Diversity,novelty,antimicrobial activity,and new antibiotics of cultivable endophytic actinobacteria isolated from psammophytes collected from Taklamakan Desert
Ting Wang,Feina Li,Qinpei Lu,Gang Wu,Zhongke Jiang,Shaowei Liu,Xugela Haben,Elizaveta A.Razumova,Ilya A.Osterman,e,Petr V.Sergiev,e,Olga A.Dontsova,e,f,Xinxin Hu,Xuefu You,Chenghang Sun,*
aInstitute of Medicinal Biotechnology,Chinese Academy of Medical Sciences&Peking Union Medical College,Beijing,100050,China
bBeijing Key Laboratory of Antimicrobial Agents,Institute of Medicinal Biotechnology,Chinese Academy of Medical Sciences&Peking Union Medical College,Beijing,100050,China
cCollege of Life Science,Xinjiang Normal University,Urumchi,830054,China
dLomonosov Moscow State University,Moscow,119992,Russia
eCenter of Life Sciences,Skolkovo Institute of Science and Technology,Moscow,143025,Russia
fShemyakin-Ovchinnikov Institute of Bioorganic Chemistry,Russian Academy of Sciences,Moscow,119992,Russia
Keywords:
Taklamakan desert
Endophytic actinobacteria
Novelty
Diversity
Streptogramin-type antibiotic
A B S T R A C T
Three hundred and twenty endophytic actinobacterial strains were isolated from psammophytes collected from Taklamakan Desert and identified.Among them,three strains already had been identified as new species of two genera and sixteen isolates showed relatively low 16S rRNA similarities<98.6% to validly described species.Seventy-five of the isolates were selected as representative strains to screen antibacterial activity and mechanism.Forty-seven strains showed antagonistic activity against at least one of the indicator bacteria.Two Streptomyces strains produced bioactive compounds inducing DNA damage,and two Streptomyces strains produced bioactive compounds with inhibitory activity on protein biosynthesis.Notably,the strain Streptomyces sp.8P21H-1 that demonstrated both strong antibacterial activity and inhibitory activity on protein biosynthesis was prioritized for exploring new antibiotics.Under the strategy of integrating genetics-based discovery program and MS/MS-based molecular networking,two new streptogramin-type antibiotics,i.e.,acetyl-griseoviridin and desulphurizing griseoviridin,along with known griseoviridin,were isolated from the culture broth of strain 8P21H-1.Their chemical structures were determined by HR-MS,and 1D and 2D NMR.Desulphurizing griseoviridin and griseoviridin exhibited antibacterial activities by inhibiting translation.
1.Introduction
Actinobacteria residing in special environments have a unique capacity to produce bioactive novel compounds and become irreplaceable sources to inhibit antibiotic-resistant pathogens[1,2].Facing the dilemma of rediscovery of known antibiotics,novel strategies and innovative techniques have been developed to speed up the discovery of new and efficient antibiotics[3,4],such as integrating new and classical biological screening model[5],the“bottom-up”genetics-based bioinformatics tools on behalf of genome mining[6,7],as well as “top-down”approach of molecular networkingandothercheminformaticstechniques.Inthisstudy,the diversity,novelty,antimicrobial activityand potential for producing new secondary metabolites from cultivable actinobacteria from plants of Taklamakan Desert were investigated.Culture-dependent method utilizing a variety of media was employed to select actinomyces.Apartfrom screening antibacterialactivitiesagainst“ESKAPE”strains(Enterococcus faecium,Staphylococcus aureus,Klebsiella pneumoniae,Acinetobacter baumannii,Pseudomonas aeruginosa,andEnterobacterspecies),high-throughputscreeningusing a double fluorescence reported system was also implemented to recognize strains producing inhibitors of ribosome or/and DNA biosynthesis[8,9].Dereplication strategies which integrated anti-SMASH platform analysis and MS/MS-based molecular networking were applied to find newantibiotics fromStreptomyces sp.8P21H-1.Finally,two new streptogramin-type antibiotics,i.e.,acetyl-griseoviridin and desulphurizing griseoviridin,along with known griseoviridin,were structurallyelucidated by HR-MS,and 1D and 2D NMR(Fig.1).

Fig.1.Chemical structure of the isolated compounds.(A)Acetyl-griseoviridin,(B)desulphurzing griseoviridin,and(C)griseoviridin.
2.Materials and methods
2.1.Collection of plant samples
A total of fifteen plant samples were collected in Taklamakan Desert,located in Xinjiang,Uygur Autonomous Region,China.The sampling information in detail is listed in Table S1.All the samples were packed in sterilized polyethylene bags and carried back to the laboratory at the earliest possible time.
