Circulating MicroRNAs as Potential Biomarkers in the Diagnosis of Neurosyphilis: A Case Control Study
Kai-Qi Wu1,2,#, Su-Fang Zhang1,3,#, Chao-Hui Bao4, Xin Zou4, Xin Gu1, Cui-Ni Wang1,Wei-Ming Gong1, Mei Shi1, Yong-Liang Lou2, Jian Huang4,∗, Ping-Yu Zhou1,3,∗
1 Sexually Transmitted Disease Institute, Shanghai Skin Disease Hospital, Shanghai 200050, China; 2Key Laboratory of Laboratory Medicine (Ministry of Education), School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou,Zhejiang 325035, China; 3Shanghai Skin Disease Hospital, Clinical School of Anhui Medical University, Shanghai 200050, China;4Key Laboratory of Systems Biomedicine (Ministry of Education) and Collaborative Innovation Center of Systems Biomedicine,Shanghai Center for Systems Biomedicine, Shanghai Jiao Tong University, Shanghai 200240, China.
Abstract
Objective: Laboratory diagnosis of neurosyphilis (NS) remains a great challenge. This study was the aimed to identify miRNA candidates as biomarkers to distinguish between NS, non-neurosyphilis, and healthy controls (HCs).
Methods: We analyzed miRNA expression profiles in peripheral blood mononuclear cells (PBMCs) from six patients with NS, eight patients with secondary syphilis (SS), and five HCs using microarray technology. The differentially expressed miRNAs were validated in 33 NS samples, 31 SS samples, and 30 HC samples using TaqMan miRNA realtime qPCR (qRT-PCR).
Results: Thirty-nine miRNAs were differentially expressed in SS and NS patients compared with HCs. Thirteen miRNAs were randomly selected to validate their expression levels in the same samples used in microarray assay by qRTPCR. All miRNAs were upregulated in SS and NS samples compared with HC. qRT-PCR analysis of the expression of the 13 miRNAs in a second cohort (76 samples) showed that the average expression levels of nine miRNAs were higher in SS than inNS(SS: 0.185, NS: 0.136, P=3.8E-10), while the expressions of the other four miRNAs were lower in SS than inNS(SS: 0.000757, NS: 0.000873, P=0.022).ROCcurve analysis of the 13 miRNAs showed the area under the curve value to be 1.00 for distinguishing SS patients from HCs, 1.00 for distinguishing NS patients fromHCs, 1.00 for distinguishing SS and NS patients from HCs, and 0.968 for distinguishing NS from SS patients.
Conclusion: The present study is the first one that identified differentially expressed miRNAs in PBMCs from patients with NS. Our results suggest that the 13 candidate miRNAs in PBMCs may be novel noninvasive biomarkers for NS diagnosis.
Keywords: diagnosis, microRNAs, neurosyphilis, peripheral blood mononuclear cells, syphilis
Introduction
The incidence of syphilis has increased dramatically in recent years, not only in developing countries,1-2but also in developed countries.3This disease has become a public health problem in China in the last two decades.4–6The epidemiology of neurosyphilis (NS) has largely paralleled that of active syphilis.5As NS is caused by Treponema pallidum (T. pallidum) invasion of the Central Nervous System (CNS), which can occur at any stage of the infection. Early diagnosis is critical for prompt therapy of NS, which is necessary to prevent irreversible sequelae.6The diagnosis of symptomatic NS is usually based on clinical manifestations and cerebrospinal fluid (CSF)abnormal findings, such as elevated white blood cell count, elevated CSF protein, or reactive CSF Venereal Disease Research Laboratory (VDRL) test. The diagnosis of patients with asymptomatic NS, which is most common in early stages, is based on CSF findings. However,collection of CSF requires a lumbar puncture (LP), an invasive operation that is difficult for both doctors and patients to decide to pursue. If patients do not have symptoms, it is difficult to determine whether LP is necessary; however, if left untreated, a patient with NS may develop irreversible sequelae. Therefore, identifying novel, non-nvasive and sensitive biomarkers from peripheral blood for diagnosis of NS is critical.
MicroRNAs (miRNAs) are short (-22 nucleotide),endogenous RNAs that negatively regulate gene expression in animals and plants by translational repression or mRNA cleavage at the post-transcriptional level,7and can modulate the innate and adaptive immune responses to pathogens by affecting mammalian immune cell differentiation and the development of diseases of immunological origin.8-9Several studies have indicated that miRNAs can be used as diagnostic and prognostic markers in cancers.10-11Notably, some studies found that diseased tissues of heart or brain release miRNAs into the circulatory blood, and these miRNAsmay be new biological markers of cardiovascular and cerebral diseases.12-13However, the miRNA expression profiles in NS are unknown.
In the present study, we used miRNAmicroarray analysis to identify the miRNA expression profile of peripheral blood mononuclear cells (PBMCs) in patients with NS compared with patients with secondary syphilis (SS) and healthy controls (HCs). The differentially expressed miRNAs were further validated in a larger sample of cases and controls using quantitative real-time reverse transcription PCR(qRT-PCR). Finally, using receiver operating characteristic (ROC) curve analysis, we estimated the value of circulating miRNAs as biomarkers for diagnosing NS.
