Current diagnostic and therapeutic strategies for COVID-19
Binbin Chen,Mengli Liu,Chengzhi Hung
aCollege of Chemistry and Chemical Engineering,Southwest University,Chongqing,400715,China
bSchool of Chemistry&Molecular Engineering,East China University of Science and Technology,Shanghai,200237,China
cKey Laboratory of Luminescence Analysis and Molecular Sensing,Ministry of Education,Southwest University,Chongqing,400715,China
dKey Laboratory of Luminescence and Real-Time Analytical System,Chongqing Science and Technology Bureau,College of Pharmaceutical Science,Southwest University,Chongqing,400715,China
Keywords:
Diagnosis
Treatment
COVID-19
Coronavirus
SARS-CoV-2
A B S T R A C T
The outbreak and spread of novel coronavirus disease 2019(COVID-19)with pandemic features,which is caused by severe acute respiratory syndrome coronavirus 2(SARS-CoV-2),have greatly threatened global public health.Given the perniciousness of COVID-19 pandemic,acquiring a deeper understanding of this viral illness is critical for the development of new vaccines and therapeutic options.In this review,we introduce the systematic evolution of coronaviruses and the structural characteristics of SARS-CoV-2.We also summarize the current diagnostic tools and therapeutic strategies for COVID-19.
1.Introduction
The outbreak and spread of novel coronavirus disease 2019(COVID-19)pose the most serious threat to global public health[1-5].According to the World Health Organization(WHO),this disease was designated as a“public health emergency of international concern”on January 30,2020.The corresponding virus was of ficially designated as 2019-nCoV by the Chinese Center for Disease Control and Prevention,and later revised to severe acute respiratory syndrome coronavirus 2(SARS-CoV-2)due to a good sequence homology towards SARS-CoV genome[6].Genomic sequencing demonstrated that SARS-CoV-2 was 96.2% identical to a bat coronavirus and shared a 79.5% sequence similarity to SARSCoV,indicating that this virus is most likely to originate from bats[7].
COVID-19 has become a major concern globally,with over 70 million confirmed cases and 1.6 million lives lost as of December 15,2020[8].Moreover,millions of people’s lives are at great risk because COVID-19 can be easily transmitted through direct contact with an infected person or indirect exposure to contaminated items[8].Health officials have approved some evidences of a transmission chain with four “generations”,and proposed a sustained human-to-human transmission[9].It has been speculated that a person infected with SARS-CoV-2 may infect more than three healthy individuals(the average reproductive number is approximately 3.28)[10].COVID-19 also has a major impact on the global economy and affects the day-to-day life of individuals.Businesses have shut down,schools have been suspended and stores have closed as a result of this disease.Therefore,COVID-19 will likely continue to become a global health challenge and have far-reaching economic impacts if no control measures are in place[11].
The development of diagnostics and therapeutics is in progress,but there is currently no vaccine or antiviral drug for the treatment of COVID-19.Diagnostic tools have a crucial role in the battle against COVID-19,which allows for the rapid implementation of control measures to suppress SARS-CoV-2 transmission via case identification,contact tracing,and isolation of positive cases[12].The mainstay for the clinical management of COVID-19 patients is symptomatic treatment instead of curative treatment,where critically ill patients are treated with ventilatory support in the intensive care unit[13].Other common treatment strategies include antiviral therapy,antibiotic therapy,and immunomodulating therapy[14].In this review,we first introduce the systematic evolution of coronaviruses(CoVs)and the structural characteristics of SARS-CoV-2.We then focus on known diagnostic tools and strategies for detecting SARS-CoV-2 in order to suppress its spread.Finally,we discuss current therapeutic methods,potential drugs,and vaccines.The overall intent of this review is to provide diagnostic and treatment strategies for dealing with COVID-19 pandemic through a summary of current findings.
2.Coronavirus family
CoVs are types of RNA viruses with an enveloped,positivestrand,and single-stranded genome.They belong to the family Coronaviridae,the subfamily Orthocoronavirinae,and the order Nidovirales[15,16].The 5′-terminal and 3′-terminal of CoV genome have a methylated cap structure and a poly(A)tail,respectively,and the sizes vary from 27 kb to 32 kb(the largest known RNA virus genome)[17].CoVs only infect vertebrates and the symptoms are similar to those of many human and animal diseases,such as the diseases of the respiratory tract,digestive tract,and nervous system[18,19].