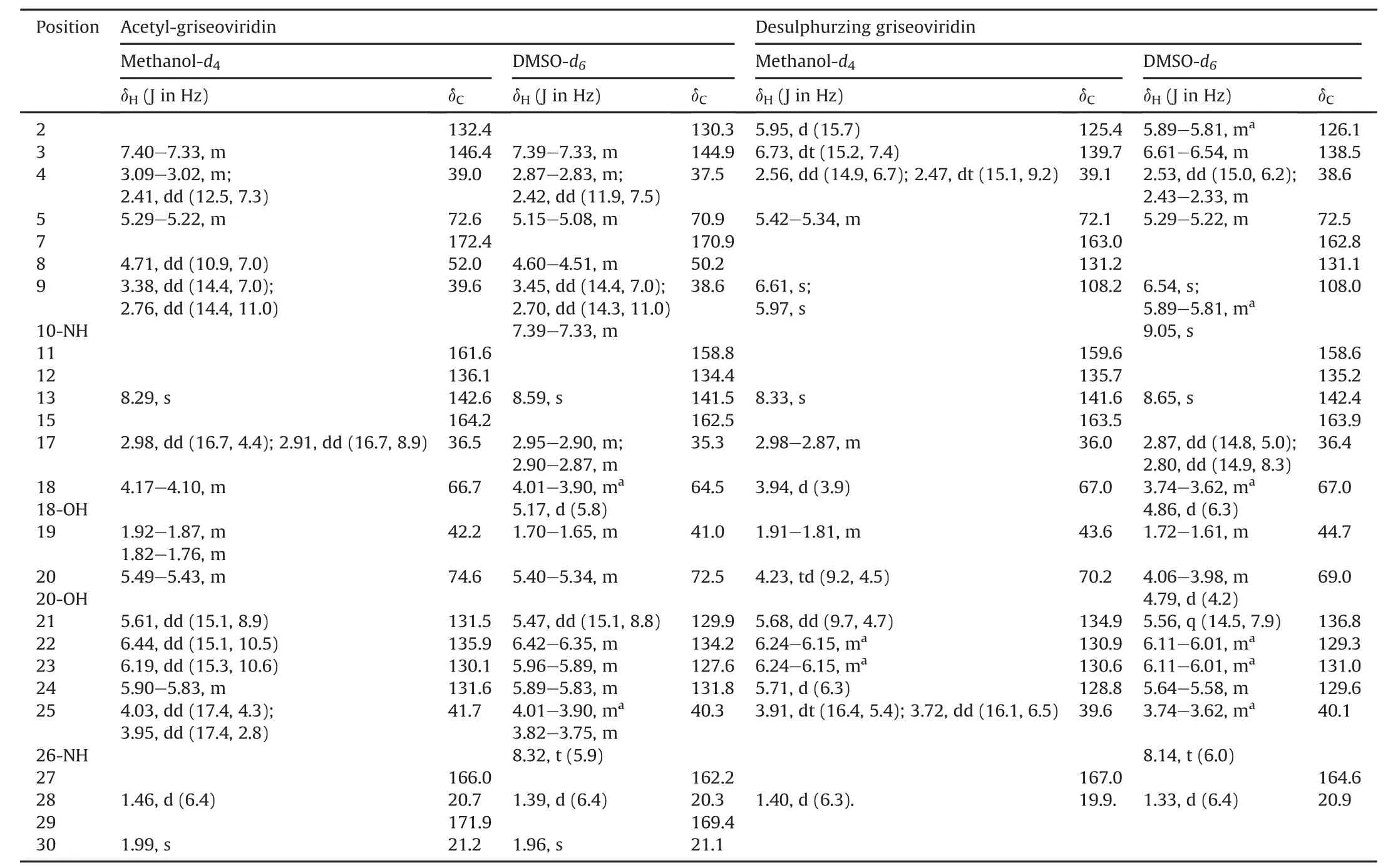
Table 1 The1H NMR(600 MHz)and13C NMR(150 MHz)data for acetyl-griseoviridin and desulphurzing griseoviridin.
2.2.Surface sterilization and processing
All samples wereimmediatelyprocessed for surface sterilization after being air-dried.The samples were subjected to a six-step surface sterilization procedure[10].In order to check the surface sterilization results,the 200μL of last rinse solution was spread on ISP2 agar plates and incubated at 28°C for 1 week to check the microbial growth.After being dried in the laminar flow hood for 2 days,the successfully surface-sterilized samples were ground into powders by using a micromill.
2.3.Endophytic actinobacteria isolation and maintenance
For the selective isolation of actinobacteria,12 isolation media were prepared(Table S2)and supplemented with nalidixic acid(20 mg/L),cycloheximide(50 mg/L)and potassium dichromate(50 mg/L)to prevent the growth of Gram-stain negative bacteria and fungi.After the plant powders were dispersed in the isolation media,the plates were incubated at 28°C for 2 months.Single colony was picked up and streaked on the freshly prepared YIM38 medium[11]to obtain the pure isolates.The pure cultures were preserved in glycerol suspensions(20%,V/V)at-80°C.
2.4.identification of endophytic actinobacteria
Extractions of genomic DNA and PCR amplification of 16S rRNA gene were performed as described by Jiang et al.[12].The PCR products were purified and sequenced on the ABI PRISMTM3730XL DNA Analyzer(Thermo Fisher Scientific,USA).The obtained sequences were compared with available 16S rRNA gene sequences in the EzBioCloud(https://www.ezbiocloud.net/)to determine an approximate phylogenetic affiliation of each strain[13].Phylogenetic tree was constructed by the algorithms of neighbor-joining in MEGA 7.0[14-16].Evolutionary distance was estimated by the Kimura’s two-parameter model[17].The bootstrap analysis was performed with 1000 replications[18].

Table 2 Information on genera distribution of actinobacterial strains in this study.
2.5.Antimicrobial screening
The agar well diffusion method was used to screen the antimicrobial activity of isolated actinobacterial strains.The indicator bacteria used for antibacterial assay consisted of 12 bacteria,which were divided into two groups based on consisted of drug-sensitive or drug-resistant of“ESKAPE”bacteria.The detailed information is listed below:Enterococcus(ATCC 33186,310682),Staphylococcus aureus(ATCC 29213,ATCC 33591),Klebsiella pneumoniae(ATCC 10031,ATCC 700603),Acinetobacter baumannii(2799,ATCC 19606),Pseudomonas aeruginosa(ATCC 27853,2774)and Escherichia coli(ATCC 25922,ATCC 35218).For each type of bacteria,the strain listed in the former was drug-sensitive strain and the latter was drug-resistant strain.All bacteria were obtained from American Type Culture Collection(ATCC),expect for Enterococcus 310682(resistant to vancomycin)and Pseudomonas aeruginosa 2774(resistant to aminoglycosides and carbapenems)from the clinic.Small-scale fermentation of selective actinobacterial strains and antibacterial assays were performed as described earlier[11].
2.6.High-throughput screening using a double fluorescent protein reporter system
Ribosome and DNA biosynthesis inhibitors were screened bythe double fluorescent protein reporter system with reporter strain JW5503-pDualrep2(also abbreviated as the pDualrep2 reported system)[8,9,19].The reporter strain JW5503-pDualrep2 was constructed by the strain JW5503(ΔtolC)of E.coli transformed by a report plasmid,pDualrep2,to screen compounds for antimicrobial activity with simultaneous classification by their mechanisms of action.Briefly,0.1 mL of ethyl acetate extracts was dried in laminarflow hood and 100 μL of DMSO was added to each sample.2 μL of each sample was applied to agar plate containing a lawn of the reporter strain.After overnight incubation at 37°C,the plate was scanned by ChemiDoc(Bio-Rad)system with two channels consisting of “Cy3-blot”(553/574 nm,green pseudocolor)for RFP triggered by DNA biosynthesis inhibitors and “Cy5-blot”(588/633 nm,red pseudocolor)for Katushka2S triggered by ribosome inhibitors.Levofloxacin and erythromycin were used as positive controls for DNA biosynthesis and ribosome inhibitors,respectively.The procedure was also applied to Section 3.9.