Materials and methods
Subjects
This case control study was performed at the Shanghai Skin Disease Hospital between May 2014 and October 2015.The syphilis patients were referred by dermatologists,neurologists, and psychiatrists after careful examination and evaluation. Patients were diagnosed by a combination of compatible history, clinical features, and the results of nontreponemal and treponemal tests of serum and CSF samples. The exclusion criteria included positive human immunodeficiency virus (HIV), hepatits B viral (HBV), and hepatitisCvirus(HCV) infection; prior history of syphilis or history of syphilis treatment; history of systemic inflammation, autoimmune disease, diabetes, or other underlying acute or chronic disease; receiving anti-inflammatory medications; immunocompromised conditions; or use of antibiotics or immunosuppressive medications in the four weeks before study inclusion.
Two groups of patients were included in this study: (1)the NS group (including subjects with asymptomatic NS and general paresis); and (2) the SS group, with normal CSF white blood cells count and protein concentration and negative CSF-VDRL. PBMCs from healthy donors, who visited Shanghai Skin Disease Hospital voluntarily for a medical check-up for Sexually Transmitted Diseases(STDs) prevention, were used as controls. All healthy subjects were negative for HIV, HBV, HCV, and serological tests for syphilis (ie, both serum rapid plasma reagin [RPR] and T. pallidum particle agglutination[TPPA] negative) and did not have any clinical symptoms consistent with T. pallidum infection.
All subjects provided written informed consent. This study was approved by the Ethics Committee of the Shanghai Skin Disease Hospital (No. 2013-05).
Serological tests
TPPA in CSF and serum was determined by a passive particle agglutination test kit (Fujirebio, Shinjuku-ku,Tokyo, Japan) according to the manufacturer’s instruction. TPPA was considered reactive if the titer was ≥1:80.CSF-VDRL test was carried out using BD VDRL Antigen kits (Becton Dickinson, Sparks, MD, USA). Serum RPR was measured using the RPR reagent from the Shanghai Rongsheng Biotech Co (Shanghai, China)
Diagnostic criteria for SS and NS
SS was defined by positive serum RPR test and TPPA test along with the clinical manifestations and medical history according to the national guidelines.14The diagnosis of NS included a reactive CSF-VDRL and a reactive CSF-TPPA in the absence of substantial contamination of CSF with blood.15In symptomatic NS patients, the clinical neurological or psychiatric manifestations were consistent with NS without other known causes for such abnormalities.
Blood collection and RNA extraction
Peripheral venous blood (10mL) from each subject was collected in an Ethylene Diamine Tetraacetic Acid anticoagulant tube and processed immediately. PBMCs were isolated on Ficoll gradients through density gradient centrifugation,immediately mixed with Trizol reagent (Invitrogen, Grand Island, NY, USA), and stored at –80 °C until use. RNA was extracted from cells using the miRNeasy Mini Kit (Qiagen,Valencia, CA, USA). The RNA quantity and quality were assessed using the NanoDrop spectrophotometer 2000(Thermo Fisher Scientific, MA, USA) and Agilent 2100 bioanalyzer systems (Agilent Technologies, Santa Clara, CA,USA) according to the manufacturers’ instructions. Samples with an RNA integrity above 7 were used in the study.
MiRNA microarray hybridization and data submission
MiRNA microarray assays were performed using the Agilent HumanmiRNA microarray platform (version V21.0, Agilent Technologies) at Shanghai Bohao biotechnology company(SBC, Shanghai, China). The microarray contains probes for 2549 miRNAs from the miRBase (release 21.0, Sanger Institute, Cambridge, UK).MiRNA was extracted from total RNA (100ng) using the mirVanaTM miRNA Isolation Kit without phenol (Cat #AM1561, Ambion, Austin, TX, USA),following the manufacturer’s instructions. RNA integration was determined by an Agilent Bioanalyzer 2100 (Agilent Technologies). The miRNA labeling, microarray hybridization, andwashing were performed based on the protocol in the Agilent miRNA microarray system. Briefly, total RNA was labeled with Cy3 by themiRNA Complete Labeling and Hyb Kit (Cat # 5190-0456, Agilent Technologies). The labeled RNAs were hybridized onto the microarray. After washing and staining, the arrays were scanned using the Agilent Microarray Scanner(Cat # G2565CA, Agilent Technologies).The scanned images were analyzed using Feature Extraction software 10.7.1.1 (Agilent Technologies). Microarray data submission for human arrays isMIAME-compliant.Rawdata werenormalized byQuantile algorithm,Gene Spring Software 12.6 (Agilent Technologies).
qRT-PCR
Blood samples from 33 patients with NS, 31 patients with SS,and 30 HCs were used to validate 15 miRNAs randomly selected fromthemicroarray results.TotalRNAswere isolated from the PBMCs using the miRNA Easy Mini Kit (Qiagen).Complementary DNA was synthesized from total RNA using the Reverse Transcription TaqMan microRNA Reverse Transcription Kit and miRNA specific stem-loop primers(Applied Biosystems Inc. Waltham, Massachusetts, USA)according to the manufacturer’s instructions. Real-time PCR was performed using the BioRad Real-Time PCR System. The 5× RT primers (miRNA specific stem-loop primers) and 20×miRNA-specific PCR primer/probe mix were supplied by the TaqMan microRNA Assays (Applied Biosystems Inc.). We calculated 2-ΔΔCtto determine relative gene expression. The reference gene was U6 small nuclear RNA (snRNA).
Statistical analysis
Statistical analyses were carried out using the Statistical Package of Social Sciences (SPSS Inc., Chicago, IL, USA)for Windows software program version 22.0. Expression levels of miRNAs were compared using the test. We calculated the miRNA relative expression levels by Kruskal–Wallis test to determine relative gene expression levels. The reference gene was U6 snRNA. ROC curves and the area under theROC curve(AUC)were analyzed to assess specificity and sensitivity of a single miRNA and combinations using binary logistic analysis. A statistical test was considered significant at a P value<0.05.