Based on the phylogenetic tree,CoVs are classified into four genera:alpha-,beta-,delta-and gamma-coronaviruses(α-,β-,δand γ-CoVs,respectively),of which β-CoV is further subclassified into four independent subgroups:β-CoV A,B,C and D(Fig.1)[20].Among them,both α-CoV and β-CoV usually infect humans and mammals,whileδ-CoV andγ-CoV mainly infect birds[21].To date,seven kinds of CoVs have been identified to infect humans,including HCoV-229E,-HKU1,-NL63,-OC43,SARS-CoV,-CoV-2 and MERS-CoV.The HCoV-229E and-NL63 are known asα-CoV,while HCoV-HKU1,-OC43,SARS-CoV,and MERS-CoV are classified asβ-CoV.HCoV-HKU1 and-OC43 belong toβ-CoV A,SARS-CoV belongs to β-CoV B,and MERS-CoV belongs toβ-CoV C.

Fig.1.Phylogenetic tree of the CoVs[20].Used with permission from Ref.[20].Copyright 2020 Chinese Center for Disease Control and Prevention.
Human CoVs,including HCoV-229E,-HKU1,-NL63 and-OC43,often cause mild-to-moderate upper respiratory tract infections.With an apparent seasonality,infection from common human CoVs generally occurs in winter and spring.The incubation period is about two to five days,during which time others are more prone to infection or being “vulnerable”if they are exposed to an infected individual[22].However,Middle East respiratory syndrome(MERS)and severe acute respiratory syndrome(SARS)are more severe viral respiratory diseases caused by MERS-CoV and SARSCoV,respectively.SARS was first identified in Guangdong(China)in 2002 and rapidly swept across the globe,resulting in the deaths of 774 people(the fatality rate reached 9.6%)[23,24].Patients with SARS were the most important source of infection,while those in the incubation and recovery periods were not infectious.The emergence of MERS was first reported in Saudi Arabia and then spread to other countries with a high fatality rate of 37% globally[25,26].Both MERS-CoV and SARS-CoV originated in bats before spreading to an intermediate animal host and laterhuman[27-29].MERS-CoV has limited capacity for person-to-person transmission.The single-humped camel is a major intermediate host of MERSCoV and is an important source of infection for humans.Similar to these two CoVs,SARS-CoV-2 predominantly attacks the lower respiratory tract,thus leading to viral pneumonia.Besides,it may also cause damage to the gastrointestinal tract,kidney,heart,liver,and/or central nervous system,thereby resulting in multiple organ dysfunction.The fatality rate of COVID-19 is currently estimated at<4%.This indicates that the new CoV has a lower fatality rate than MERS-CoV and SARS-CoV[30],but is more contagious or transmissible than SARS-CoV[31].
Currently,the zoonotic sources of SARS-CoV-2 have not been verified;however,bats have been identified as the key reservoir via sequence-based analysis[32].Further homologous recombination analysissuggests that the receptor-binding S protein of SARS-CoV-2 is originated from a SARS-CoV(CoVZC45 or CoVZXC21)and a previously unidenti fiedβ-CoV[33].Numerous observations have revealed that SARS-CoV-2 has person-to-person transmission capacity,through direct contact with an infected individual and Specifically exposure to airborne SARS-CoV-2 particles and/or aerosols droplets expelled during breathing,coughing,sneezing and speaking[34].These aerosols can enter the lungs by means of nasal or oral inhalation[34].
3.Structural properties of SARS-CoV-2
Similar to that of other CoVs,the genome(about 29.8 kb in size)of SARS-CoV-2 comprises 14 open reading frames(ORFs)that encode for 27 proteins(Fig.2).Fifteen non-structural proteins are responsible for viral DNA replication,which are mainly encoded by ORF1 and ORF2 at 5′-terminal genome regions.Four structural proteins,namely,envelope(E),membrane(M),nucleocapsid(N)and spike(S)proteins,which responsible for virus assembly and infection,are located at 3′-terminal genome regions[33,35].