2.7.identification of biosynthesis gene clusters for secondary metabolite
The procedures of genomic DNA extraction,whole-genome sequencing and subsequently assembly were performed according topreviouslydescribedmethodsbyLietal.[20].Thesequenceswere examined using the bioinformatics tool antiSMASH 5.0.0(https://antismash.secondarymetabolites.org/).The bacterial database was setasthereferencedepository,and the FASTA filewithunannotated nucleotide sequence was uploaded to antiSMASH platform(version 5.0.0).The gene cluster basic local alignment search tool(BLAST)comparativeanalysisand“ActiveSiteFinder”functionwereselected withrelaxeddetectionparametersthatdetectnotonlywell-defined clusterscontainingallrequiredpartsbutalsopartialclustersmissing one or more genes.The characteristics of each predicted secondary metabolite wereinvestigated bysearching related literature.On the basis of the results of the high-throughput screening platform,the putative secondary metabolites that might target ribosome were prioritized for further analysis.
2.8.Molecular networking
The crude extract(1 mg)of Streptomyces sp.8P21H-1 cultured in ISP2 broth was dissolved in methanol(1 mL)and filtered with 0.22μm filter membrane.Then the sample was analyzed by UPLCHRESI-MS/MS(Waters Xevo G2-XS QTof;ACQUITY UPLC BEH C18column,2.1 mm ×100 mm,1.7μm).MS method was acquired by data dependent acquisition performed in positive ion mode for Global Natural Products Social Molecular Networking(GNPS)analysis.The full MS survey scanwas performed for 0.1 s time in the range of 100-1600 Da,and MS/MS scanned over a mass range of 50-1600 Da with the same scan time.The raw data obtained were converted digitally to mzxml format files using freely available MSConvert software(http://www.proteowizard.sourceforge.net)[21]and were uploaded subsequently to the online workflow at GNPSplatform (http://gnps.ucsd.edu).TheMS/MSmolecular networking was generated using the GNPS molecular networking workflow(METABOLOMICS-SNETS-V2),in which the MS-Cluster was activated.The generated molecular networking was visualized using Cytoscape 3.7.1[22]and searched for clusters of m/z data that could be used toclarify the compounds generating the clusters.

Table 3 The sequence analyses based on almost full-length 16S rRNA gene of 16 potential new species and 3 new species.
2.9.Large-scale fermentation,extraction and isolation workflow
Streptomycessp.8P21H-1wasgrowninseedbroth(ISP2medium)for two days at 28°C,and then an aliquot(50 mL)was transferred to inoculate1000mLoffermentationmediumin5-Lshake flasks,which were cultivated at 180 rpm and 28°C for 6 days.The whole culture broth(20 L)was extracted with equal volumes of ethyl acetate three times(20L/time)toaffordacruderesidue(0.96g)afterconcentration in vacuo.In order to optimize the isolation workflow toward previouslypredictedcompounds,2μLofthewholeextractsolution(1mg/mL)was analyzed by UPLC-HRMS/MS in positive ion mode.All the other crude residue was subjected to Sephadex LH-20 column chromatography and eluted with MeOH to yield 35 fractions(Frs.1-35).Frs.1to4containingtargetedcompoundsweremergedtogetherand then chromatographed over an ODS column chromatography with a gradientsystemofMeOH-H2O(20:80,40:60,60:40,80:20,95:5,V/V)to yield seven subfractions(Subfrs.A-G).Subfr.B was successively purified by Zorbax SB-C18column(9.4 mm×250 mm,5μm,Agilent Technologies Inc.,Santa Clara,USA)with MeOH-H2O(28:72,V/V)as mobile phase at the flow rate of 2 mL/min in semi-preparative HPLC(Agilent 1200,Agilent Technologies Inc.,Santa Clara,USA)to obtain griseoviridin(30.5mg,Rt=22.2min).Subfr.Cwasseparatedbysemipreparative HPLC with a gradient mobile phase(MeOH-H2O,47% MeOH isocratic conditions for 37 min,then 47%-90% linear gradient over 15 min; flow rate of 2 mL/min)to yield acetyl-griseoviridin(1.8 mg,Rt=26.3 min)and desulphurzing griseoviridin(1.38 mg,Rt=46.6 min),respectively.
2.9.1.Acetyl-griseoviridin
White,amorphous powder;1H NMR and13C NMR data,see Table 1; HR-ESI-MS m/z 542.1578 [M+Na]+(calcd.for C24H29N3O8SNa,542.1573).
2.9.2.Desulphurzing griseoviridin
White,amorphous powder;1H NMR and13C NMR data,see Table 1; HR-ESI-MS m/z 468.1749 [M+Na]+(calcd.for C22H27N3O7Na,468.1747).
2.10.Antibacterial activities of isolated compounds
The minimum inhibitory concentration(MIC)values of the compounds were evaluated for antibacterial activities using an agar dilution method according to the Clinical and Laboratory Standards Institute recommended guidlines[23].Levofloxacin was used as the positive control(final concentration ranged from 0.03μg/mL to 128μg/mL).The finaltestconcentrationsofgriseoviridinrangedfrom 0.03 μg/mL to 64 μg/mL.Final concentrations of acetyl-griseoviridin anddesulphurzinggriseoviridinrangedfrom0.03μg/mLto32μg/mL.The test bacteria were cultured in Mueller-Hinton(MH)broth mediumat35°Cfor8handthenwereadjustedtoaturbidityof0.5using the McFarland standard.The bacterial suspensions were inoculated onto the drug-supplemented MH agar plates using a multipoint inoculator.After being incubated at 35°C for 16 h,the MICs were definedasthelowestconcentrationsthatpreventedvisiblegrowthof the bacteria.
3.Results and discussion
3.1.Isolation of endophytic actinobacteria
In the present study,a total of 428 purified isolates were obtained from 15 samples collected from Taklamakan Desert.The 16S rRNA gene sequence analyses revealed that 320 strains belonged to actinobacteria,which were assigned to 23 genera in 14 families(Fig.2).Streptomyces was the dominant genus(119 strains,37.2%,Table 2),which was in line with the previous reports[24,25].In addition to Streptomyces,other six genera,i.e.,Micromonospora,Nocardia,Nonomuraea,Nocardiopsis,Amycolatopsisand Modestobacter,were widespread in desert.Besides,other rare actinobacterial genera from desert plants have been identified such as Labedella,Rathayibacter,Leucobacter,Frigoribacterium,Kineococcus and Aeromicrobium.