Results
Clinical characteristics of the patients
We recruited 94 individuals, including 33 patients with NS, 31 patients with SS, and 30 HCs. All subjects were of Han ethnicity in China. The demographic and clinical characteristics of all subjects are summarized in Table 1.The mean ages (mean±SD) of NS patients, SS patients and HCs were (51.1±12.6), (44.6±14.6), and (42.4±10.2)years, respectively. Patients were divided into two cohorts:cohort I comprised 19 biologically independent samples from patients (Table S1, http://links.lww.com/JD9/A3),including five HCs, eight SSs, and six NSs, while cohort II comprised 75 biologically independent samples, including 25 HCs, 23 SSs, and 27 NSs.
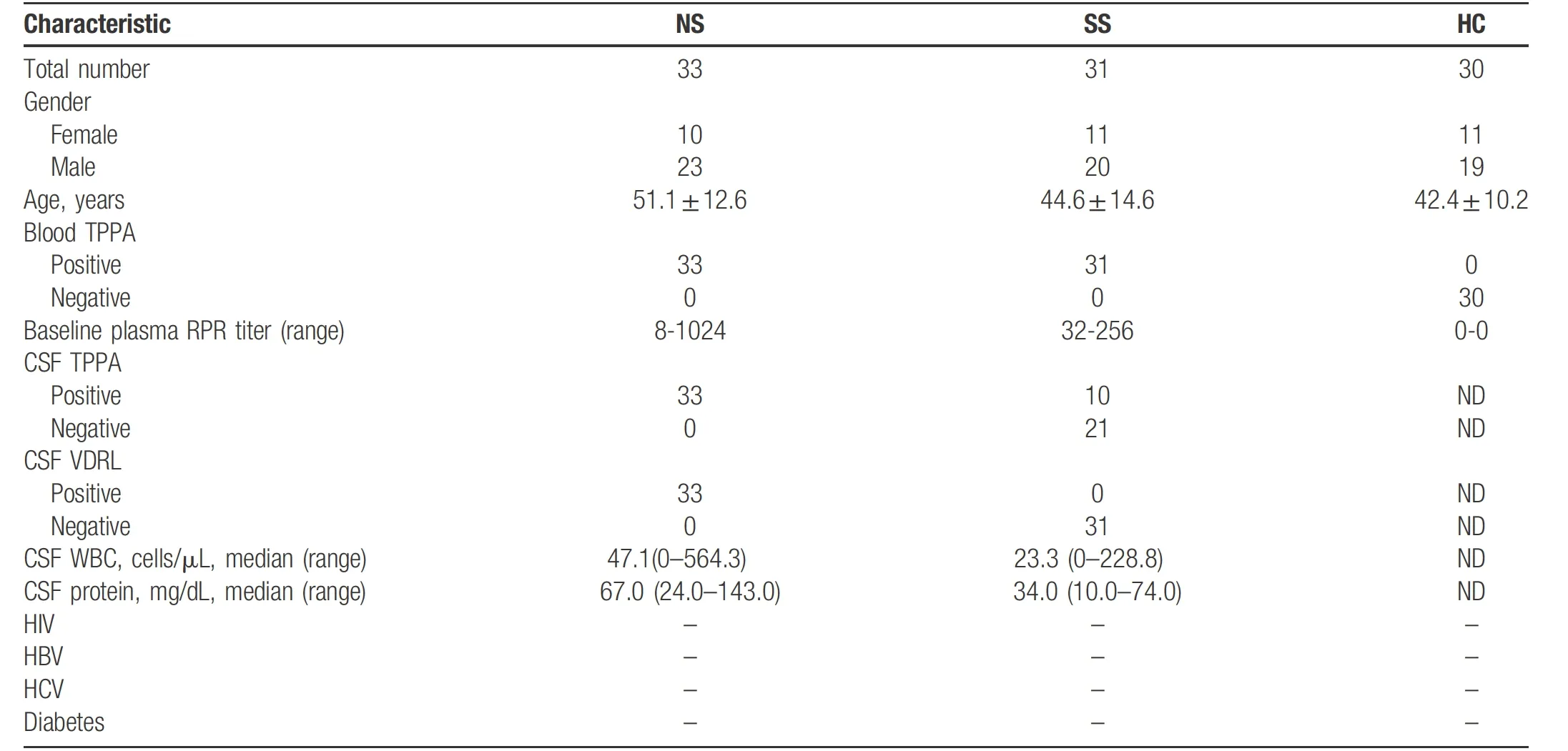
Table 1 Characteristics of study participants with neurosyphilis (NS), secondary syphilis (SS) and healthy controls (HC).
Expression profiles of miRNAs in PBMCs from different groups
RNA samples from cohort I, including six patients with NS, eight with SS, and five HCs, were used in the microarray assay (Table S1, http://links.lww.com/JD9/A3). Thirty-nine miRNAs were significantly differentially expressed in SS patients and NS patients compared with HCs (Fig. 1). The 39 differentially miRNAs were divided into three groups (G1, G2, and G3) according to their expression profiles (Fig. 1 and Table S2, http://links.lww.com/JD9/A8). The seven miRNAs in G3 were upregulated in both SS and NS patients, the nine miRNAs in G2 were only significantly upregulated in NS patients, and the 23 miRNAs in G1 were downregulated in NS patients(Table S2, http://links.lww.com/JD9/A8). Interestingly,the eight samples from SS patients were clustered into two groups: in four samples, the expression profile was similar to that of HCs, while the expression profile of the other four samples was similar to that of NSs (Fig. 1). This finding suggests that the SS patients with similar miRNA expression profiles to HCs might not have CNS involvement, while SS patients with similar miRNA expression profiles to NS may have CNS T. pallidum invasion.Moreover, the miRNA expression profile of one patient with SS was consistent with the miRNA expression profile of NS patients (Fig. 1).
