Comprehensive metabolic profiling of Alismatis Rhizoma triterpenes in rats based on characteristic ions and a triterpene database
Lu Wng , Sen Li , Jixin Li , Zhongzhe Cheng , Yulin Feng , Hui Ouyng ,Zhifeng Du ,**, Hongling Jing ,*
a Tongji School of Pharmacy, Huazhong University of Science and Technology, Wuhan, China
b Department of Pharmacy, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
c Jiangxi University of Traditional Chinese Medicine, Nanchang, China
d School of Pharmacy, Weifang Medical University, Weifang, China
Keywords:Alismatis rhizoma Triterpenes Metabolites HPLC-QTOF-MS/MS
ABSTRACT Alismatis Rhizoma (AR) is widely used in Chinese medicine, and its major bioactive components, triterpenes, reportedly possess various pharmacological activities. Therefore, it is very important to study the metabolism of triterpenes in vivo. However, the metabolism of AR triterpene extract has not been comprehensively elucidated due to its complex chemical components and metabolic pathways. In this study, an ultra-performance liquid chromatography quadrupole time-of-flight mass spectrometry method, which was based on the characteristic ions from an established database of known triterpenes,was used to analyze the major metabolites in rats following the oral administration of Alismatis Rhizoma extracts (ARE). As a result, a total of 233 constituents, with 85 prototype compounds and 148 metabolites,were identified for the first time.Hydrogenation,oxidation,sulfate and glucuronidation conjugation were the major metabolic pathways for triterpenes in AR.In addition,the mutual in vivo transformation of known ARE triterpenes was discovered and confirmed for the first time. Those results provide comprehensive insights into the metabolism of AR in vivo, which will be useful for future studies on its pharmacodynamics and pharmacokinetics. Moreover, this established strategy may be useful in metabolic studies of similar compounds.
1. Introduction
Alismatis Rhizoma (AR), a traditional Chinese medicine (TCM)made from the dried rhizome of Alisma orientale Juzepzuk (Alismataceae), has been used widely for treating various diseases such as dysuria, hyperlipidemia, inflammation, and cancer [1-3].Protostane-type triterpenes, the major bioactive and pharmacodynamic ingredients of AR,are considered to be chemotaxonomic markers of the genus [4-6]. For example, recent studies have reported that protostane-type triterpenes may have hypoglycemic activity by inhibiting alpha-glucosidase activity and promoting glucose uptake [7]. Triterpenes can also lower the serum lipid level in HFD-induced hyperlipidemic mice, and thus they have the lipid-regulating properties observed in previous research[8].To the best of our knowledge,more than 90 triterpenes have been isolated and identified in AR,such as alisol O,alisol S 23-acetate,and alisol H [9-11]. It is generally known that the metabolism plays an essential part in pharmaceutical research on TCMs because it is helpful for the discovery of potentially toxic or active metabolites and it forms the material basis of the metabolic process in vivo[12,13].Although various pharmacological effects of triterpenes have been reported, there is no report about the metabolic profile of ARE triterpenes in vivo, even though there have been a few reports on the metabolism of some single triterpene components in vitro[14-17].
Generally, identification of metabolites from TCMs is a challenge because of the complicated and trace chemical components, interference from endogenous substances, and the complex transformation and unpredictable metabolic pathways in vivo [18-20]. With the development of research instruments[21-23], ultra-high performance liquid chromatography coupled with quadrupole time-of-fight mass spectrometry(UHPLC-QTOFMS/MS)can detect the exact mass and give the unique formula of the compounds, and it has become an important analytical instrument in metabolite identifications of TCMs.
According to previous research [11], the triterpenes of AR belong to the protostane type; most of them have a parent nucleus of cyclopentanoperhydrophenanthrene and a side chain on the C17of the parent nucleus,and the major triterpenes in AR can be classified into three categories according to the different characteristic ions produced during mass spectral fragmentation.Type I triterpenes have a basic prototane triterpenoid skeleton,and type II triterpenes are different from type I because of the presence of a carbonyl at C16.Moreover,type III triterpenes have a C16, C23-oxido six-membered ring on the basic skeleton [11].Previous research on the in vitro metabolism of alisol A and alisol G has shown that most metabolites maintain the structural features of the prototype compounds [15,17]. This research has laid the foundation for identifying unknown triterpene metabolites in AR in vivo. In addition, it is well known that the log P value of a compound, which is the logarithm of its partition coefficient between n-octanol and the water log (coctanol/cwater), is a wellestablished measurement of the compound’s hydrophilicity. The Clog P values,which are the calculated values of the log P,are then usually used to identify the isomers of these compounds[24].For example, Liang et al. [22] identified the metabolites of Paeoniae Radix Rubra decoction in rats by comparing the base peak chromatograms and ClogPs based on the databases of parent compounds, known metabolites and characteristic neutral losses using the HPLC-DAD-ESI-IT-TOF-MSntechnique.
In this study, a four-step strategy was established to identify the prototype compounds and metabolites in rat bile, plasma,urine and feces rapidly and systematically after the oral administration of ARE based on UHPLC-QTOF-MS/MS and the characteristic ions in the mass spectral fragmentation.To the best of our knowledge, this is the first report about the in vivo metabolic profile of triterpenes in ARE, and it will provide some new insights for future pharmacodynamic and pharmacokinetic research on those compounds.
2. Experimental
2.1. Chemicals, reagents and materials
HPLC-grade acetonitrile and methanol were purchased from Fisher Scientific (Fair Lawn, NJ, USA). Formic acid (≥96%) and carboxymethylcellulose sodium (CMC-Na) were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Ultrapure water was produced using a Milli-Q water system (Millipore, Bedford, MA, USA). Oasis solid phase extraction (SPE) cartridges were purchased from Waters Corp.(Milford,MA,USA).All the other solvents and chemicals were of reagent grade or better.
Dried AR materials were obtained from Sichuan Province,China,and identified and authenticated by Professor Keli Chen of Hubei University of Chinese Medicine, China. The reference standards included alisol A-24 acetate, alisol A-23 acetate, 24-deacetyl alisol O, alisol G and 25-O-methylalisol A, with a purity of ≥98%, and they were isolated in house from the dried AR. In addition, reference standards of alisol B-23 acetate, alisol C-23 acetate,16-oxo-alisol A and alisol F with a purity of ≥98% were obtained from Shanghai Yuanye Bio-Technology Co., Ltd.Shanghai, China.
