic link between intestinal microbiota and iron metabolism,it is necessary to investigate the correlation between intestinal microbiota and biochemical indexes of IDA rats.After 4 weeks of iron supplementation(week 8),the composition of gut microbiota was altered;especiallyLactobacillusandBifidobacteriumwere markedly increased in the AOS-iron groups compared with the AM group(Fig.4).This result was similar to the report of Lin et al.[37],they studied the effect of ferrous-chelating hairtail peptides on the intestinal flora of IDA rats and found that theLactobacillusandBifidobacteriumin the IDA rats supplemented with ferrous-chelating hairtail peptides increased slightly compared to that in IDA rats.Previous study has reported that IDA rats supplemented with AOS-iron have high oxidative capacity[9],which was related to the increase in the number ofLactobacillusandBifidobacterium.BecauseLactobacillusandBifidobacteriumcan scavenge free radicals by producing antioxidant enzymes,which effectively inhibit the production of oxidative stress[38,39].The increase in the concentration of butyrate is beneficial to intestinal health,such as anti-inflammatory effects[40]and regulation of intestinal barrier dysfunction[41].Interestingly,the probiotic strain,AnaerrotruncusandAnaerostipesbelonging to the Firmicutes,can express the enzymes required for butyrate production[42].In this study,AOS-iron intervention effectively improved the abundance ofAnaerrotruncusandAnaerostipes,both of which positively correlated with blood metabolic parameters,iron contents,GSH-PX and T-AOC levels and body weight,and negatively correlated with MDA content(Fig.5).On the other hand,the supplementation of AOS-iron significantly reduced the relative abundance of opportunistic pathogens,includingStreptococcus,Proteus,Morganella,FusobacteriumandSerratia[43–46],and correlation analysis suggested thatMorganella,FusobacteriumandSerratiahad negative effects on the biochemical parameters of IDA,such as serum RBC and Hb levels,iron contents and body weight(Fig.5).In addition,we found that theEnterococcus,gram-positive cocci,had a growth advantage in even feeding low-iron diet because of limited demand for iron,and the advantage was lost during iron supplementation(Fig.4),which was consistent with the findings of Dostal et al.[27].
5.Conclusions
In this study,we studied the effects of AOS-iron on intestinal tissue and microbiota in IDA rats and explored the relationship between intestinal microbiota to iron metabolic disorders by Spearman’s correlation analysis.After supplementation of AOS-iron for 4 weeks,the medium and high dose of AOS-iron effectively repaired cecum and colon tissues lesion.The low dose of AOS-iron was able to increase intestinal microbiota diversity to the baseline,and the beneficial microbiota was enriched while the potential opportunistic pathogens reduced.Spearman’s correlation analysis indicated that biochemical parameters,expect MDA content,were positively correlated withSMB53,Anaerotruncus,AnaerostipesandCoprobacillusbut negatively correlated withMorganella,FusobacteriumandSerratia.The improvement of gut microbiota recovered the iron metabolism disorder to the normal levels.Altogether,this study suggests that AOS-iron conferred additional health benefits to the host by stimulating gut microbiota.
Declaration of Competing Interest
The authors declare no competing financial interests.
Acknowledgments
This research has been co-financed by the General Program of the National Natural Science Foundation of China(No.31271913),Fujian Regional Development Project(2016N3004),Scientific and Technological Innovation Fund of Fujian Agriculture and Forestry University(No.CXZX2018059).
- 食品科学与人类健康(英文)的其它文章
- Effect of xanthan gum on the quality of low sodium salted beef and property of myofibril proteins
- Food science and COVID-19
- Inhibitory effects of oxyresveratrol on ERK and Smad1/2 phosphorylation and HSC activation in preventing carbon tetrachloride-induced rat liver fibrosis
- Effect of proteolytic starter culture isolated from Chinese Dong fermented pork(Nanx Wudl)on microbiological,biochemical and organoleptic attributes in dry fermented sausages
- In vitro and in silico analysis of dual-function peptides derived from casein hydrolysate
- Ganoderma lucidum spore oil(GLSO),a novel antioxidant,extends the average life span in Drosophila melanogaster