Validation of miRNA expression using qRT-PCR
Tovalidate the results of the microarray assay,we randomly selected 13 miRNAs (Table 2) and examined expression levels in cohort I, the same samples used in microarray assay, using qPCR. The expression profiles of the 13 miRNAs showed the same trend as the microarray results(Fig. 2), which confirms the results of the microarray assay.Wethen detected the expression of the 13 miRNAs in cohort II using qPCR. The average expression levels of the 13 miRNAs in SS andNSsamples were higher than that in HCs(Fig. 3A and 3B). The average expression levels of nine miRNAs in SSs were higher than that in NSs (Fig. 3C and 3D), and the expressions of the remaining four miRNAs were lower in SSs than in NSs (Fig. 3E and 3F).
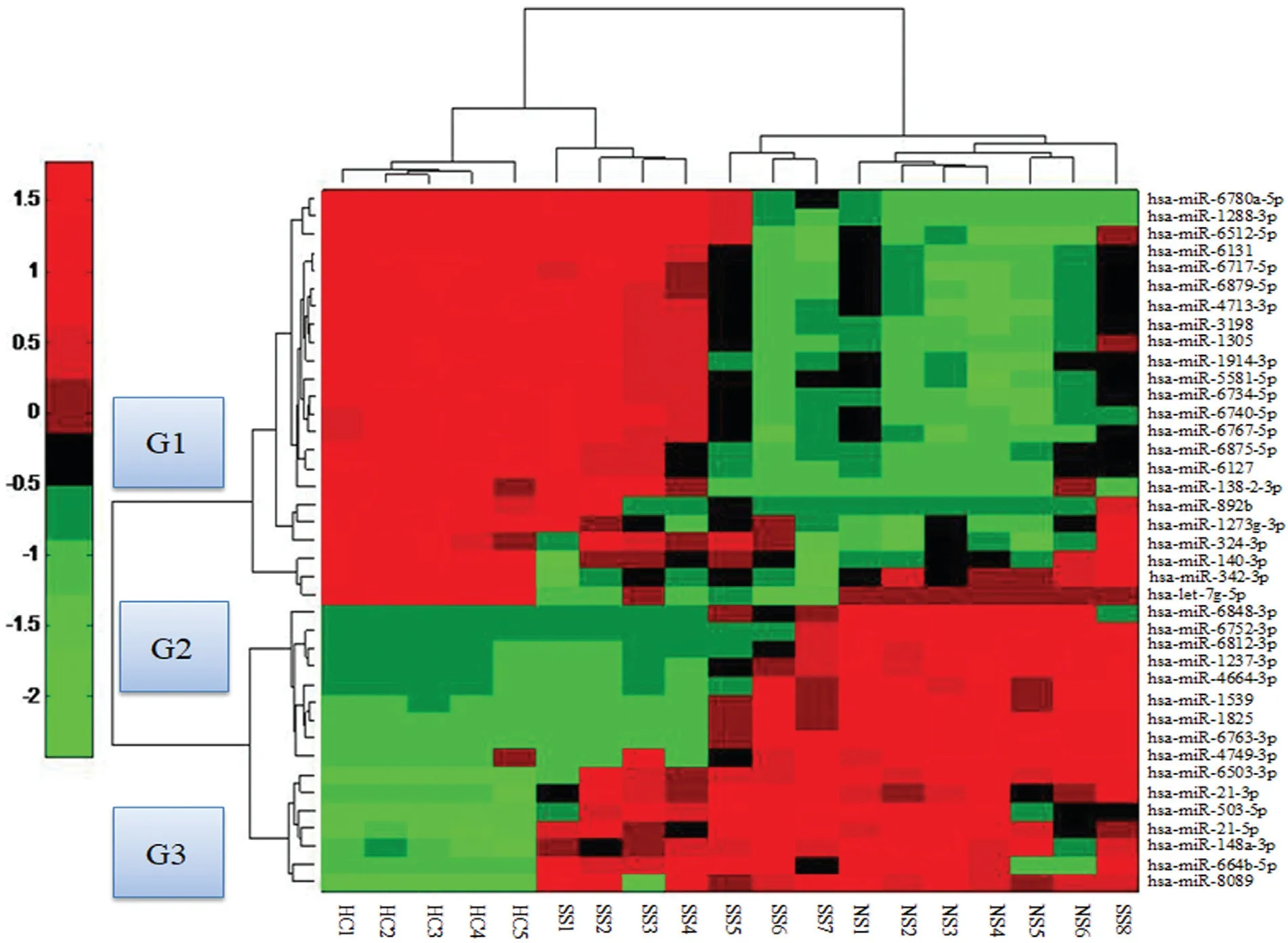
Figure 1. Heat map showing 39 differentially expressed miRNAs in peripheral blood mononuclear cells (PBMCs) from neurosyphilis (NS; n=6),secondary syphilis (SS) patients (n=8), and healthy controls (HCs; n=5). These miRNAs are clustered into three groups: G1, G2, and G3. Rows represent miRNA species, and columns represent individual blood samples. The relative miRNA expression is depicted according to the color scale. Red indicates upregulation; green indicates downregulation. The numbers with NS denote neurosyphilis; numbers with SS denote secondary syphilis; and numbers with HC denote healthy controls.