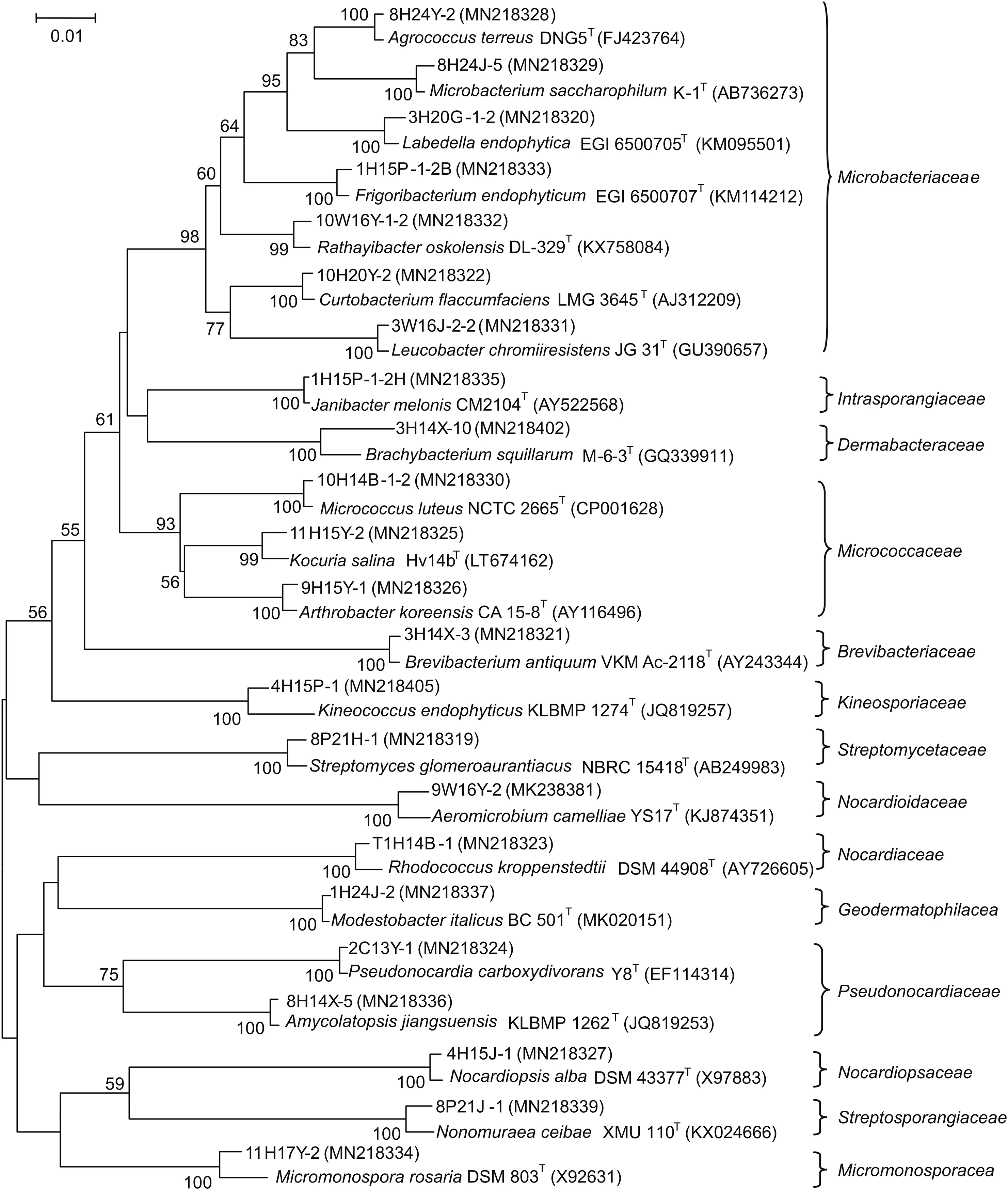
Fig.2.Phylogenetic tree based on the 16S rRNA gene sequences using neighbor-joining method for 23 representative actinobacterial strains and their closely related type strains.Numbers at nodes indicate the level of bootstrap support>50% based on 1000 replications.Bar,2 nt substitutions per 100 nt.
The distributions of the 320 strains isolated from 12 different media and 5 different plant tissues are shown in Figs.3A and B,respectively.Among the 12 different isolation media,both M10 and M3 were the most successful media for isolating actinobacterial strains.The highest number(65 strains,20.3%)and the lowest number(2 strains,0.6%)of the endophytic actinobacteria obtained were isolated from S6(Phragmites australis)and S8(Reaumuria soongorica),respectively.The result of calculating the average number of actinobacteria from each plant tissue showed that the actinobacteria isolated from roots had the highest diversity(30 strains in 5 genera from one sample).Although the diversity was relatively low,actinobacteria isolated from barks were relatively more than those from branches,leaves and flowers.Our findings supported the previous reports that many endophytes originate from the rhizosphere of plants[26].
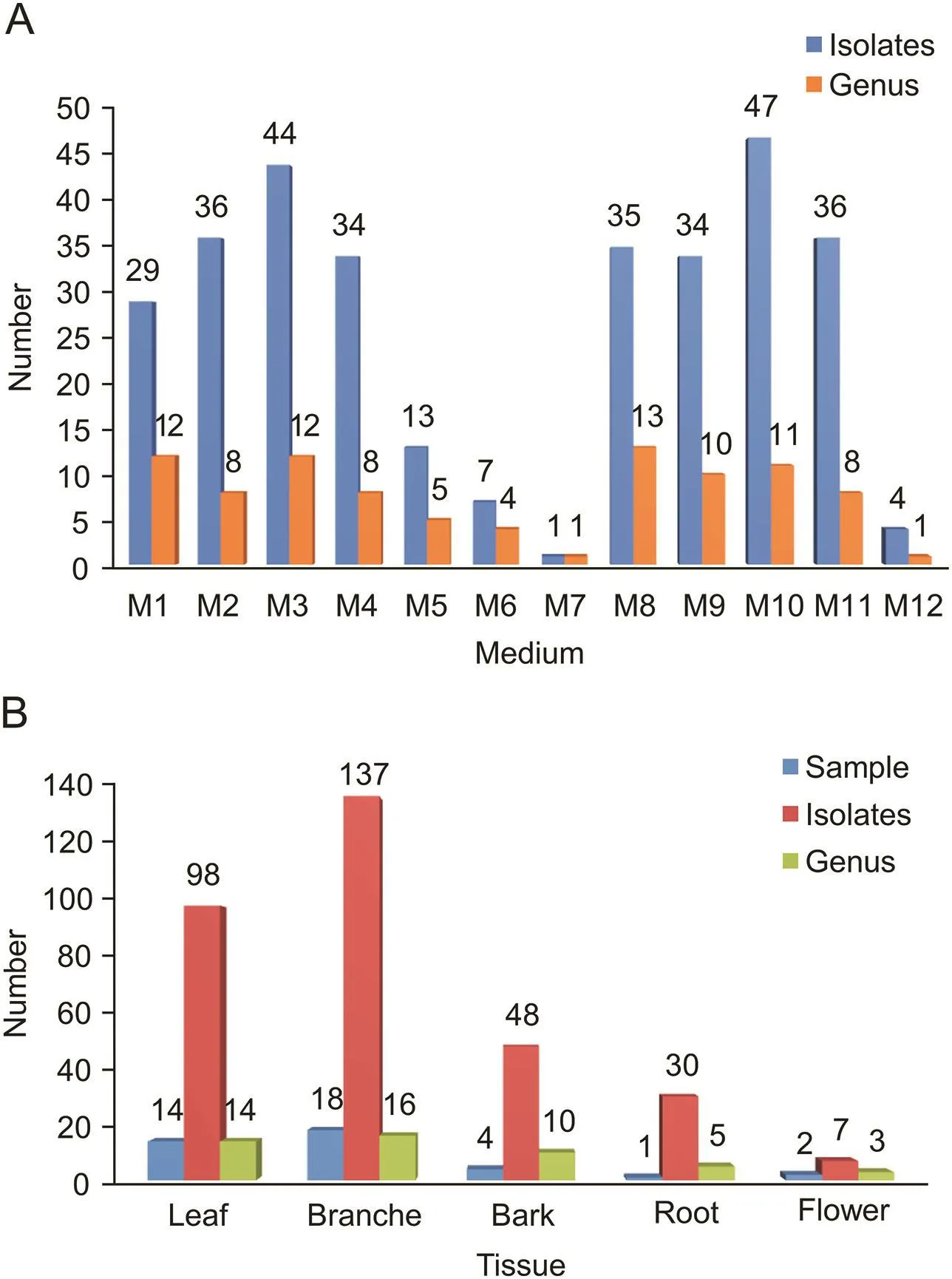
Fig.3.Diversity of cultivable actinobacteria from psammophytes collected in Taklamakan Desert.(A)Number of actinobacterial isolates recovered from the different culture media;(B)number of actinobacterial isolates from different plant tissues.
3.2.Novelty analysis of endophytic actinobacteria
Among the 320 actinobacterial strains,16 strains exhibited low sequence similarities<98.65%,the threshold for differentiating two species[27]with validly described species based on the results of BLAST search in EzBiocloud database(http://www.ezbiocloud.net/)(Table 3).The results indicated that these isolates might represent hitherto unrecognized new species.In the phylogenetic dendrogram based on almost full-length 16S rRNA gene sequences(Fig.4),these potential novel strains fell into three genera,i.e.,Brachybacterium(9 strains),Kineococcus(4 strains),and Microbacterium(3 strains).Strains 