2.2. Solution preparation
2.2.1. Preparation of standard solution
The appropriate amounts of three standards(alisol F,alisol C-23 acetate, and alisol B-23 acetate) were dissolved in 2.0 mL of 0.5%CMC-Na solution to achieve better solubility[16,25],and the final concentration of each standard solution was equal to 2 mg/mL for oral administration. In addition, the appropriate amounts of nine standards(alisol F,alisol C-23 acetate,alisol B-23 acetate,alisol G,alisol A-23 acetate,alisol A-24 acetate,24-deacetyl alisol O, 25-O-methylalisol A, and 16-oxo-alisol A) were dissolved in 200 μL of methanol,and the final concentration of every standard in mixed standard solution was equal to 500 ng/mL. A 10.0 μL of each mixed standard solution was injected for UHPLC-QTOF-MS/MS analysis.
2.2.2. Preparation of ARE
The dried AR was ground into powder (24 mesh), and the pulverized powder samples (60.0 g) were extracted three times with 200 mL of acetonitrile in an ultrasonic bath for 30 min at room temperature.The supernatant of these samples was mixed and evaporated to dryness under a vacuum at 50°C following individual filtration and centrifugation at 17,000 g for 5 min.The residue was dissolved in 30 mL of 0.5%CMC-Na solution,and the final concentration of the AR suspension was equal to 2.0 g/mL.Moreover,10.0 μL of the acetonitrile AR extract at 2.0 g/mL was injected for UHPLC-QTOF-MS/MS analysis.
2.3. Metabolic study in rats
2.3.1. Animals
Male Sprague-Dawley rats (200-260 g) were obtained from the Animal Center of Tongji Medical College at Huazhong University of Science and Technology(Wuhan,China),and this study was approved by the Animal Ethics Committee of this institute.The rats were kept in a controlled environment for a week before the experiments were started, and they were fasted overnight before the test.
2.3.2. Drug administration and sample preparation
The ARE and three representative standards were given to the rats orally at 10 g/kg and 10 mg/kg, respectively, based on a previous study on the pharmacokinetics,pharmacodynamics and toxicology of AR triterpenes and the metabolism of similar compounds[2,8,16,26-31].Blood(0.2 mL)was collected at 0.25,0.5,1,2, 4, 6, 8,12,16, 20, and 24 h from the angular vein post-dosing and was centrifuged immediately at 1,000 g for 10 min at 4°C to obtain the plasma.For the plasma samples,an aliquot of 1 mL of mixed plasma from six rats was mixed with 5 mL of methanol to precipitate the protein. The plasma samples were then vortexed for 5 min and centrifuged at 17,000 g for 10 min at 4°C.The supernatant was blown to dryness with a nitrogen stream, and the residue was dissolved with 1000 μL of acetonitrile:water(1:1,V/V) and filtered through a 0.22-μm microporous membrane before injection. Blank plasma biosamples were collected before oral administration and the processing procedure is mentioned above.
To collect urine and fecal samples,rats were kept in DXL-D rat metabolism cages. Their feces were collected at 0-24 and 24-36 h post-dosing. Blank fecal biosamples were collected before oral administration.The fecal samples from three rats were mixed, and 1 g of mixed feces was extracted with 20 mL of methanol using ultrasonication for 60 min. The supernatant was blown to dryness with a nitrogen stream, and the residue was dissolved with 1000 μL of acetonitrile:water(1:1,V/V)and filtered through a 0.22-μm microporous membrane before injection.
Urine was collected at 0-4, 4-8, 8-12,12-24, and 24-36 h post-dosing. Blank urine biosamples were collected before oral administration at the same time as fecal biosamples. Urine and bile samples were processed similarly using SPE cartridges(Oasis HLB 3 mL; Waters). An aliquot of 3 mL of mixed bile or urine biosamples was loaded onto pretreated SPE cartridges. The cartridges were washed with 3 mL of water and then eluted with 3 mL of methanol. The eluent was collected and then blown to dryness with a nitrogen stream, and the residue was dissolved with 1000 μL of acetonitrile:water(1:1,V/V)and filtered through a 0.22-μm microporous membrane before injection.
The rats were anaesthetized and a PE-10 (polyethylene-10)bile duct was inserted into each one before the bile biosamples were collected. The rats were allowed to recover from the anesthesia before the blank bile biosample was collected. The bile samples were then collected at 0-12,12-24 and 24-36 h postdosing. The samples were processed as mentioned above. All the prepared samples were stored at -20°C until analysis. An aliquot of 10 μL of the supernatant from each sample was injected into the UHPLC-MS/MS system for analysis. The reproducibility and robustness of the method were evaluated using parallel samples of each matrix and nine standards of AR triterpenes as the quality control samples.
2.4. UHPLC-QTOF-MS/MS conditions
A UHPLC-MS/MS system consisting of a Shimadzu UHPLC(Shimadzu, Kyoto, Japan) coupled with a Triple TOF™5600 system (AB SCIEX, Foster City, CA, USA) was applied to identify the metabolites. The UHPLC system was equipped with an LC-30AD binary pump, a SIL-30AC autosampler, a CTO-30A column oven,a DGU-20A3degasser and a CBM-20A controller. Chromatographic separation was performed on an Ultimate XB-C18column(2.1 mm × 100 mm,1.8 μm, Welch, China) at 40°C. The elution gradient program for the mobile phase consisting of water containing 0.1% formic acid (A) and acetonitrile (B) and was as follows: 0-5 min,15%-35% B; 5-23 min, 35%-77% B; 23-24 min,77%-95% B; 24-27 min, 95% B; 27-27.1 min, 95%-15% B; and 30 min,15%B.The flow rate was 0.3 mL/min and the autosampler temperature was maintained at 4°C.The mass spectrometer was operated in positive ion mode, and the MS conditions were as follows:IS,4500 V;TEM,450°C;CUR,40 psi;GS1 and GS2,50 psi;and DP,60 V.The collision energy was set at 35 eV with a collision energy spread of 15 eV for the MS/MS experiments. The mass range was set at m/z values from 200 to 900 for a TOF-MS scan and m/z values of 50-900 for a TOF-MS/MS scan.
2.5. Data processing
The data obtained by UHPLC-QTOF-MS/MS were analyzed by Metabolite Pilot Software (version 2.0, AB Sciex) and Peakview(version 1.2, AB Sciex). The Clog P values were calculated using ChemDraw 12.0 in this experiment.
