Identification of a novel planthopper resistance gene from wild rice(Oryza rufipogon Griff.)
Meng Yng,Jiein Lin,Ling Cheng,Hilin Zhou,Shu Chen,Fng Liu,Rongi Li,Yongfu Qiu,*
aState Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, Agricultural College, Guangxi University, Nanning 530005,Guangxi,China
bCollege of Agriculture,Yangtze University,Jingzhou 434025,Hubei,China
Keywords:Oryza rufipogon Griff.Planthopper resistance Next-generation sequencing-based bulked-segregant analysis Gene mapping Resistance mechanism
A B S T R A C T Rice planthoppers, including brown planthopper (BPH) and white-backed planthopper(WBPH), are the most destructive pests in Asian rice cultivation regions. Planthopper resistance genes that have been mapped and characterized advance our understanding of underlying resistance mechanisms and facilitate the breeding of resistant varieties,thereby contributing to an efficient pest management strategy. In this study, a novel resistance gene Bph38 derived from the wild rice species Oryza rufipogon Griff.was found to confer high resistance to BPH and WBPH. Conventional mapping was performed to identify regions associated with BPH and WBPH resistance,and two mapping efforts led to the same region on chromosome 4 flanked by markers RM16563 and RM16763. Bulked-segregant analysis and next-generation sequencing were performed using the same population to detect the resistance gene.Conventional mapping narrowed the region to a 12.3-Mb segment,and fine mapping using BC1F2 recombinants identified a 79-kb segment flanked by markers YM112 and YM190.Near-isogenic lines(NILs)carrying Bph38 in the 9311(indica)and BR54(japonica)genetic backgrounds were developed by crossing and backcrossing with marker-assisted selection. The agronomic traits and BPH and WBPH resistance of the NILs were similar to those of the recurrent parents.Mandatory feeding and host-choice tests revealed that Bph38 showed both antibiotic and antixenotic effects in both insects, with stronger effects in indica-background lines. Further fine mapping and characterization of the major gene may result in map-based cloning of the gene and allow its application in breeding insectresistant rice varieties.
1. Introduction
Rice (Oryza sativa) is an essential food crop for about half of the world's population. Rice yield is compromised by various pathogenic bacteria and pests. Of these threats, brown planthopper (BPH; Nilaparvata lugens Stål) and white-backed planthopper (WBPH; Sogatella furcifera Horvath) have become the most destructive insect pests of rice, causing major yield reductions and economic losses worldwide, especially in the Asia–Pacific region [1–3]. Both insects belong to the family Delphacidae(order Hemiptera),but they have distinct feeding habits. BPH is monophagous and feeds only on rice plants,whereas WBPH is oligophagous and feeds on several gramineous plants[2,4,5].Planthopper outbreaks have become more frequent and severe in recent decades. Chemical pesticide control has long been an effective management strategy, but it causes environmental pollution and ecological damage as well as “rampant” rice planthopper infestations [6]. Hostplant resistance is one of the most effective and economical approaches for managing rice planthoppers [7,8]; however, it requires the identification of resistant rice lines, mapping of resistance genes,and incorporation of these resistance genes.Although many planthopper resistance genes have been detected in various rice species, only a few have been characterized and applied, especially for WBPH resistance.Moreover, resistance genes may lose their efficacy when insect biotypes change or evolve novel virulence [3]. Controlling these evolved biotypes requires continually identifying new resistance genes for breeding insect-resistant rice.
BPH-resistance germplasm has been sought since 1969,and respectively 25 and 12 major resistance genes have been identified in cultivated O. sativa and wild rice species. The latter include O. australiensis, O. eichingeri, O. latifolia, O.officinalis, O. rufipogon, O. glaberrima, and O. minuta [9–12].Eight major resistance genes have been reported for rice WBPH. Wbph7 and Wbph8, which were derived from introgression lines of O. officinalis with the CC genome, were mapped to the same chromosomal regions as the BPH resistance genes Bph14 and Bph15, respectively [13]. Bph14 was found to confer resistance to both BPH and WBPH [14].The other six WBPH-resistance genes were identified in different cultivars. After gene identification and mapping,major resistance genes await cloning. At least eight BPHresistance genes (BPH14, BPH3, BPH26, BPH29, BPH18, BPH9,BPH32, and BPH6) have been cloned in the past ten years[15,16],but no WBPH-resistance genes have been successfully cloned.