10H14J-1,2H14Y-4,8H14J-1-1,8H14X-2,3H14X-6,1H14B-1,6H14B-1 and 3H14B-1-1 were isolated from Populus euphratica at sample S4.These 8 potential novel strains formatted a monophyletic in clade,which may represent a novel strain of genus Brachybacterium.In the same way,strains 1H15P-2,8H15P-1,4H15P-1 and 3H15P-2 were isolated from Populus euphratica in sample S5.These 4 potential novel strains formatted a monophyletic clade,which may represent a novel strain of genus Kineococcus.These strains will be further identified with a polyphasic approach to determine their taxonomic positions.In addition,three strains with relatively high 16S rRNA sequence similarities,i.e.,11W25H-1T,8H24J-4-2T,and 9W16Y-2T,had been identified as type species and named as Labedella phragmitis,Labedella populi,and Aeromicrobium endophyticum,respectively[28,29].In this study,novelty analysis suggested that desert plants have the potential as excellent sources of novel actinobacterial species.
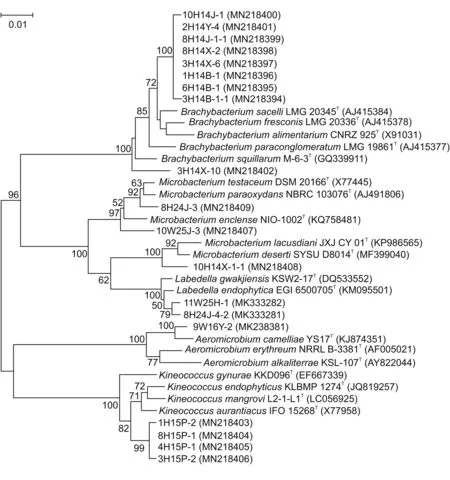
Fig.4.Phylogenetic neighbor-joining tree of 16 potential novel strains and 3 new species based on almost full-length 16S rRNA gene sequences.Numbers at nodes indicate the level of bootstrap support>50% based on 1000 replications.Bar,1 nt substitutions per 100 nt.
3.3.Antimicrobial activities
Seventy-five strains were selected for antimicrobial assay based on analyses of partial 16S rRNA gene sequences and phenotypic characteristics.The antibacterial profilesofthe selected strains against“ESKAPE”bacteria are shown in Fig.5 and Table 2.Among the 75 strains selected for antimicrobial assay,47 strains showed antagonistic activity against at least one of the indicator bacteria.They were distributed in 20 different genera,and the predominant active strains were affiliated to genus Streptomyces,which was in accordance with the reported data[30],since members of the genus Streptomyces usually possess a numberofbiosyntheticgeneclustersthatencodemultifunctional biosynthetic enzymes[31,32].Twenty Streptomyces strains showed inhibitory activity against at least one of Grampositive bacteria,and seven of them also showed activity against at least one of Gram-negative bacteria(Table S3).