3. Results and discussion
3.1. Optimization of HPLC separation and MS conditions
For the optimization of the UHPLC conditions and mass spectrometry parameters, considering that there is no reference standard for the unknown metabolites, nine typical triterpenes(Fig. S1 and Table S1) were selected to optimize the parameters according to a previous study [11].
To obtain better separation, different mobile phase compositions, including 0.1% and 0.01% formic acid, 0.5 and 5 mM of ammonium formate, and various gradients were tested using representative AR standards.After a comprehensive comparison,better separation and sensitivity were obtained by using acetonitrile and 0.1% aqueous formic acid as the eluting solvent(Fig. S1). Some new isomers of known triterpenes in ARE can be separated under this chromatographic condition(Figs.S2 and S3).For example,prototype compound P65 with a[M+H]+ion at m/z 515.372 (C32H51O5+, calculated m/z 515.374) and P66 with a[M+H]+ion at m/z 515.372 not only have similar precursor ions and fragmentation pathways,but also have similar retention time of 23.32 and 23.63 min,respectively,as shown in Fig.S3.P65 was separated and identified for the first time. Similarly, prototype compounds P32 and P34 and P82 and P83 were two pairs of isomers separated well for the first time, and the detailed information is shown in Fig. S3 and Table S2.
Mass spectrometry parameters such as the collision energy and decluttering potential were optimized using acetonitrile solutions of pure AR standards by direct infusion. Moreover, the positive ion mode was more sensitive than the negative ion mode for most of the standards in the mass spectrometer, so the positive ion mode was chosen.
3.2. Analytical strategy for systematic analysis of triterpene metabolites
As shown in Fig.1,the first step of the analytical strategy was data acquisition.The data acquisition was performed by multiple mass defect filtering (MMDF) and dynamic background subtraction (DBS) based on UHPLC-QTOF-MS/MS.
The second step was to construct an in-house AR database for metabolite identification. First, the information containing the molecular formula, retention time, precursor ion, characteristic ion,major product ion and Clog P values of the AR triterpenes was summarized based on previously published literature [9-11] and the characteristic triterpene chemical profile of ARE (Fig. S2).According to previous research[11],all triterpenes can be divided into three types: type I triterpenes, which are represented by alisol B-23 acetate with characteristic ions of m/z 383.3 and 339.3(Fig. S4A); type II triterpenes, which are represented by alisol C-23 acetate with a characteristic ion of m/z 353.3 (Fig. S4B); and type III triterpenes, as represented by alisol F with characteristic ions of m/z 381.3 and 339.3 (Fig. S4C). It is worth noting that all the characteristic ions were derived from the parent nucleus.Considering that there have been no reports about the in vivo metabolism of triterpenes in the literature,the in vivo metabolic profile of three representative single triterpene standards (alisol B-23 acetate, alisol C-23 acetate and alisol F) was characterized and summarized. After that, an in-house database combining all the information was constructed in the present study.

Fig.1. Strategy for the systematic identification of the metabolites from Alismatis Rhizoma.
The third step was to characterize the prototypes and different types of metabolites in vivo. First, the raw data from triterpene prototypes of ARE were imported into Metabolite Pilot 2.0 software to screen the potential metabolites in vivo. Second, refiltering the preliminary screened data according to characteristic ions using Peakview 2.2 software and comparing them with the established database allowed for the rapid identification of the prototypes in vivo (Fig.1). It is well known that the metabolites transformed from the prototype compound have similar fragmentation patterns as that of the prototype.Therefore,for the in vivo metabolic profile of three representative single triterpene standards, when the metabolic site is on the side chain of the triterpenes, the characteristic ions of the type I, II and III triterpenes in AR will be identical to that of their prototypes.However, when the metabolic site is on the parent nucleus, the characteristic ions will be changed.For example,when the parent nucleus of three types of triterpenes is oxidized in vivo, extra dehydration occurs during fragmentation compared with the prototype,and their characteristic ions will be changed due to the formation of a double bond at the dehydration site. The type I triterpenes changed from m/z 339.3 to 337.3,type II changed from m/z 353.3 to 351.3,and type III from m/z 381.3 and 339.3 to 379.3 and 337.3. Thus, the type of metabolites can be rapidly characterized.
The last step was to identify the triterpene metabolites in AR based on the established database. Through the fragmentation patterns and in vivo metabolic profile of the three representative single triterpene standards, the conjugation site for some metabolites could be inferred, and then the proposed structure of the metabolites in vivo was further confirmed. Moreover, the Clog P values could be used to distinguish among metabolic isomers. Generally, metabolites with larger Clog P values were eluted later and had longer retention time in a reverse-phase chromatographic system [24]. For example, a pair of isomers,alisol A-23 acetate (ST5, Table S1) with Clog P 5.73 and alisol A-24 acetate (ST6, Table S1) with Clog P 6.34, have retention time of 18.40 and 20.08, respectively. The analytical strategy is summarized in Fig. 1.
3.3. Characterizing the metabolic profile of AR triterpene standards in rats by UHPLC-QTOF-MS/MS
Considering that there is no literature on the in vivo metabolic profile of triterpenes in AR, three representative standards of triterpenes, namely alisol B-23 acetate, alisol C-23 acetate and alisol F, were chosen as representatives of type I, II and III triterpenes, respectively, for a preliminary study on metabolism in vivo.
3.3.1. Metabolites of alisol B-23 acetate in rats
Alisol B-23 acetate is one of the most abundant compounds in AR,and it is also the most typical compound in type I triterpeneswith characteristic ion m/z values of 383.3 and 339.3,according to a previous study [11]. Fig. S4A shows the chemical structure and MS/MS spectra of alisol B-23 acetate.In the present study,a total of 30 compounds(B0-B29,Table S3),including the prototype and metabolites, were identified. Among the identified metabolites,only 3 metabolites were found in plasma,15 metabolites in bile,18 metabolites in urine and 13 metabolites in feces(Table 1).The major metabolic pathways include dehydrogenation, oxidation,deacetylation, glucuronidation and combined multiple-step metabolism. All the specific information about the formula,retention time, observed m/z, calculated m/z, error, and major product ions of the prototype and metabolites are shown in Table S3.

Table 1 Summary of the amount of metabolites after oral administration of three representative standards of triterpenes in AR characterized by UHPLC-QTOF-MS/MS.