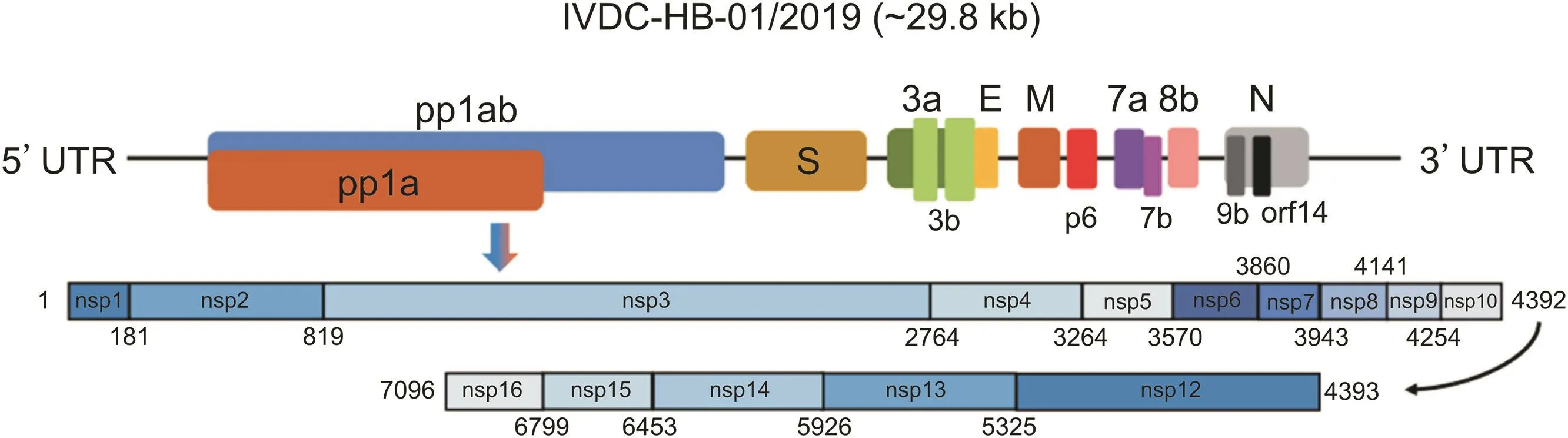
Fig.2.Characterization of the genome composition of SARS-CoV-2.Pp1ab protein is encoded by the longest open reading frame(orf1ab),which consists of fifteen nsps(nsp1 to nsp16,except for nsp11).Meanwhile,pp1a protein is encoded by orf1a gene,which comprises ten nsps(nsp1 to nsp10).The remaining structural proteins are encoded by different structural genes,namely,E(envelope),M(membrane),N(nucleocapsid)and S(spike).The accessory genes are disseminated throughout the entire genome.Used with permission from Ref.[35].Copyright 2020 Cell Press.
Fig.3A delineates the structural domains of the construct used for protein expression.Fig.3B illustrates the 3D structures of prefusion SARS-CoV-2 spike ectodomain at 3.5 Å resolution,with one receptor-binding domain(RBD)in the up conformation and two RBDs in the down conformations[36].The spike protein is composed of two functional subunits,namely,S1 and S2,which play an essential role in mediating the attachment of viral proteins to the host cell surface.S1 subunit is involved in the binding to a host-cell receptor,while S2 subunit is associated with the fusion between the viral membrane and the host-cell membrane[37-39].The binding of S1 subunit to a host-cell receptor can destabilize the prefusion trimer,which in turn sheds the subunit and converts S2 subunit into a rigid post-fusion structure[17,40].The RBD of S1 subunit displays an intrinsic hinge-like conformational movement as a means to bind to the host-cell receptor[36].The two states of S1 subunit are characterized by the “up”and “down”conformations,which correspond to the receptor-accessible(less stable)and receptor-inaccessible states,respectively[36].The overall structure of SARS-CoV-2 S protein is comparable to that of SARS-CoV S protein,but a key difference between these two structures is the RBD location at the “down”conformation[36].Thus,understanding the structural and functional properties of S protein may contribute to the development of new vaccine,therapeutic antibodies,and/or diagnostic tools.

Fig.3.Cryo-electron structures of SARS-CoV-2 spike protein in the prefusion conformation.(A)Structural domains of the expression construct.CH:central helix;CD:connector domain;CT:cytoplasmic tail;FP:fusion peptide;HR1:heptad repeat 1;HR2:heptad repeat 2;S2′:S2′protease cleavage site;SS:signal sequence;TM:transmembrane domain.Arrow denotes protease cleavage site.(B)Side and top views of prefusion SARS-CoV-2 spike ectodomain.The protomer with receptor-binding domain ‘up’state is corresponded to the structural representation in(A),while the other two protomers with receptor-binding domain ‘down’state are colored gray or white[36].Used with permission from Ref.[36].Copyright 2020 American Association for the Advancement of Science.
The entry mechanism of CoVs is closely related to cellular proteases that cleave the S protein and allows it to undergo conformational changes[41].The residue 394(glutamine)in SARS-CoV-2 RBD can bind to the residue 31(lysine)in human angiotensinconverting enzyme 2(hACE2)receptor[42].The entry processes ofSARS-CoV-2pathogen,includingattachment,penetration,uncoating,replication,assemblyand virion release,are described in Fig.4[34].