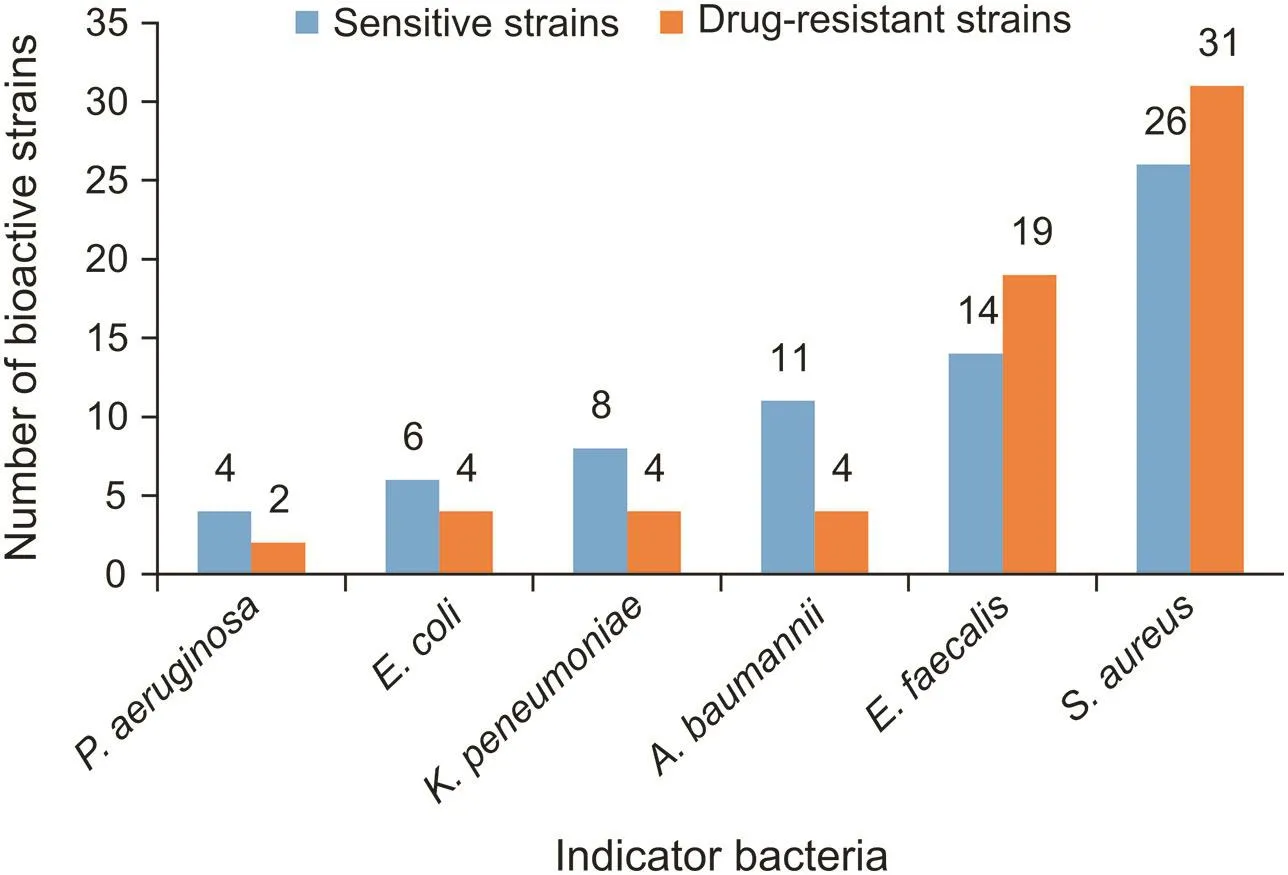
Fig.5.The antibacterial profiles of the actinobacteria against“ESKAPE”bacteria.
Rare actinobacteria are also important sources for the discovery of novel antibiotics[33].Recently,saccharothriolides A-C[34]and saccharothriolides D-F[35],a group of new secondary metabolites,have been found from Saccharothrix sp.A1506 isolated from a soil sample,which further indicates the rare actinobacteria deserve to be studied extensively to find new antibiotics.In the present study,active strains distributed in 18 rare genera such as Brachybacterium,Leifsonia,Arthrobacter,Labedella and Leucobacter were obtained and have been rarely studied for their bioactive secondary metabolites.Notably,both strains 11W25H-1Tand 8H24J-4-2Tas two new species in rare genus Labedella,showed promising activity against S.aureus(Table S3).Thus,these rare actinobacteria deserved further studies on their bioactive secondary metabolites.
3.4.High-throughput screening using a double fluorescent protein reporter system
The pDualrep2 reporter system is a very sensitive screening model for detection of compounds that inhibit protein translation and DNA biosynthesis.According to the results of primary screening,twoStreptomycesstrains,3H14X-7 and 8P21H-1,induced Katushka2S expression as erythromycin did.Meanwhile,two Streptomyces strains,2H20Y-2 and 3H20G-2,induced SOS-response as levofloxacin did(Fig.6).Furthermore,taking results of antibacterial activities of these four Streptomyces strains into consideration,strain 8P21H-1 was prioritized as an example for chemical research to find new antibiotics targeted on ribosome.
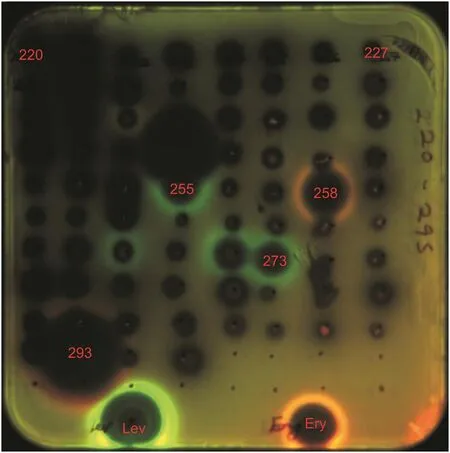
Fig.6.Induction of a two-color dual reporter system sensitive to inhibitors of the ribosome progression or inhibitors of DNA replication,respectively.Spots of erythromycin(Ery),levofloxacin(Lev),and tested samples were placed on the surface of an agar plate containing E.coliΔtolC cells transformed with the pDualrep2 reporter plasmid.The shown image is the fluorescence of the lawn of E.coli cells scanned at 553/574 nm(green pseudocolor)for red fluorecent protein fluorescence and 588/633 nm(red pseudocolor)for Katushka2S fluorescence.Induction of expression of Katushka2S was triggered by translation inhibitors,while RFP was upregulated by induction of DNA damage SOS response.255:2H20Y-2;258:3H14X-7;273:3H20G-2;293:8P21H-1.
3.5.identification of biosynthesis gene clusters for secondary metabolite
The assembled draft genome of strain 8P21H-1 was 6.65 Mbp.A total of 71 biosynthetic gene clusters were identified by antiSMASH platform(https://antismash.secondarymetabolites.org/).Single gene clusters encoded the biosynthetic pathway for“griseoviridin/viridogrisein”non-ribosomal peptide synthases-polyketide synthases(NRPS-PKS),indole,nucleoside,lanthipeptide,lassopeptide,melanin,and LAP,respectively.There were four gene clusters for bacteriocins,four for siderophores,eight for terpenes,twenty-three for PKS,and twenty-four for NRPS.Furthermore,one gene cluster showed no predicted product family.AntiSMASH analysis also showed putative gene clusters previously identified for micromonolactam,nanchangmycin,bleomycin,griseoviridin/viridogrisein and diisonitrile antibiotic SF2768 biosynthesis in strain 8P21H-1.Among these putative compounds,especially for known antibiotics,the“griseoviridin/viridogrisein”demonstrated antibacterial activity by inhibiting ribosome[36].Presumably,this group of antibiotics was the active components of the strain as the results of bioinformatics analysis indicated.