Metabolite B0 with a [M+H]+of 515.373 (C32H51O5+, calculated m/z 515.374),retention time of 23.68 min and product ions of m/z 383.295 and 339.269 was identified as a prototype of alisol B-23 acetate by comparing it with the reference standard (ST4,Table S1 and Fig. S1). Metabolites B1-B20 were phase I metabolites,and it is worth noting that phase I metabolites B1-B13 were found to be known triterpenes that had been transformed from alisol B-23 acetate according to the established database. For example,metabolite B1(Fig.2A&a)with[M-H2O+H]+at 515.374 as well as a retention time of 19.46 min and product ion m/z values of 383.294 and 339.268 was identified as alisol A-24 acetate by comparing them with the reference standard(Fig.2B&b and ST6, Table S1), and it was presumed that B1 was converted from B0 through the transfer of an acetic acid ester from C-23 to C-24 after the hydration of the side chain at C24/C25.In addition,metabolite B3(Fig.2C&c and Table S3)was identified as 16-oxoalisol A-24 acetate isomers by comparing the retention time,[M+H]+and abundant product ion with that of the ARE components (Fig. 2D & d and P82, Table S2). In addition, metabolite B2 was an isomer of B3.Additionally,metabolites B5 and B6,B8 and B9 were also identified as alisol O and alisol B isomers, respectively. The other metabolites B4, B7, B10, B12 and B13 were identified as alisol C-23 acetate,alisol A,11-deoxy-alisol A,16,23-oxido-alisol B isomers and alisol C, respectively.
Metabolites B14-B20 were phase I metabolites produced by multiple-step metabolism. Metabolite B14 was preliminarily screened with Metabolite Pilot 2.0 software.With a precursor ion[M+H]+of 545.347 (C32H49O7+, calculated m/z 545.348) and a retention time of 14.77 min, it was subsequently inferred to be a dioxidation-dehydrogenation product because its molecular weight was 30 Da more than that of alisol B-23 acetate(Table S3).Moreover,the abundant product ions with m/z values of 395.269 and 351.232 indicated that the dioxidation and dehydrogenation simultaneously occurred on the parent ring in comparison with the corresponding characteristic ions of alisol B-23 acetate at m/z 383.3 and 339.3 (Fig. S4A). However, since the fragmentation pattern of the parent ring was irregular, it was difficult to determine the metabolic site on the parent ring from MS/MS data alone. The identification of metabolite B17 with a precursor ion[M+H]+of 487.342 (C30H46O5+, calculated m/z 487.342) and product ions at m/z 397.272 and 353.250 is similar to that of B14,and it was identified as the oxidation and dehydrogenation product of alisol B on the parent ring.
Metabolite B15 with [M+H]+at 487.341 was eluted at 9.83 min. Its major product ion with m/z 339.267 was in accordance with that of prototype alisol B-23 acetate, namely, the metabolic reaction occurred at the side chain. Another product ion of m/z 397.273 was 14 Da greater than the product ion of m/z 383.3 for alisol B-23 acetate (Fig. S4A); therefore, the metabolic site of B15 can be further inferred. Considering the 28 Da difference between the [M+H]+of B15 and the prototype (Table S3),two possible structures were suggested, 22-hydroxy-16,23-oxido-alisol B with a Clog P of 4.72 or 11-dehydro-22-hydroxyalisol F with a Clog P of 4.58 (Scheme S1). However, metabolite B16 with a [M+H]+at m/z 487.340 and product ions at m/z 397.273 and 339.268 and a retention time of 12.16 min was the isomer of B15. Therefore, metabolites B15 and B16 were finally identified as 11-dehydro-22-hydroxy-alisol F and 22-hydroxy-16,23-oxido-alisol B, respectively, by comparing their retention time and Clog P values.
Metabolite B18(Fig.2E&e)with an m/z of 489.358 was 26 Da less than that of B0, and it was eluted at 13.11 min with product ions at m/z 381.271 and 337.252, indicating that the oxidation occurred on the parent ring according to a comparison with the product ions of m/z 383.3 and 339.3 from alisol B-23 acetate(Fig. S4A). In addition, B18 is the same compound as metabolite C13 (Fig. 2F & f, Table S3) based on the [M+H]+, fragmentation pattern and retention time. By combining these data, metabolite B18 was identified as 16-hydroxy-alisol B (Clog P 5.16) (Scheme S1). Then, as the isomers of B18, metabolites B19 and B20 had a[M+H]+of m/z 489.36 and were eluted at 10.34 and 12.41 min,respectively. Their product ions with m/z 339.3 indicated that oxidation occurred on the side chain. Considering their fragmentation patterns, four possible structures (Scheme S1) were predicted: 22-oxo-24-deoxy-alisol A (Clog P 4.86), 22-hydroxyalisol B(Clog P 5.04),22-oxo-25-deoxy-alisol A(Clog P 5.74),and 24-oxo-alisol A(Clog P 6.34).Clog P values can be used to assign metabolites B19 and B20 by comparing the Clog P values of possible structures with that of B18, which should be the largest among those three isomers.In addition,the retention time of B19 and B20 indicated that B20 metabolites should have higher Clog P values. Thus, metabolites B19 and B20 were preliminarily speculated to be 22-oxo-24-deoxy-alisol A (Clog P 4.86) and 22-hydroxy-alisol B (Clog P 5.04), respectively.

Fig.2. The extracted ion chromatograms and MS/MS spectra of the compounds.(A),(C),(E)and(G)The metabolites B1,B3,B18 and B16 transformed from alisol B-23 acetate in rats;(B) alisol A-24 acetate in a mixture of nine representative standards (ST6, Table S1 and Fig. S2); (D) 16-oxo-alisol A-24 acetate in ARE (P82, Table S2); (F) the metabolite C13 transformed from alisol C-23 acetate in rats; (H) the metabolite F11 transformed from alisol F in rats; and (a-h) indicate the MS/MS spectra of the compounds in (A-H).
Metabolites B21-B29 were identified as phase II metabolites,and glucuronidation was the major phase II metabolic reaction.Metabolite B25 with a[M+H]+of 707.400(C38H59O12+,calculated m/z 707.401) was chosen as an example to illustrate the identification of phase II metabolites. First, its precursor ion at m/z 707.400 was 192 Da greater than that of alisol B-23 acetate,suggesting that it could be a glucuronide conjugate.Subsequently,the abundant product ion at m/z 531.366 confirmed the neutral loss of a glucuronic acid moiety(176 Da).The product ions at m/z 531.366 and 337.253 then showed that the oxidation reaction occurred on the parent ring compared with the precursor ion and characteristic ions at m/z 515.373 and 339.267 of the prototype.For the rest of the phase II metabolites, detailed information is shown in Table S3.