Fig.4.Entry processes of SARS-CoV-2 to human host cell.Upon binding to ACE2,the S protein facilitates viral entry into target cells via the endocytic pathway.A virion is subsequently released into the host cell.Pp1ab and pp1a are formed by genome translation and then cleaved into smaller,functional products.Subgenomic mRNAs are synthesized by a discontinuous transcription mechanism,and eventually translated into viral polyproteins.The proteins are assembled into a virion in the host ER and Golgi,transported by a vesicle and ultimately released through endocytosis.ACE2:angiotensin-converting enzyme 2;ER:endoplasmic reticulum;ERGIC:ER-Golgi intermediate compartment[34].Used with permission from Ref.[34].Copyright 2020 Elsevier.
4.Diagnosis of COVID-19
Fast and accurate diagnostic tests are needed to confirm COVID-19 infection,trace its origin,and suppress its transmission[43,44].The current diagnostic strategies for COVID-19 include both clinical and laboratory diagnosis.
4.1.Clinical diagnosis
The symptoms of COVID-19 patients may develop graduallyover one week or longer, first with mild symptoms,and then(in some cases)progress to dyspnea and even shock[30].Common symptoms at disease onset are fever(98%),cough(76%),and fatigue or myalgia(44%);while less-common symptoms are sputum production(28%),headache(8%),hemoptysis(5%),and diarrhea(3%)[30].Guan et al.[45]investigated the clinical features of COVID-19 patients by extracting the data of 1099 cases with laboratory confirmed COVID-19 from 552 hospitals.Patients’median age was 47 years and the median incubation period was estimated at four days.Their most common symptom was fever,followed by cough(67.8%),whereas their least common symptoms were vomiting or nausea(5.0%)and diarrhea(3.8%).Moreover,Zhou and coworkers[46]assessed the clinical features of 191 patients with COVID-19.Approximately half of these patients had comorbidities,of which hypertension,diabetes and coronary heart disease were the most frequent.Multivariable regression indicated the increased risk of in-hospital mortality was probably attributed to the older age.
The main method currently used to clinically diagnose COVID-19 is chest imaging,including chest X-ray and computed tomography(CT)scans.This method is also essential for diagnosing pneumonia and assessing its severity.Most patients with COVID-19 have abnormalities in chest imaging,usually with bilateral lung involvement,ranging from a ground-glass shadow at the recovery stage in patients with mild symptoms to a consolidation of lung lobes in patients with severe symptoms[47-50].
Chest X-ray examination typically shows interstitial alterations and multiple patchy shadows[30],which are remarkable in the lung periphery[50]of pneumonia cases at the early stage.Patients with severe symptoms may present with multiple ground-glass opacities(GGOs),infiltrative shadows and pulmonary consolidation,together with pleural effusions[9].Chest CT images(Fig.5)of patients with COVID-19 show multilobar ground glass opacities with a peripheral distribution[51].
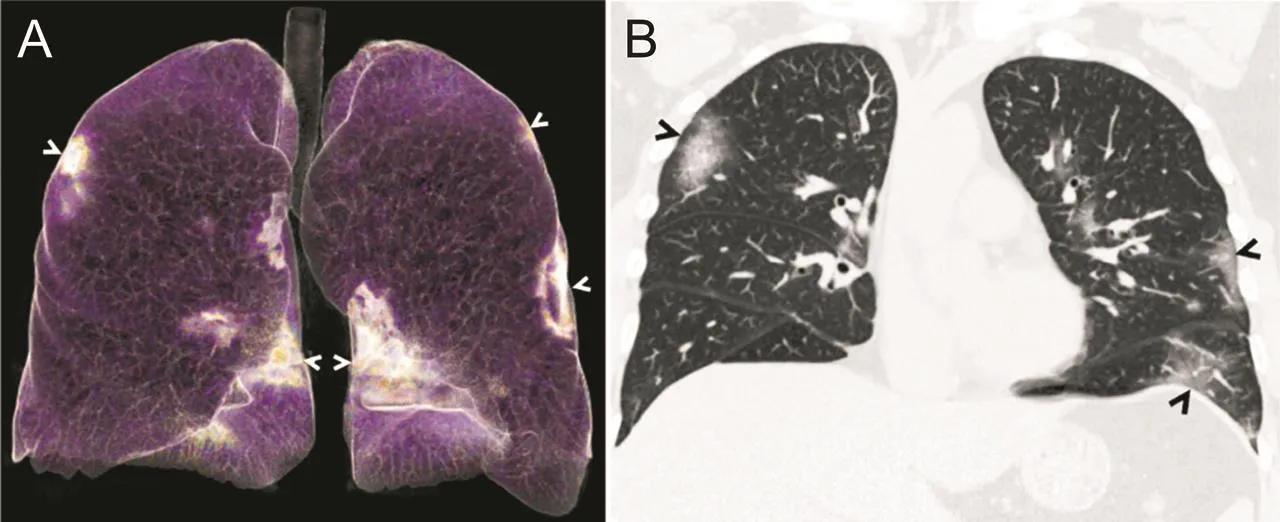
Fig.5.(A)Three-dimensional volume rendering of chest CT image reveals groundglass opacities in both lungs,with a predominantly peripheral distribution(arrows).(B)Coronal reconstructed image from a CT scan demonstrates ill-defined ground-glass opacities in both lungs,with a predominantly peripheral distribution(arrows)[51].Used with permission from Ref.[51].Copyright 2020 the Radiological Society of North America.