3.6.Molecular networking
One major advantage of the molecular networking strategy is the function of identifying known compounds and potential analogs,thereby allowing for the prioritization of the isolation workflow and guiding the user toward unknown molecules[4].A molecular networking was established to investigate the sample of Streptomyces sp.8P21H-1 using the GNPS platform(https://gnps.ucsd.edu/).The crude extract was subjected to LC-MS/MS analysis,a major chromatographic fraction with nominal mass of 477 and similar UV characteristic spectrum profiled to that of griseorividin.Based on the analysis of molecular networking(Fig.7A),the major component was identified to be griseoviridin and fell into the cluster(Fig.7B).This result was consistent with that of the anti-SMASH analysis that griseoviridinwas a potential compound of one gene cluster.Other nodes in cluster(Fig.7B)had clear connections with griseoviridin and were predicted as its analogs.These analogs were not identified to the previously reported compounds according to the dereplication tool at GNPS platform[37],thus suggesting that they were potential newcompounds.Given the limit of the amounts of analogs present in the cluster(Fig.7B),only the putative griseoviridin-like acetyl-griseoviridin([M+H]+ion peak at m/z 520.275)and desulphurzing griseoviridin([M+H]+ion peak at m/z 446.269)were suitable for further purified and structure elucidation by NMR analysis.
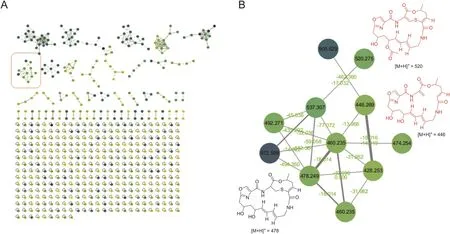
Fig.7.(A)Molecular networking of Streptomyces sp.8P21H-1.(B)Molecular networking of griseoviridin analogs.
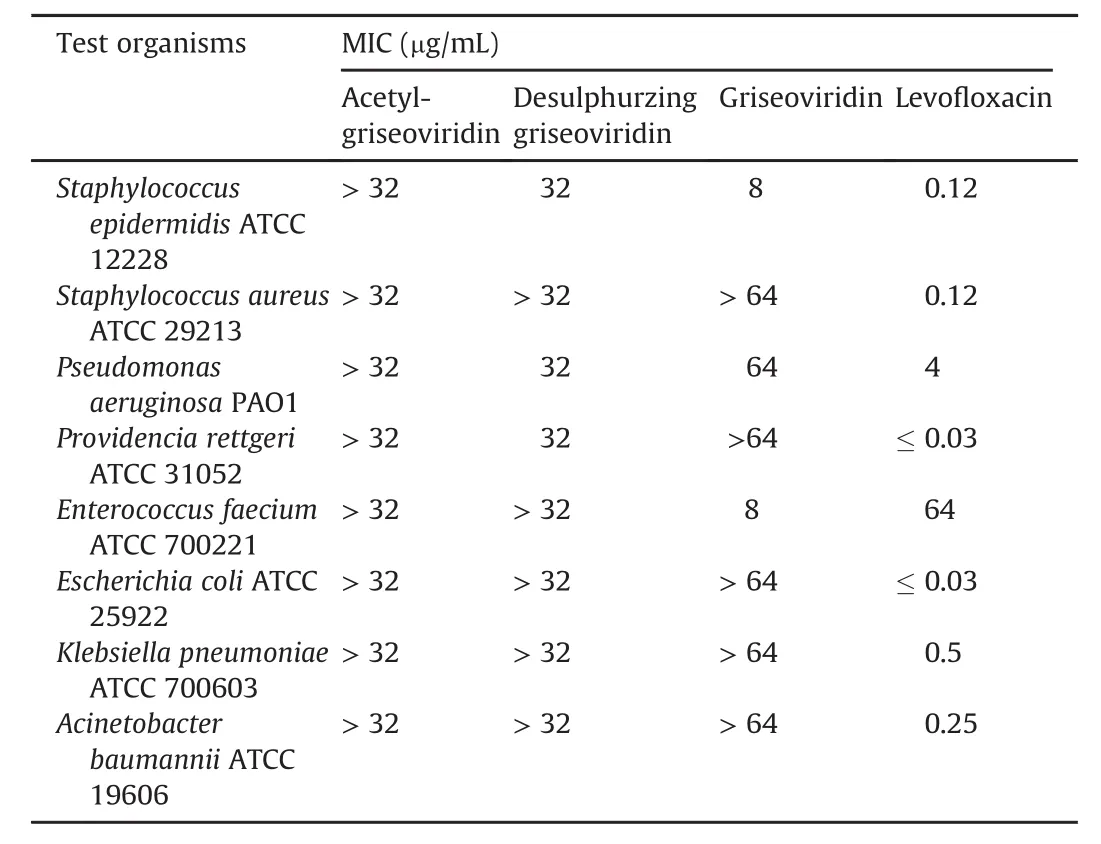
Table 4 Antimicrobial activities of acetyl-griseoviridin,desulphurzing griseoviridin,and griseoviridin.
3.7.Structure elucidation of compounds
Acetyl-griseoviridin was isolated as a white amorphous powder.The molecular formula was determined as C24H29N3O8S on the basis of HR-ESI-MS(Fig.S1)(m/z 542.1578[M+Na]+,calc.for 542.1573),requiring 12°of unsaturation.The 1D NMR data(Table 1)of acetyl-griseoviridin suggested the presence of seven quaternary carbons,ten methines,five methylenes,and two methyl groups.Careful analysis of the NMR data(Figs.S2-S12,Table 1)indicated that acetyl-griseoviridin was an analogue of griseoviridin[38,39]but with an acetoxy rather than a hydroxyl group attached to C-20(δC74.6).The heteronuclear multiple bond correlation(HMBC)(Fig.8)from H-20(δH5.49-5.43)to C-29(δC171.9),and from H3-30(δH1.99)to C-20 and C-29 indicated the presence of the acetoxy group linked to C-20 as well.

Fig.8.Summary of1H-1H COSY and HMBC experiments of acetyl-griseoviridin and desulphurzing griseoviridin in methanol-d4.