The extracted ion chromatograms (XICs) of the detected compounds are shown in Fig. S5, and the proposed metabolic pathways of alisol B-23 acetate are shown in Fig. S6. There were 20 phase I metabolites, most of which were found in urine and feces, and 9 phase II metabolites, which were primarily found in bile. However, only 3 metabolites were found in plasma, indicating that most metabolites of alisol B-23 acetate were not distributed in the plasma. Those results are summarized in Table 1.
3.3.2. Metabolites of alisol C-23 acetate in rats
Alisol C-23 acetate represents type II triterpenes. It has a characteristic ion m/z of 353.3(Fig.S4B),and it was selected as a typical compound to study the metabolism of type II triterpenes.A total of 24 metabolites(C1-C24)were identified in the plasma,bile,urine and feces of the rats following the oral administration of alisol C-23 acetate, from which 19 metabolites were found in the bile, 13 metabolites were found in the urine, and 6 and 4 metabolites were detected in the feces and plasma, respectively(Table 1). The XICs of all the metabolites and the prototype are shown in Fig.S7.In addition,the primary metabolic pathways of alisol C-23 acetate (Fig. S8) were similar to those of alisol B-23 acetate, except for sulfation.
Metabolite C0 with a [M+H]+at m/z 529.353 (C32H49O6+,calculated m/z 529.353) and a retention time of 14.98 min was regarded asa prototype ofalisol C-23acetate upon comparisonwith the reference standard(ST1,Table S1).For phase I metabolites,C1-C8 represented the conversion of the prototype into other known triterpenes in a comparison with the established database. For example, metabolite C1 with [M+H]+at m/z 547.362 (C32H51O7+,calculated m/z 547.363) corresponding to the hydrogenationoxidation product of alisol C-23 acetate was identified as a 16-oxo-alisol A-24 acetate isomer through a comparison with the ARE compounds(P82,Table S2).Similarly,metabolite C2 was also identified as 16-oxo-alisol A-24 acetate isomers. Moreover, metabolites C3, C4, C5 and C6 were alisol C isomers that had been convertedfromprototypeC0(TableS4).Metabolites C7and C8were identified as 16-oxo-alisol A and 23-dehydro-16-oxo-alisol A isomers,respectively,from a comparison of the prototypes in ARE(P50 and P47,Table S5).In addition,metabolites C9-C18 were primarily oxidative and multiple-step reaction products. The identification process was similar to that of alisol B-23 acetate and the result is summarized in Table S4.However,C19-C24 were phase II metabolites.Among them,C20-C23 primarily underwent glucuronidation,and only C19 and C24 were sulfate conjugates. For example,metabolite C19 with a[M+H]+at 625.303(C32H49O10S+,calculated m/z 625.305)was 96 Da larger than that of alisol C-23 acetate,and therefore it was characterized as an oxidation-sulfation metabolite of alisol C-23 acetate.The oxidation occurred on the parent ring as notedbycomparingthe major product ion of351.23 forC19 with the characteristicion of353.30foralisolC-23acetate(Fig.S4B).Detailed information on the other metabolites is shown in Table S4.
As shown in Table 1, 24 metabolites were characterized following the oral administration of alisol C-23 acetate in rats.Among these metabolites,there were 18 phase I metabolites and 6 phase II metabolites.Moreover,it is worth noting that all of the phase II metabolites could only be found in bile samples, and a smallest number of metabolites were found in plasma samples.
3.3.3. Metabolites of alisol F in rats
Alisol F represents type III triterpenes.It has characteristic ions at m/z 381.3 and 339.3 (Fig. S4C), and it was chosen as a representation to study the metabolism in vivo. A total of 28 components, including prototypes (F0) and metabolites (F1-F27), were characterized after the oral administration of alisol F by UHPLCQTOF-MS/MS. Among them, 21 metabolites were found in bile,17 metabolites were in urine, and 11 and 6 metabolites were found in feces and plasma,respectively,as shown in Table 1.The primary metabolic pathways are still common metabolic reactions such as oxidation, dehydrogenation,glucuronidation and sulfation.
F0 (Table S5) with a [M+H]+at m/z 489.360 (C30H49O5+,calculated m/z 489.358) and a retention time of 15.24 min was identified as a prototype by comparing it with the reference standard(ST2,Table S1).The identification process of metabolites F1-F27 is similar to that of alisol B-23 acetate, and detailed information is summarized in Table S5.Additionally,the XICs of the metabolites and metabolic pathways of alisol F in rats are shown in Fig.S9 and Fig.S10.The distribution of metabolites for alisol F is similar to that of alisol B-23 acetate, that is, most of the phase II metabolites were observed in the bile and urine (Table 1).
3.3.4. In vivo biotransformation of three representative standards
Based on the results of three representative triterpene standards, the mutual transformation of known triterpenes was confirmed in vivo for the first time.The transformations of three representative standards are shown in Fig. 3. Alisol B-23 acetate(B0),one of the most common components in AR belonging to the type I triterpenes, was selected as an example to illustrate the transformation of triterpenes in vivo. When the ester of acetic acid at C-23 is deacetaldehyde and it forms a single oxygen-sixmembered heterocycle with the C-16 of the parent ring, alisol B-23 acetate can be converted into type III triterpenes, such as metabolites B6 and B12 (Fig. 3 and Table S3), which were identified as an alisol O isomer (P59 and P60, Table S2) and a 16,23-oxido-alisol B isomer (P16, Table S2) by comparing them with components of ARE (Fig. S2). In addition, when carbonyl substitution occurs at the C-16 of the parent ring,alisol B-23 acetate can be transformed into type II triterpenes, such as metabolite B4(Fig. 3 and Table S3), which was identified as alisol C-23 acetate through a comparison with the reference standard(ST1,Table S1).
Notably, different AR standards can be transformed into the same compound in vivo.As shown in Fig.3,the 16-oxo-alisol A-24 acetate isomers (B2, B3, C1, and C2) can be transformed from alisol B-23 acetate(B0)or alisol C-23 acetate(C0).Unlike previous one-to-multiple metabolic processes for single standard triterpenes, this transformation is a multiple-to-one process. It is noteworthy that the multiple-to-one process is very useful for the identification of metabolites in vivo.For example,metabolite B16(Fig. 2G & g and Table S3), which has the same retention time,precursor ion and fragmentation pattern as those of F11(Fig.2H&h and Table S5), was identified as the same metabolite, 22-hydroxy-16,23-oxido-alisol B. The in vivo biotransformation found herein further showed the complexity of TCM metabolism,which will be illuminating for the study of in vivo pharmacodynamics.