Thus,the combined application of chest imaging with laboratoryand clinical findings can facilitate the early diagnosis of COVID-19 pneumonia.
Overall,CTscans seem to be moresensitive[52]and clearer than X-ray examinations for the diagnosis of COVID-19,but normal CT manifestations may not be able to distinguish COVID-19[53].An investigation of the CTscans of COVID-19 patients(n=21)revealed that three(14%)patients presented with normal CT scans,twelve(57%)presented with GGOs only,and six(29%)presented with GGOs and consolidation[54].Another study of CTscans found pure GGOs in 77% of cases,GGOs with interlobular and/or interstitial septal thickening in 75% of cases,and GGOs with consolidation in 59% of cases[55].Furthermore,the GGOs were bilateral in 88% of cases,involved the posterior lungs in 82% of cases,and the peripheral lungs in 85% of cases[55].The imaging characteristics of COVID-19 patients were comparable to those of patients infected with MERS or SARS,which is not surprising since they all belong to CoVs.
4.2.Laboratory diagnosis
4.2.1.Nucleic acid testing
Nucleic acid testing,including reverse-transcription polymerase chain reaction(RT-PCR),RT loop-mediated isothermal amplification(RT-LAMP)and real-time RT-PCR(rRT-PCR),is currently the most effective and accurate method for diagnosing COVID-19[56-58].Many RT-PCR assays have been designed and prepared for detecting SARS-CoV-2 genetically.The first step in RT-PCR is the conversion of SARS-CoV-2 RNA into single-strand complementary DNA(cDNA)through retroviral DNA polymerases,followed by PCR amplification of the target cDNA regions[59,60].The protocol generally involves two major steps:(i)multiple sequence alignment and primer/probe design,and(ii)optimization and validation of RT-PCR assays[12].
Through analysis of the related SARS genome sequences[61-63],Corman et al.[64]designed various primer/probe sets for detecting SARS-CoV(Table 1).Three regions have conserved sequences in SARS-related viral genomes:(i)E gene,(ii)N gene,and(iii)RdRp gene(RNA-dependent RNA polymerase gene)(Fig.6).Although both RdRp and E gene assays can detect SARS-CoV-2 without binding site polymorphisms,Corman et al.[64]also designed in vitro-transcribed RNA standards to match SARS-CoV-2 sequences for absolute viral load quantification and limit of detection(LOD)evaluation.Both E and RdRp gene assays were highly sensitive in detecting SARS-CoV-2(LOD=3.9 and 3.6 copies per reaction,respectively),whereas N gene assay was less sensitive(8.3 copies per reaction).RdRp gene assay contained two probes:a wide-range probe that reacts with SARS-CoV-2 and SARS-CoV and an extra probe that only interacts with SARS-CoV-2.After designing primer/probe sets,the next step is to optimize assay conditions,such as reaction time,temperature and reagent conditions,followed by PCR amplification[65].Laboratory testing that involves oropharyngeal and nasopharyngeal swab tests has become a standard procedure for the diagnosis of patients infected with COVID-19 based on the latest diagnostic criteria reported by the China National Health Commission.

Fig.6.Genome positions of the amplification regions of SARS-CoV-2 and SARS-CoV(GenBank NC_004718)[64].E:envelope;M:membrane;N:nucleocapsid;RdRp:RNA-dependent RNA polymerase;S:spike.Used with permission from Ref.[64].Copyright 2020 European Centre for Disease Prevention and Control.

Table 1 List of primer/probe sets used for real-time RT-PCR assay.