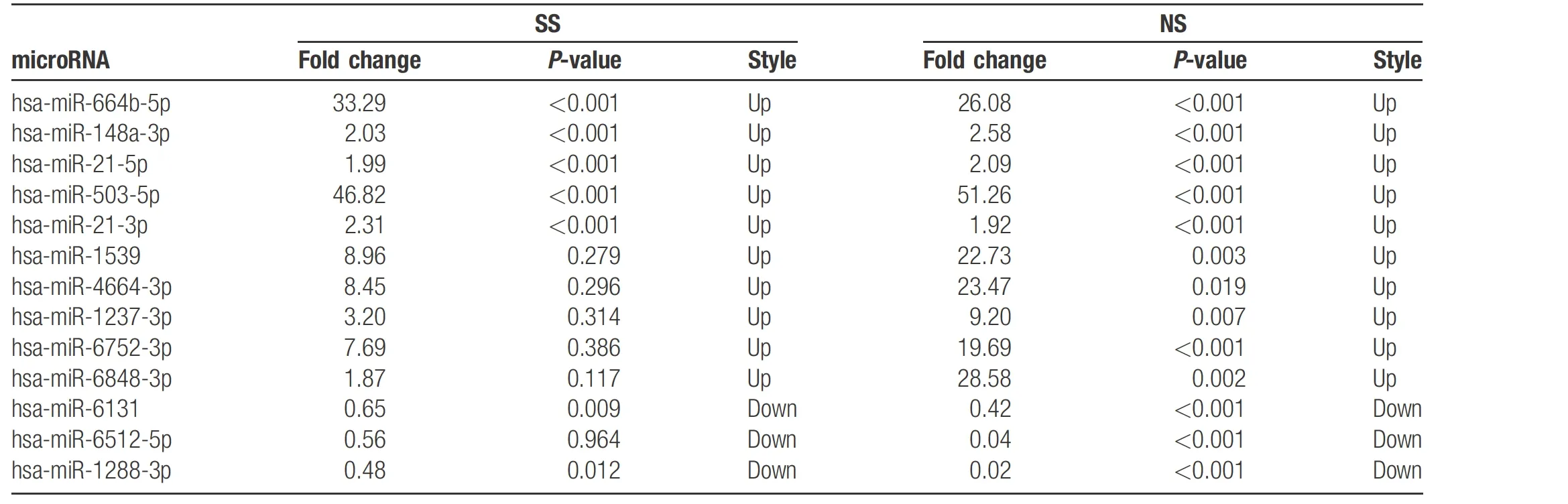
Table 2 Differentially expressed microRNAs identified by qPCR in PBMCs from SS and NS patients versus controls.
MiRNAs as potential biomarkers
To evaluate whether the 13 miRNAs may be used for diagnostic biomarkers, ROC curve analysis was carried out to determine the potential diagnostic value of the 13 miRNAs for syphilis and/or NS, which included the nine upregulated miRNAs and four downregulated miRNAs in SS samples compared with NS samples. The AUC value was 1.00 (95% confidence interval [CI], 1.000–1.000] for HCs and SS; 1.000 (95% CI, 1.000–1.000) for HCs and NS; 1.000 (95% CI, 1.000–1.000) for HCs and SS plus NS; and 0.968 (95% CI, 0.927–1.000) for SS and NS(Fig. 3G–J and Table 3). These results indicated that the 13 miRNAs are suitable biomarkers for diagnosing NS or SS patients from HCs.
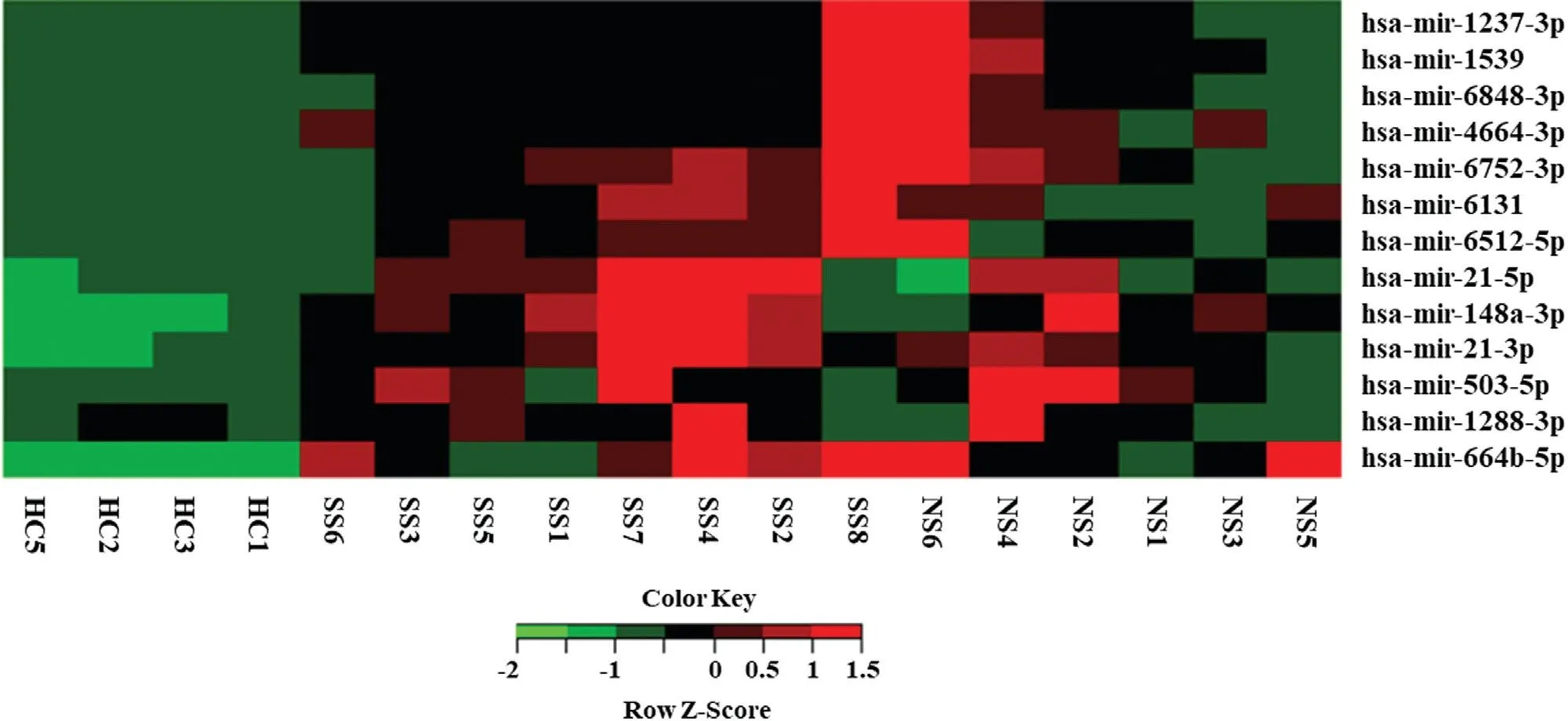
Figure 2. Quantitative real-time reverse transcription PCR (qRT-PCR) validation of miRNA expression levels in PBMCs from NS (n=6), SS(n=8), and HCs (n=4). The 2-ΔΔCt method was used to normalize the relative gene expression data in