Fig. 3. Proposed mutual transformation of known triterpenes following the oral administration of three representative AR standards in rats. Red letters indicate three triterpene standards for oral administration. Black and blue letters indicate the numbers and names of metabolites. Sul: sulfate; GluA: glucuronic acid residue.
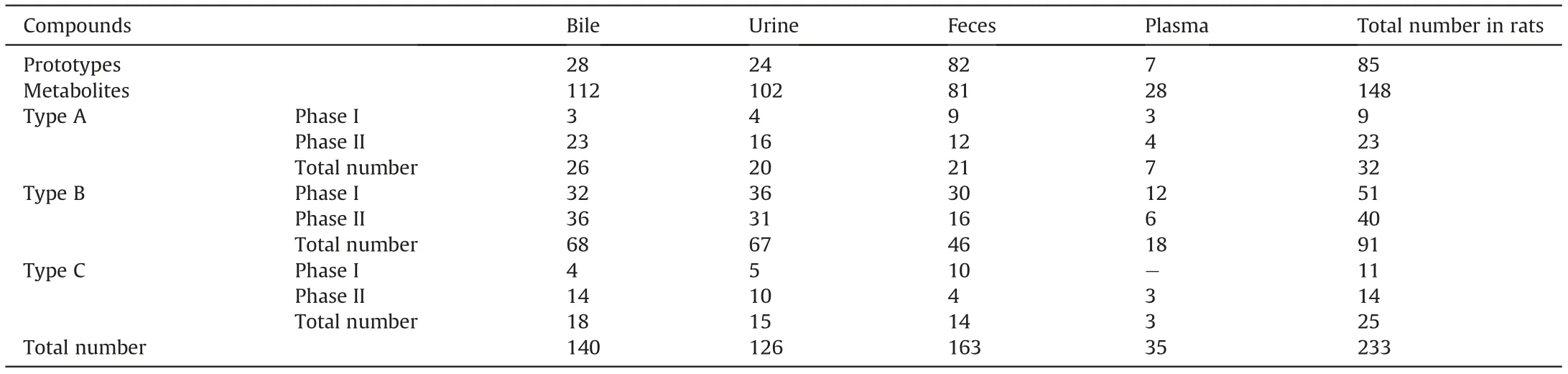
Table 2 Summary of the number of metabolites after oral administration of ARE characterized by UHPLC-QTOF-MS/MS.
The metabolism of three triterpene standards is simpler and more intuitive for understanding the metabolic process of triterpenes in vivo. In general, most of the metabolites of the three representative triterpene standards were identified in bile, urine and feces, with a few found in plasma. Interestingly, a previous study [16] showed that most of the AR triterpenes were distributed in the intestine, stomach, liver and kidney tissue after oral administration. There may be an underlying correlation. Moreover, the administration of a single triterpene standard makes it possible to discover the mutual transformation of known triterpenes, which is helpful for the further study of triterpene metabolism in TCMs. In addition, studying the metabolites of three standards laid the foundation to establish the database and the characterization of the ARE metabolic profile in rats.
3.4. Characterization of ARE metabolic profile in rats by UHPLCQTOF-MS/MS
3.4.1. Identification of prototypes from ARE
There are three primary steps in the identification of prototype compounds after the oral administration of ARE in rats.First,the prototypes identified in ARE by using the established database were entered in Metabolite Pilot 2.0 to screen the potential metabolites.Second,all the compounds screened by Metabolite Pilot 2.0 were filtered again according to the characteristic ions of known triterpenes in Peakview 2.0 to search for the possible prototype compounds. Third, these compounds were compared with the components of ARE (Fig. S2) and could thus be divided into two types: prototype compounds and potential metabolites with the same characteristic ions as the prototype compounds.Finally, the prototype compounds were identified and classified by their product ions according to their fragmentation pattern.As shown in Table 2, 85 prototype compounds were identified in vivo,among which 28 and 24 compounds were observed in bile and urine samples,82 in feces samples and only 7 were found in plasma samples. Among them, 46 prototype compounds were discovered only in feces and the other compounds could be found in two or more different biological samples (Table S2).
According to the established database, 22 type I triterpenes(P6-P8, P20-P29, P41-P43, P63-P66, P74, and P75, Table S2), 34 type II triterpenes (P9-P12, P30-P36, P44-P56, and P76-P85,Table S2) and 29 type III triterpenes (P1-P5, P13-P19, P37-P40,P57-P62, and P67-P73, Table S5) were identified. Among them,20 new isomers of known compounds of ARE(P6,P14,P18,P20-22,P25-28,P32,P48,P51,P53,P56,P57,P65,P80,P83,and P85)were identified in vivo after comparing them with previous findings [10,11].
The XICs of the prototype compounds in Fig.S11 were similar to those of the components of ARE in Fig.S2.Detailed information on these prototype compounds is shown in Table S2.
3.4.2. Identification of metabolites from ARE
This is the first profile and characterization of the metabolites of ARE in vivo,and a total of 148 metabolites including 71 phase I metabolites and 77 phase II metabolites were identified based on the UHPLC-QTOF-MS/MS analysis. As shown in Table 2,112 metabolites were identified in bile,102 metabolites were detected in urine,and 81 and 28 metabolites were found in feces and plasma,respectively.Detailed information on the formula,retention time,observed m/z, calculated m/z, error and major product ions is summarized in Table S6, and the XICs of all the metabolites are shown in Fig. 4. The phase I metabolic pathways consisted of hydroxylation, dehydrogenation, dioxidation and others. Additionally, the phase II metabolic pathways included sulfation and glucuronidation conjugation. Moreover, multiple-step metabolism such as oxidation-sulfation, deacetylation-oxidation, and oxidation-glucuronidation was also found. In this study, all the identified metabolites of ARE can be divided into three types(A,B,and C) according to the parent ring structure since the metabolites and their corresponding prototype compounds have similar fragmentation patterns.