4.2.2.Protein testing
The detection of viral protein antigens and antibodies produced during SARS-CoV-2 infection plays a major role in the diagnosis of COVID-19.The variation of viral load in the infected patients may make it difficult to detect viral proteins.However,antibody tests can be used for the effective surveillance of COVID-19 patients because the antibodies formed in response to viral proteins can provide a large window of opportunity[12].Research on serology testing for COVID-19 has been recently undertaken by Zhang et al.[66].The authors analyzed immunoglobulin G(IgG)and immunoglobulin M(IgM)in the serum samples of COVID-19 patients using an ELISA kit.Rp3 nucleocapsid protein is employed as an antigen,which has more than 90% amino acid identical to that of other related coronaviruses.They found that the positive rate of IgM was increased from 50% to 81%,while that of IgG was increased from 81% to 100%.Protein testing is beneficial to the diagnosis of COVID-19 after recovery,as opposed to nucleic acid testing.This allows clinicians to trace both infected and recovered cases in order to estimate the accumulated number of SARS-CoV-2 infections.
4.2.3.Point-of-care testing
In the absence of laboratory infrastructure,point-of-care testing can be applied to diagnose patients more simply and quickly.A typical detection[12]uses a paper-like membrane strip coated with 2 lines.One line is conjugated with antibody-gold nanoparticles and the other line captures antibodies.The patient’s sample is dropped onto the membrane,and the targets are drawn up by capillary action into all the strips.When the sample moves along the first line,the proteins(antigens)may bind to antibody-gold nanoparticle conjugates,and the resulting complex will flow to and fuse with the membrane.When the complex reaches the second line,the complex is captured by the immobilized antibodies,and a blue or red line is visualized.The dispersed gold nanoparticle is red in color,while the aggregated gold nanoparticle is blue because of the coupling of the plasmon band.Unfortunately,a major obstacle for this test is the need to improve its sensitivity.
Another strategy to be used at the point-of-care is fluorescencebased biosensors.Elledge et al.[67]developed the split luciferase antibody biosensors for a fast and low-cost detection of antibodies in whole blood,plasma,serum,and saliva.This point-of-care strategy,which obtained quantitative results within 5 min,greatly reduced the complexity and improved the scalability of COVID-19 diagnostic testing.Point-of-care testing can serve as an inexpensive,easy-to-use,and handheld technique for diagnosing COVID-19 patients at areas outside a centralized facility,such as community centers,in order to reduce the burden of COVID-19 on clinical laboratories.
5.Treatments for COVID-19
5.1.Antiviral agents
Currently,no Specific anti-SARS-CoV-2 agent has been approved for treating COVID-19 patients.With only in vitro findings(for the SARS-CoV-2 and/or related viruses)and a lack of clinical experiences,severalantiviralagentshavebeen used underthe“compassionate use”principle in the United States as they undergo testing in clinical trials.
5.1.1.Lopinavir and ritonavir
Lopinavir and ritonavir have been approved by the U.S.Food and Drug Administration(FDA)for treating human immunodeficiency virus(HIV)infection[68,69].The agents have also been used in the empirical treatment of SARS[70],and they are currently under study for the treatment of MERS[71]because lopinavir is a promising agent that suppresses the protease activities of CoVs in vitro.In China,these two antiviral agents have been used to treat some COVID-19 patients in concomitant with alpha interferon[72].Han and co-workers[73]reported a case study of a 47-year-old man with a 1-week history of cough,fever,and bosom frowsty.The patient had a historyof stage two hypertension and type II diabetes.To resolve phlegm,relieve asthma and suppress virus replication,this patient was treated with 800 mg of lopinavir,200 mg of ritonavir,10 million IU of interferon alfa-2b recombinant,400 mg of moxifloxacin hydrochloride,60 mg of ambroxol hydrochloride,and 40 mg of methylprednisolone each day.The next day,the patient had low-grade intermittent fevers(36.0°C-37.2°C).Moreover,many symptoms such as cough with production of phlegm/mucus,runny or stuffy nose,fatigue and vertigo were considerably alleviated.Due to the repeated negative testing results on days six and seven and in addition to the partial absorption of lung lesions,the patient was no longer infectious and discharged from hospital on day 10,suggesting that the treatment strategy could be effective.A recent review[74]found that the anti-CoV effect of lopinavir/ritonavir combination therapy mainly occurred with early treatment window and included the reduction of patient fatality and glucocorticoid administration.However,if the ideal window was missed,the treatment might have no significant effects on patient outcomes.Therefore,further studies that explore the clinical effects of the early use of lopinavir/ritonavir combination therapy for treating SARS-CoV-2 pneumonia are needed.