the qPCR assay. U6 snRNA was set as the reference gene. The miRNA expression value was used for the cluster analysis. HC: healthy controls; NS: neurosyphilis; PBMCs, peripheral blood mononuclear cells; SS: secondary syphilis.
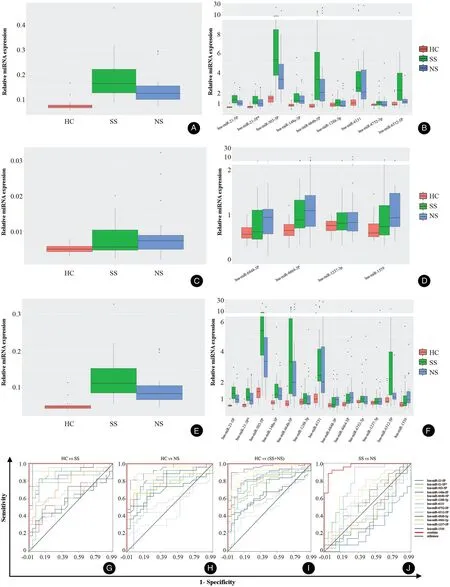
Figure 3. Expression levels and receiver operating characteristic (ROC) curve of the 13 miRNAs. A: The expression of nine miRNAs in NS, SS and HC groups. B: The expression of nine miRNAs in each of the NS, SS, and HC groups. C: The expression of four microRNAs in NS, SS, and HC groups. D: The expression of four microRNAs in each of the NS, SS, and HC groups. E: The expression of 13 microRNAs in NS, SS, and HC groups. F: The expression of 13 microRNAs in each of the NS, SS, and HC groups. G: ROC curve of the 13 differentially expressed miRNAs for HCs versus SS. H: ROC curve of the 13 differentially expressed miRNAs for HCs versus NS. I: ROC curve of the 13 differentially expressed miRNAs for HCs versus SS + NS. J: ROC curve of the 13 differentially expressed miRNAs for NS versus SS. HC: healthy controls; NS:neurosyphilis; SS: secondary syphilis.