3.4.2.1. Type A metabolites. Type A metabolites have a similar fragmentation pattern to that of type I triterpenes. Alisol B-23 acetate (P66, Table S2) and its deacetylation product alisol B(P23, Table S2), alisol A-24 acetate (P63, Table S2) and its deacetylation product alisol A (P22, Table S2), and alisol G (P7,Table S2) were the major type I triterpenes in ARE (Fig. S2).Therefore, type A metabolites could be primarily transformed from these type I triterpenes in vivo. In this study, 32 type A metabolites (M1-M32, Table S6), including 9 phase I metabolites (M1-M9) and 23 phase II metabolites (M10-M32), were identified after the oral administration of ARE. The possible metabolic pathways are shown in Scheme S2. Metabolite M1 with precursor ion [M+H]+of m/z 473.362 and product ions of m/z 339.268, 383.295 and 419.295 was in accordance with the data for alisol B (P24, Table S2), and therefore, it was identified as an alisol B isomer. Similarly, metabolites M4-M6 with a precursor ion of 491.37, which was 18 Da larger than that of alisol B, were identified as alisol A isomers (Scheme S2). M2 with a protonated molecular ion [M+H]+at m/z 475.377 was proposed to be the deoxidation product of alisol A or the hydrogenation of alisol B. The major product ions of m/z 339.265 indicated that a metabolic reaction occurred on the side chain,and the product ion of m/z 383.293 indicated that it might be 24-deoxy-alisol A (Clog P 5.99) or 25-deoxy-alisol A (Clog P 6.19), (Scheme S1). However, M3, the isomer of M2 that had a longer retention time than M2 (Table S6), was identified as 25-deoxy-alisol A,and thus M2 was identified as 24-deoxy-alisol A accordingly. Metabolite M7 has a [M+H]+ion of m/z 499.376 and a formula of C32H50O4, and is 16 Da less than that of alisol B-23 acetate (P65, Table S2). The product ion of m/z 341.279 suggested that the deoxidation occurred on C-11(Fig.S4A),and therefore, M7 was identified as 11-deoxy-alisol B-23 acetate.The[M+H]+ion of M8 was observed at m/z 507.368,which was 16 Da greater than that of alisol A. Its product ions of m/z 381.278 and 337.251 indicated that the oxygen was introduced into the parent ring of alisol A (Scheme S1). Unlike M8,metabolite M9, the isomer of M8 (Table S6) with product ions at m/z 339.267 and 381.278 (Scheme S1), suggested that the oxidation occurred on the C-22 of side chain according to the characteristic ions at m/z 383.3 and 339.3 of alisol A. Most type A metabolites were phase II metabolites, and all of them could be identified in bile samples (Table 2). The structural characterization of the representative metabolites is described as follows. M10 had a [M+H]+ion at m/z 553.321 and its molecular formula was C30H48O7S.The neutral loss of 80 Da indicated that M10 was a sulfate conjugate. Furthermore, a similar fragmentation pattern was observed between M10 and alisol B,indicating that M10 was the sulfate conjugate of alisol B.Similarly, M11 was the sulfate conjugate of alisol B-23 acetate.The conjugation site of sulfate on alisol B-23 acetate could be accurately located because there was only one hydroxyl at C-11.Thus, M11 was alisol B-23 acetate-11-O-Sul. Metabolite M12 with a[M+H]+at m/z 631.386 and a product ion at m/z 455.354 has a neutral loss of 176 Da (C6H8O6), which showed the conjugation of glucuronide. Subsequently, with product ions of m/z 455.354, 383.297 and 339.270, M12 was identified as 11-dehydro-alisol G + GluA (Scheme S2) by comparing it with the precursor ion and the product ions of m/z 473.3, 383.3 and 339.3 for alisol G. Metabolite M13 was the isomer of M12. In addition, all the other phase II metabolites were glucuronidation conjugates. Similar to M12, they were characterized by 176 Da neutral loss and characteristic ions. The detailed information is summarized in Table S6.

Fig.4. XICs of metabolites detected in rats after the oral administration of ARE.(A)Plasma;(B)bile;(C)urine;and(D)feces.(1)Metabolites with relatively high abundance and(2)metabolites with relatively low abundance.
3.4.2.2. Type B metabolites. Type B metabolites were derived from type II triterpenes, and they displayed similar fragmentation patterns,with product ions at m/z 353.3,m/z 355.3 or m/z 369.3.Alisol C-23 acetate(P72,Table S2)and its deacetylation products alisol C(P32,Table S2),alisol M -23 acetate(P81,Table S2),16-oxo-alisol A (P52, Table S2) and 13,17-epoxy-alisol B (P36,Table S2) were major type II triterpenes in ARE (Fig. S2). In this study,type B metabolites were the major triterpene metabolites following the oral administration of ARE in rats.A total of 91 type B metabolites were identified through a comparison with the established database. There were 51 phase I metabolites and 40 phase II metabolites. The detailed results for all the type B metabolites are displayed in Table S6, and their possible metabolic pathways are proposed in Scheme S2. The identification of representative type B metabolites (M37, M38, M84, M85 and M117)is elucidated as follows.M37 and M38 displayed a[M+H]+ion at m/z 473.36,and their formula was calculated as C30H48O4.Their product ion of m/z 355.26 could have come from two types of structures, with one being the deoxidation on the C-11 or hydrogenation on the parent ring of type II triterpenes (Scheme S1) by comparing with their characteristic ion of m/z 353.3(Fig. S4B). Another possibility is that oxidation occurs on the moiety of characteristic ion m/z 339.3 in type I triterpenes or type III triterpenes; for example, dehydrogenation at C-11 for type I triterpenes (Scheme S1). In addition, the product ion of M37 at m/z 399.3 (Table S6) indicated that hydroxyls were on the C-23 and C-24/25 (Scheme S1), while the product ion of M38 at m/z 397.3(Table S6)indicated that hydroxyls were on the C-22 and C-23(Scheme S1).Therefore,M37 could be tentatively assigned as 11,25-dideoxy-16-oxo-alisol A, 11,24-dideoxy-16-oxo-alisol A,11-dehydro-25-deoxy-alisol A or 11-dehydro-24-deoxy-alisol A.M38 was tentatively assigned as 11,24,25-trideoxy-22-hydroxy-16-oxo-alisol A or 11-dehydro-22-hydroxy-24,25-dideoxy-alisol A.However,these isomers could not be distinguished by MS/MS only.Metabolites M84 and M85 with retention time of 16.47 and 16.93 min were isomers of C30H47O7S,and they had a[M+H]+at m/z 551.301 and 551.307.They can be inferred to be alisol H or 11-deoxy-alisol C sulfate conjugates according to the major product ions of m/z 453.33 and 355.26. In addition, the Clog P values of 11-deoxy-alisol C-23-O-sulfate and alisol H-25-O-sulfate were 4.94 and 5.03. Therefore, M84 and M85 were identified as 11-deoxy-alisol C-23-O-sulfate and alisol H-25-O-sulfate, respectively. Metabolite M117 was formed by multiple-step metabolism simultaneously including phase I and phase II metabolism.M117 was eluted at 5.29 min,with a[M+H]+at m/z 697.380 and a product ion of m/z 521.348 derived from a neutral loss of 176 Da, which indicated that it was a glucuronide conjugate metabolite. After a 176 Da loss, it could be an oxidation product of 16-oxo-alisol A according to a comparison with the molecular ion of the prototype compounds. Moreover, its product ions at m/z 521.348, 395.259 and 353.250 showed that the oxidization occurred on the C-22 of the side chain of 16-oxoalisol A(Scheme S1).Thus,metabolite M117 was identified as 16-oxo-22-hydroxy-alisol A +GluA.