5.1.2.Remdesivir
Remdesivir,a monophosphoramidate prodrug of C-adenosine nucleoside analog,can be incorporated into viral RNA chains and thereby initiate the premature termination of RNA replication[75].It displays a significant anti-CoV activity in vitro[75].Previous studies[76-78]showed that remdesivir could inhibit the replication of SARS-CoV,MERS-CoV,and bat CoV strains inprimaryhuman airway epithelial cells and regulate cell entry through hACE2 receptor.Remdesivir acts during early-stage infection and dose dependently reduces RNA levels,which parallels a decrease in virus titers[79].Remdesivir displays an EC90(90% effective concentration)value of 1.76μM towards SARS-CoV-2 in Vero E6 cells,indicating that it is effective in non-human primates[80].Besides,it is noted that SARS-CoV-2 requires RdRp gene to replicate,which can be covalently bound to remdesivir,hence terminating chain elongation[81].Moreover,remdesivir also suppresses virus infection in human liver cancer Huh-7 cells,a cell line that is susceptible to SARS-CoV-2[80].Furthermore,remdesivir has been given on a compassionate use basis to COVID-19 patients[82],and the results indicated that a 10-day course of the antiviral drug(200 mg on day 1,followed by 100 mg daily for 9 days),might exhibit potential clinical benefits for these patients.
5.1.3.Chloroquine
Chloroquine,apotentbroad-spectrum antiviralagent,is commonly utilized as an autoimmune disease or anti-malarial agent[83-85].Chloroquinemay stopa virusinfection by elevating the endosomal pH necessary for virus-cell fusion and disrupting the terminal glycosylation of hACE2 receptor[86].Recent work has shown that chloroquine inhibits SARS-CoV-2 infection at entry and post-entry stages in a Vero E6 cell line[80].Afteroral administration,chloroquine is distributed throughout the body,especially in the lungs.Apart from its antiviral activity,chloroquine also exhibits immunomodulatory activities,which in turn leads to a synergistic enhancement of its antiviral effect in vivo[80].Chloroquine has been considered as a potential drug for the treatment and prevention of COVID-19 pneumonia[87,88].However,a recent study has found that chloroquine is ineffective for the treatment of COVID-19[89].Given the controversy about the effectiveness of this antiviral agent,it is important to determine whether chloroquine has potential applicability for SARS-CoV-2 treatment and prevention.
5.1.4.Arbidol
Arbidol is a broad-spectrum antiviral agent that blocks influenza A and B viruses by inhibiting virus-cell membrane fusion[90].Lowlevel evidence shows that arbidol taken alone or concomitantly with other antiviral drugs produces therapeutic benefits for COVID-19 pneumonia[91-93].In China,many randomized control trials are currently under way to assess the efficacy of arbidol on COVID-19 pneumonia.Zhang et al.[94]performed a retrospective cohort trial on healthcare workers(n=124)and family members(n=66 members in 27 families)who have been exposed to a confirmed case of COVID-19.The authors found that arbidol could decrease the risk of SARS-CoV-2 infection in both hospital-care and familycare settings.Zhu et al.[95]compared the efficacy of arbidol and lopinavir/ritonavirin 50 patientswith laboratory-confirmed COVID-19,and the results demonstrated that COVID-19 patients treated with arbidol recovered more rapidly than those treated with lopinavir/ritonavir(P<0.01).
5.2.Immunity-based therapy
Immunity-based therapy,including convalescent plasma therapy and immunoglobulin therapy,is also an important strategy in the treatment of COVID-19.Convalescent plasma therapy is one of the classic adaptive immunotherapies used for the treatment and prevention of numerous infectious diseases such as SARS and MERS.Because of the similarity of clinical and virological characteristics among COVID-19,MERS and SARS,convalescent plasma therapy has emerged as a choice of rescue for COVID-19 patients[96].Duan and co-workers[97]investigated the effectiveness of convalescent plasma therapy against COVID-19.All symptoms(e.g.,cough,fever and difficulty breathing)of the 10 patients with COVID-19 disappeared or largely improved within one-to-three days of convalescent plasma therapy.Furthermore,all of the patients displayed varying degreesof absorption of pulmonarylesions after convalescent plasma transfusion.Among the convalescent plasma samples of 40 recovered COVID-19 cases,39 demonstrated high antibody titers of more than 1:160,and only one exhibited a lower antibody titer of 1:32.Thus,these results indicate that convalescent plasma therapy is well tolerated and can enhance the health outcomes of patients with severe COVID-19 by neutralizing viremia.