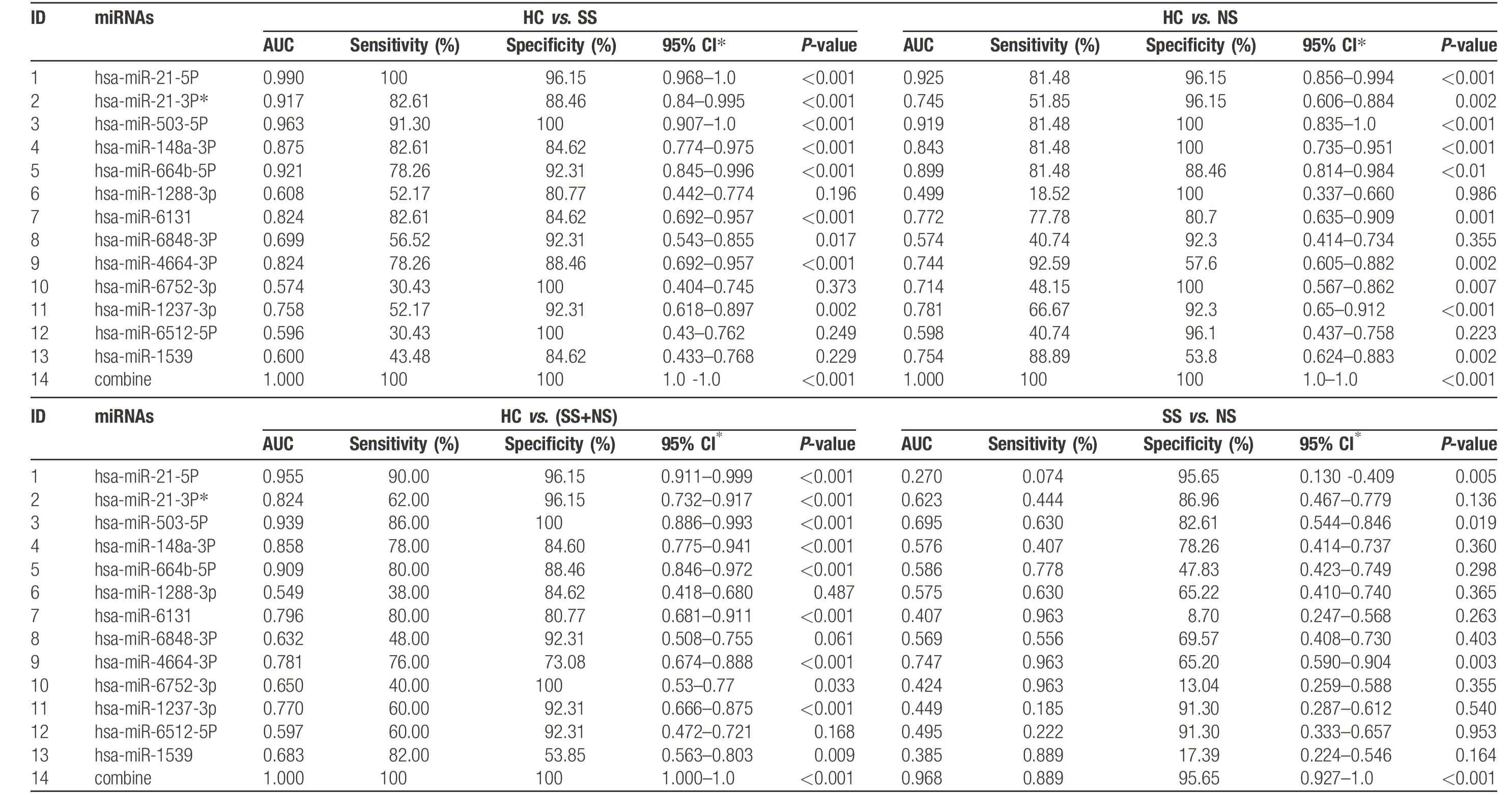
Table 3 Summ ary of re sult s analy z ed by ROC cu rv e fo r HC vs .SS,NS,SS+NS and SS vs.NS usin g 13miR NA s.
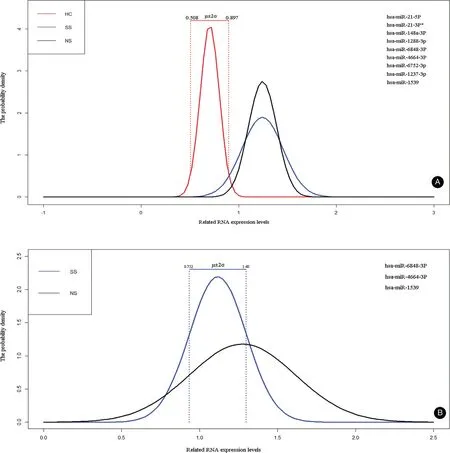
Figure 4. Analysis of miRNAs as potential biomarkers. A: Nine of thirteen miRNAs with lower dispersion (standard deviation <2) were selected to calculate their normal range (μ±2σ) in 25 HC samples using dnorm function based on R language. B: Three of four miRNAs with lower dispersion (standard deviation <2) were selected to calculate their normal range (μ±2σ) in 25 HC samples using dnorm function based on R language. HC: healthy controls.
Furthermore, to clearly distinguishNS from SS,we selected nine miRNAs (Table S3, http://links.lww.com/JD9/A9) with lower dispersion(standard deviation<2) and calculated their normal value range (μ±2σ) in 25HC samples using dnorm function based on R tool. The results showed that μ±2σ ranged from 0.508 to 0.897 (Table S3, http://links.lww.com/JD9/A9 and Fig. 4A), which suggests that samples in which the relative expression level was higher than 0.897 would be defined as abnormal samples, including SS and/or NS.Using the samemethod,we selected threemiRNAs(Table S4,http://links.lww.com/JD9/A10) and calculated their normal value range (μ±2σ) in 23 SS samples. The μ±2σ ranged from 0.752 to 1.480 (Table S4, http://links.lww.com/JD9/A10 and Fig. 4B). In general, the samples in which the relative expression level was higher than 1.480 would be defined as NS samples. These findings require verification in future studies.
Discussion
Given the fact that the incidence of syphilis has increased dramatically worldwide in recent years and that NS has largely paralleled active syphilis, delayed diagnosis and treatment of NS could be a serious health problem,5-6which highlights the urgent need for early diagnosis of NS.However, successful early diagnosis of NS remains challenging because most early NS cases do not show symptoms and the confirmed diagnosis of NS is largely based on CSF findings, which requires LP, an invasive operation. Additionally, it is difficult to persuade patients to undergo LP when there are no CNS symptoms.Therefore, the development of novel biomarkers for NS diagnosis in peripheral blood is urgently needed.
MiRNAs regulate a wide range of biological processes7 by mediating post-transcriptional regulation of proteincoding genes and are associated with the occurrence and development of a variety of diseases.8–11In this study, we used microarray analysis to identify dysregulated miRNAs in the PBMCs of syphilis patients and compared the differential expression of miRNAs between HCs, SS patients and NS patients to identify potential biomarkers for the diagnosis of NS.