3.4.2.3. Type C metabolites. Type C metabolites were transformed from type III triterpenes based on their similar fragmentation patterns. Alisol F (P19, Table S2) and 16,23-oxido-alisol B (P16,Table S2) were the typical type III triterpenes. Twenty-five metabolites were primarily detected in bile, urine and feces(Fig.S12D).Among them,11 phase I metabolites were identified.M125 and M135 were selected as examples to illustrate the identification of type C metabolites. The possible metabolic pathway is summarized in Scheme S2.M125 with a[M+H]+at m/z 457.368 had a molecular formula of C30H48O3,which was 32 Da less than that of alisol F. Its product ions of m/z 383.297 and 341.281 showed that deoxidation(Fig.S4C)occurred on the C-11 and the side chain of alisol F simultaneously.Therefore,M125 was identified as 11,24-dideoxy-alisol F or 11,25-dideoxy-alisol F. It was impossible to determine the structures by MS/MS only.M135 was eluted at 17.30 min with a[M+H]+at m/z 629.370,suggesting the molecular formula of C36H52O9.The major product ion at m/z 453.338, which is 36 Da less than that of alisol F, indicated the neutral loss of 176 Da(GluA).Simultaneously,product ions at m/z 381.279 and 339.269 could point to the identification of 11,25-anhydro-alisol F. Thus, M135 was identified as 11,25-anhydroalisol F-24-O-GluA.
In this study, 148 metabolites were identified after the oral administration of ARE. Most of them could be found in the bile samples, especially phase II metabolites. Eighteen metabolites(M16, M19, M20, M32, M43, M79, M81, M83, M86, M91, M92,M98,M101,M116-M118,M141,and M146)were detected only in bile samples. Four metabolites M47, M53, M94 and M123 were identified only in urine samples,and 20 metabolites(M2-M4,M7,M9, M33-M36, M40-M42, M55, M68, M124-M127, M130, and M131)were found only in feces.It was interesting that all of them were phase I metabolites. In addition, only 28 metabolites were detected in the plasma,which is less than that of other matrices.It is worth noting that many metabolites were present both in bile and urine,and most of them were phase II metabolites(Table S6 and Fig. S12C). The ratio and matrix distribution of prototype compounds found in vivo are shown in Figs. S12A and B. Generally, a total 233 compounds, including 85 triterpenes prototype compounds(P1-P85)and 148 metabolites(M1-M148)of the ARE,were identified through a comparison with the established database of known triterpenes in AR, which is based on the characteristic ions. Among them, 140 compounds including 28 prototypes and 112 metabolites were identified in the bile. According to previous studies,AR can promote the decomposition of cholesterol and inhibit the synthesis of endogenous cholesterol in the liver, and it can also lower the lipid levels in vivo [32].Therefore, the distribution of the prototypes and metabolites of AR in bile expanded on the pharmacology to some extent. Similarly,AR has long been accepted to have a diuretic effect [1,3]. In the present study,126 compounds, including 24 prototypes and 102 metabolites, were identified in urine. This finding further supported the rationality of those results. However, previous studied [16] have shown that the intestine and stomach are the most widely distributed tissues for most triterpenes in AR after oral administration. Perhaps that is why there are more prototypes and phase I metabolites in feces. There is no doubt that the fewest prototypes and metabolites are distributed in plasma.This finding may be due to the low bioavailability of AR after oral administration.In addition,it is worth mentioning that the same metabolites were identified after the oral administration of three representative standards and ARE, which supported the validity and reliability of the analytical strategy. Interestingly, the most accurate identification for metabolites was performed in comparison with the metabolite standards. However, the complexity of the composition of traditional Chinese medicine makes it difficult to obtain metabolite standards. Using synthesized standards for the metabolites with a definite structure will be an optimal method of confirming the identified metabolites. At present, relevant experiments are in progress.
4. Conclusion
The metabolic profile of AR triterpenes in vivo was characterized by UHPLC-QTOF-MS/MS in this study. A total of 233 metabolites, including 85 prototype compounds and 148 metabolites,were identified for the first time in rats following the oral administration of ARE. Oxidization, hydroxylation, dehydrogenation,glucuronidation and sulfation were the major metabolic pathways for AR triterpenes in rats. In addition, the same metabolites were identified after the oral administration of representative standards and ARE, which rationalized the identification of ARE metabolites.The metabolism of ARE in vivo was shown to be very complicated, as indicated by the mutual transformation of standards in vivo, and it is worth mentioning that the mutual transformation of known ARE triterpenes in vivo was reported for the first time. In conclusion, these results provide the scientific basis for understanding the metabolism of AR in vivo,and the established strategy may be useful for conducting metabolic studies on similar compounds with characteristic ions in mass spectral fragmentation.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China(Grant No.81803717 and U1603104).
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2020.03.010.
 Journal of Pharmaceutical Analysis2021年1期
Journal of Pharmaceutical Analysis2021年1期
- Journal of Pharmaceutical Analysis的其它文章
- Evaluation of apoptotic effects of mPEG-b-PLGA coated iron oxide nanoparticles as a eupatorin carrier on DU-145 and LNCaP human prostate cancer cell lines
- Development of an analytical method for multi-residue quantification of 18 anthelmintics in various animal-based food products using liquid chromatography-tandem mass spectrometry
- Drug target discovery by magnetic nanoparticles coupled mass spectrometry
- Analytical methodologies for sensing catechol-O-methyltransferase activity and their applications
- Applications and challenges of low temperature plasma in pharmaceutical field
- Solid phase microextraction chemical biopsy tool for monitoring of doxorubicin residue during in vivo lung chemo-perfusion