Immunoglobulin is also being investigated as a possible therapeutic option for COVID-19 patients.Cao and co-workers[98]published a case study of the treatment of severe COVID-19 using high-dose intravenous immunoglobulin(IVIg).A 34-year-old male patient from Jinyintan Hospital reported a feverof up to 38.5°C and a dry cough,and subsequently was diagnosed with severe COVID-19.The patient was treated with 25 g/day of IVIg for five days,and became afebrile on the second day of treatment,with a progressive alleviation of breathlessness.He was discharged from the hospital about a week later.
5.3.Vaccines
The S protein structures of SARS-CoV-2 have been identified previously.Since most of the existing CoV vaccines only target S protein,this discovery provides the basis for further research aimed at optimizing the design of COVID-19 vaccination strategies[99].Thus far,there are no available vaccines for COVID-19,as vaccine development is typically a long process.Gao et al.[100]developed an inactivated vaccine candidate(PiCoVacc)against SARS-CoV-2.To obtain a pilot-scale PiCoVacc for in vivo studies,different SARSCoV-2 strains were propagated in Vero cell line at a working volume of 50 L,purified,and then inactivated withβ-propiolactone.The antibodies neutralized ten representative SARS-CoV-2 strains,indicating its broader neutralizing activity towards SARS-CoV-2 strains.More recently,Moderna Therapeutics announced that they have produced an mRNA-1273 vaccine that may play a role in targeting the S protein of viruses.A clinical trial to evaluate the safety and immunogenicity of mRNA-1273 in COVID-19 patients is currently in progress(available at clinicaltrials.gov using the identifier NCT04283461)[58].In addition,there are about 18 private companies and universities in China that are developing COVID-19 vaccines.Among them,Tianjin University has successfully established an oral SARS-CoV-2 vaccine,which applies spike protein as the target and Saccharomyces cerevisiae as the carrier[58].
6.Conclusions and outlook
Currently,SARS-CoV-2 has spread to more than 240 countries worldwide,which causes illness ranging from the common cold to lung pneumonia,making it a very serious public-health threat.From the initial outbreak of COVID-19 to now(15 December 2020),the United States has greatly surpassed China as the country with the largest number of confirmed COVID-19 cases globally.Based on a WHO report[8],the number of confirmed cases per week in the United States has increased by 1.4 million,while that in China has dropped to about 790.This shows that China’s experience infighting the SARS-CoV-2 pandemic can play a huge role in reducing the prevalence of COVID-19.
In the fight against COVID-19,there are still many problems to be solved,such as the origin,transmission route and structures of SARS-CoV-2.Although bat has been identified as a natural reservoir and vector for the transmission of CoVs,the origin and transmission routes of SARS-CoV-2 are still under investigation.SARS-CoV-2 has typical features that are distinguishable from the CoV family and is classified under theβ-CoV 2b lineage.It may be transmitted from human to human via respiratory droplets or mucous membranes in the eye,nose and mouth.However,the spread mechanism of SARSCoV-2 is still uncertain and knowledge about human-to-human transmission is mainly derived from similar CoVs.Arguably,the greatest challenge for COVID-19 is the lackof availabilityof effective antiviral drugs.
To date,there is still no effective drug to treat SARS-CoV-2.Clinical treatment of COVID-19 is mainly symptomatic treatment.From the current clinical cases,the SARS-CoV-2 is more dangerous to the elderly and patients with other diseases.In COVID-19 prevention,wearing masks and reducing crowd aggregation is the most cost and effective approach.Fortunately,many scientists and enterprises are stepping upthe developmentof antiviral agents and vaccines,some of which have begun clinical trials.We believe that in a short period of time,effective Scientific measures will prevent the infectious spread of COVID-19.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
We are grateful for the financial support from the National Natural Science Foundation of China(Grant No.21976144).
 Journal of Pharmaceutical Analysis2021年2期
Journal of Pharmaceutical Analysis2021年2期
- Journal of Pharmaceutical Analysis的其它文章
- Chemically modified carbon-based electrodes for the determination of paracetamol in drugs and biological samples
- Development of chromatographic technologies for the quality control of Traditional Chinese Medicine in the Chinese Pharmacopoeia
- Design and preparation of a new multi-targeted drug delivery system using multifunctional nanoparticles for co-delivery of siRNA and paclitaxel
- The effective transfection of a low dose of negatively charged drugloaded DNA-nanocarriers into cancer cells via scavenger receptors
- A dual-signal sensor for the analysis of parathion-methyl using silver nanoparticles modified with graphitic carbon nitride
- Curcumin encapsulated dual cross linked sodium alginate/montmorillonite polymeric composite beads for controlled drug delivery