NS is caused by T. pallidum invasion of the CNS, which can occur at any stage of T. pallidum infection. SS is the early stage of T. pallidum systemic infection. Although SS is classified into the early stage, the duration of SS can range fromweeks to months, and some patients in this stagemay have CNS involvement. In this study, we found that the miRNA expression profiles in some of the SS patients were similar to that ofHCs,while profiles of other SS patients were similar to that of NS patients. In one case, the expression profile was the same as that of NS patients. This range of resultsmay have several interpretations. Wespeculate that in cases in which T. pallidum does not invade the CNS, the miRNA profile is similar to that of HCs. At the beginning of CNS invasion by the pathogen, the immune system may not have enough time to generate antibodies, such asCSF-VDRL and CSF-TPPA, and CNS inflammatory responses may also be absent. Thus, white blood cell count and proteins in CSF appear to be normal, which leads to the missed diagnosis of NS when diagnosis depends on traditional technology. As reported in many studies, even in the absence of laboratory evidence showing NS, the CNS may be infected by T.pallidum.16-17The finding that patients with SS and NS shared some of the same miRNAs profiles suggest that there are specific miRNAs that can be used as peripheral blood biomarkers for the diagnosis of NS. If such miRNAs were capable of discriminating the different stages of T. pallidum infection, especially for NS, it may not only overcome the difficulties of NS diagnosis and reduce the trauma of LP, but also lead to improved treatment because the treatment regimen of early syphilis and NS are different.
The microarray results were verified by qPCR analysis of 13 of the differentially expressed miRNAs. The expression level of the 13 miRNAs was higher in NS and SS samples than HC samples, which suggests that expression profiles of these miRNAs may be correlated with T. pallidum infection. To further evaluate whether the miRNAs could be potential diagnostic indicators, we constructed ROC curves on the basis of the 13 miRNA expression levels between two groups (HCs vs. SS, HCs vs. NS, HCs vs.NS + SS, and NS vs. SS). The results showed that either the four miRNAs with higher expression level in NS than in SS samples or the nine miRNAs with higher expression levelsin SS than in NS samples could distinguish syphilis and NS from HCs (Fig. 3). However, the value of the four miRNAs to distinguish NS from SS is limited. The AUC value was only 0.604 (95% CI, 0.442–0.766) for SS and NS(Table S5, http://links.lww.com/JD9/A11). To increase the ability of miRNAs to distinguish NS from SS, we integrated the diagnostic value of the four miRNAs and nine miRNAs using ROC curves. We found that the AUC value was increased to 0.968 (95% CI, 0.927–1.0) for SS and NS (Table 3). These results suggest that the miRNAs may serve as novel biomarkers for differentiating NS from non-NS or from HCs.
Among the differentially expressed miRNAs identified in this study, miR-21-5p is the most frequently reported miRNA. The function of miR-21 is complex and has elicited considerable interest in diverse fields including embryonic development,18tumorigenesis,19fibrosis,20and immune reaction.21The majority of studies on miRNA-21-5p are related to various human cancers such as breast cancer,22lung cancer,23and colorectal cancer.24Recently,some studies found that miRNA-21 might play an important role in chronic infections caused by microorganisms. Chen’s study indicated that miRNA-21-5p was upregulated during HCV infection, which in turn suppressed HCV-triggered type I IFN production, thus promoting HCV replication.25In another study on leprosy, upregulated miRNA-21-5p in lepromatous lesions targets multiple genes associated with the immunologically localized disease form, providing an effective mechanism to escape from the vitamin D-dependent antimicrobial pathway.26Our study showed that miRNA-21-5p was upregulated in NS patients, which suggests that miRNA-21-5p may play a negative role in the clearance of T.pallidum. miR-21-3p was reported to be involved in ovarian cancer25 and cardiac disease,27–29but no information has been reported on its association with infectious diseases. Little is known about the biological functions of the other miRNAs identified in this study. The findings that their expressions are correlated with NS provide new clues for future study of NS pathogenesis.In summary, here we report the first identification of 13 differentially expressed miRNAs in PBMCs of NS patients in comparison with SS patients and HCs. These miRNAs in PBMC may be used as potential biomarkers for NS diagnosis. Our findings also provide new directions for studying the pathogenesis of NS. A limitation of this study was the small sample size. Therefore, the utility of the differentially expressed miRNAs in diagnosis of NS should be validated with larger cohorts and more samples.
Acknowledgments
The authors greatly appreciate the clinic staff who recruited patients and we thank all participants of this study for their cooperation.
Source of funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81572039), Shanghai Science and Technology Commission (Nos. 17DZ2293300,YDZX20193100002868), Clinical Research Plan of SHDC(No. 16CR1029B), National mega project on key infectious diseases (No. 2017ZX10202102-001-007),and Shanghai Municipal Commission of Health and Family Planning (No.20164Y0260).
- 国际皮肤性病学杂志的其它文章
- NCSTN Gene Silencing Inhibits the Retinoic Acid Signaling Pathway in Human Immortalized Keratinocytes
- A Clinical Retrospective Analysis of 340 Inpatients With Malignant Skin Tumors in Western Inner Mongolia
- Subepidermal Autoimmune Bullous Disease Associated With Monoclonal Gammopathy of Unknown Significance and Unexpected Positive Direct Nikolsky Phenomenon: A Case Report
- Nicolau Syndrome Following Metamizole Injection: A Case Report
- A Study of Normal Epidermal Melanocyte Distribution
- Erosive Pustular Dermatosis of the Scalp After Excision and Skin Grafting of Scalp Squamous Cell Carcinoma