Desulphurzing griseoviridin was a white amorphous powder with a molecular formula C22H27N3O7based on the HR-ESI-MS(Fig.S13)(m/z 468.1749[M+Na]+,calc.for 468.1747).The HR-ESIMS showed the S atom was absent in desulphurzing griseoviridin.The NMR spectra(Figs.S14-S26,Table 1)of desulphurzing griseoviridin were similar to those of griseoviridin[38,39],suggesting the presence of a similar skeleton.The chemical shifts of C-9(δC39.5)and C-8(δC52.0)in griseoviridin were changed dramatically to downfield in desulphurzing griseoviridin(C-9 atδC108.2;C-8 at δC131.2),indicating that presence of an additional double bond in position C-8 and C-9 and absence of the S atom in position 2,which was further confirmed by the HMBC correlations(Fig.8)from H2-9(δH6.61,5.97)to C-7(δC163.0)and C-8(δC131.2).
Known griseoviridin was isolated as a white amorphous powder.In the HR-ESI-MS spectrum,a molecular ion peak at m/z 500.1632 [M+Na]+(calc.forC22H27N3O7SNa)provided the elemental formula C22H27N3O7S.The chemical structure was identified by comparison of the NMR data(Table S4)with those in the literature[38,39].Griseoviridin as a representative member of polyunsaturated macrolactones was first discovered in the culture of Streptomyces graminofaciens in 1953[40].
Both acetyl-griseoviridin and desulphurizing griseoviridin are classified as streptogramin antibiotics just like griseoviridin,which are a family of natural products isolated from Streptomyces.To the best of our knowledge[40-42],streptogramin family is generally divided into two structurally different subclasses of type A and type B.Acetyl-griseoviridin and desulphurizing griseoviridin are cyclic polyunsaturated macrolactones belonging to type A.Chemical modifications to various members of the streptogramins have been pursued,andthenewnaturalcompoundsprovideanew perspective for further structure modifications.
3.8.Antibacterial activities of isolated compounds
All isolated compounds were evaluated for their antibacterial activities by MIC assay.The results(Table 4)indicated that only desulphurzing griseoviridin and griseoviridin exhibited modest antibacterial activities against partial test strains.Griseoviridin showed antibacterial activities against Staphylococcus epidermidis ATCC 12228 and Enterococcus faecium ATCC 700221 with MIC value of 8μg/mL.Desulphurzing griseoviridin showed antibacterial activities against Staphylococcus epidermidis ATCC 12228,Pseudomonas aeruginosa PAO1,and Providencia rettgeri ATCC 31052 with MIC value of 32μg/mL.It had been reported that streptogramins could be inactivated by acetyl transferases,which could explain why acetyl-griseoviridin showed no antibacterial activity[41].
3.9.Mechanism of action determination for isolated compounds
The mechanism of action of griseoviridin(named as H11 when tested),desulphurizing griseoviridin(named as 1792 when tested)and acetyl-griseoviridin(named as 1791 when tested)were tested with the pDualrep2 reported system.The results are shown in Fig.9,and both desulphurzing griseoviridin and griseoviridin demonstrated antibacterial activity by inhibiting translation.This finding corresponded to the primarily screening results described in antibacterial activity and high-throughput screening.Remarkably,a thorough literature survey has revealed that this type of molecules is believed to prevent access of the active aminoacyltRNA to the ribosomal A site[41,42],thus prohibiting peptide bond formation.Importantly,members of these subclasses have shown excellent synergistically biological activities(100-fold than each individual compound)with the other subclass,cyclic octadepsipeptide.In consideration of the excellent activity and unique mechanism of action,the typical antibiotics are very valuable to further research for synergistical action and bioactivity.

Fig.9.Agar plate coated with a layer of reporter strain JW5503(ΔtolC)of E.coli transformed by plasmid pDualrep2 with the tested compounds(1791;1792;H11).Cells with tested compounds were identified by white circles and corresponding number.1791 was on behalf of acetyl-griseoviridin;1792 was on behalf of desulphurzing griseoviridin;H11 was on behalf of griseoviridin.
4.Conclusions
As an illustratively systemic study,the pipeline from isolation and identification of actinobacteria to discover bioactive compounds was successfully accomplished.Among 320 endophytic actinobacteria,three strains were identified as new species,and 16 strains were considered as potential new taxa.The strain 8P21H-1 with strong antibacterial activity targeting ribosome was selected for exploring new antibiotics.Ultimately,two new streptogramintype antibiotics,i.e.,acetyl-griseoviridin and desulphurizing griseoviridin,along with known griseoviridin,were isolated from the culture broth of the strain.Acetyl-griseoviridin possessed an acetylated hydroxyl group and desulphurizing griseoviridin obtained an exocyclic double bond.Desulphurizing griseoviridin and griseoviridin exhibited antibacterial activities by inhibiting translation.A valuable integrated strategy for tracing new and/or bioactive compounds was established in this study,which is conducive to addressing the key bottleneck of rediscovery of known antibiotics and helping to find novel secondary metabolites.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
The authors are grateful to Dr.Zhanfeng Xia from the Tarim University,Xinjiang,China,forassistanceinsamplingatTaklamakan Desert,and Dr.MohsinT.Cheema fromUniversity of the Punjab and Dr.Rongfeng Li from Johns HopkinsUniversity for useful discussion.The research work was supported by CAMS Innovation Fund for Medical Sciences(Grant Nos.CAMS 2017-I2M-B&R-08 and 2017-I2M-1-012),the PUMC Doctoral Innovation Fund Project(Grant No.2018-1007-16),the Drug Innovation Major Project of China(Grant No.2018ZX09711001-007-002),the Russian Foundation for Basic Research(Grant No.20-54-53014),and the National Natural Science Foundation of China(Grant No.82011530051).
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2020.06.004.
 Journal of Pharmaceutical Analysis2021年2期
Journal of Pharmaceutical Analysis2021年2期
- Journal of Pharmaceutical Analysis的其它文章
- Current diagnostic and therapeutic strategies for COVID-19
- Chemically modified carbon-based electrodes for the determination of paracetamol in drugs and biological samples
- Development of chromatographic technologies for the quality control of Traditional Chinese Medicine in the Chinese Pharmacopoeia
- Design and preparation of a new multi-targeted drug delivery system using multifunctional nanoparticles for co-delivery of siRNA and paclitaxel
- The effective transfection of a low dose of negatively charged drugloaded DNA-nanocarriers into cancer cells via scavenger receptors
- A dual-signal sensor for the analysis of parathion-methyl using silver nanoparticles modified with graphitic carbon nitride