Gene or quantitative trait locus (QTL) mapping is a basic method for revealing the genomic components of phenotypic variation. Over several decades, this method has been successfully used for map-based cloning of genes of interest and for marker-assisted selection (MAS) in crop breeding.However, conventional gene or QTL mapping requires genotyping and phenotyping of numerous individuals, rendering it a laborious and time-consuming process [17].Bulked-segregant analysis (BSA) is a simple and rapid approach for identifying molecular markers that are tightly linked to target genes or QTL[18].With the rapid development of high-throughput next-generation sequencing (NGS), NGSbased BSA (NGS-BSA) has become a widely adopted strategy for major gene or QTL mapping and gene identification,providing key supporting evidence. Several studies have used NGS-BSA for identifying qualitative and quantitative traits [17,19,20]. For instance, Takagi et al. [19] used NGS-BSA for mapping several QTL underlying seedling vigor and resistance to fungal blast disease in rice.Yang et al. [21]used the approach to map QTL conferring cold tolerance at the seedling stage in rice.In particular,the study mapped six QTL to chromosomes 1, 2, 5, 8, and 10, and the three most remarkable QTL were validated by comparison with previous studies that used conventional QTL mapping. Win et al. [17]used NGS and conventional methods to detect five QTL(dm2.2, dm4.1, dm5.1, dm5.2, and dm6.1) that conferred downy mildew resistance in cucumber.
Plants have evolved various physical, chemical, and physiological strategies to cope with insect pests and reduce the damage they inflict. Three physiological resistance mechanisms—antixenosis, antibiosis, and tolerance—were first proposed by Painter et al. [22] and further developed by Kennedy et al.[23].Antibiosis reduces insect survival,growth rate, or reproduction following the ingestion of host tissue;antixenosis helps repel or disturb insects, thereby reducing their colonization or oviposition; and tolerance refers to the capacity to produce a high-quality crop or high yield despite insect infestation [24]. BPH-resistance genes usually confer resistance through one or a combination of the three defense types.Identifying the mode of gene action might help improve BPH and WBPH management practices.Study of the BPH-and WBPH-resistant cultivars should yield information about resistance mechanisms of WBPH-resistance genes that can be incorporated into breeding programs for producing insectresistant rice.
RBPH327, an introgression line derived from the wild rice species O. rufipogon with, carrying the AA genome, shows resistance to BPH [25]. This study aimed to (1) perform rough mapping independently for the BPH and WBPH resistance genes using conventional methods; (2) verify the location of the resistance gene using NGS-BSA; (3) fine-map the resistance gene;and(4)characterize and apply the resistance gene using near-isogenic lines (NILs) in indica and japonica genetic backgrounds.
2. Materials and methods
2.1. Development of mapping populations and NILs
RBPH327 was crossed with the susceptible line 9311 to obtain F1progeny,which were self-pollinated twice to develop an F2:3population of 132 lines. F1was also repeatedly backcrossed with 9311 to generate BC1F1, BC2F1, BC3F1, and BC4F1generations. The F2:3lines were used for rough mapping and BC1F2and BC2F2populations for fine mapping of the resistance gene.Three BC4F1plants with agronomic traits comparable to those of 9311 were selected and self-pollinated to generate BC4F2:3lines.Homozygous BC4F2:3lines carrying the resistance gene were designated as NIL-9311. During this process of backcrossing,marker RM16720 tightly linked to the resistance gene was used to select resistant progeny for repeated backcrossing. RBPH327 was also crossed with the japonica variety BR54 to develop the same backcrossed generations described above for 9311. The NILs developed in the BR54 genetic background were designated as NIL-BR54. Both NIL-9311 and NIL-BR54 were used to characterize the resistance gene and agronomic traits. Rice lines RBPH327 and NILs were developed by our group, and BR54 and 9311 were maintained as germplasm at our laboratory.All field trials were conducted at the Rice Research Institute of Guangxi University and complied with Chinese law.
2.2. Insects and resistance evaluation
BPH and WBPH populations were collected from rice fields in Nanning(22.49′N,108.19′E)in 2011 and 2013,respectively.The insects were reared on the highly susceptible rice variety TN1 in aluminum cages (50 × 50 × 100 cm in length, width, and height, respectively) with a fine light-transmitting nylon mesh. BPH and WBPH were fed on TN1 seedlings more than 3 weeks old and about 10 days old, respectively. The insects were maintained in a greenhouse at (26 ± 2) °C and under a day/night photoperiod of 14/10 h. For the tests, second- to third-instar nymphs and emerging adult insects were used.
Insect resistance was evaluated using the standard seedbox screening test following Qiu et al. [26]. For the BPH resistance test,the seedbox(58×38×9 cm)contained a 5 cm layer of field soil and was divided lengthwise into two parts with a total of 28 rows. Germinating seeds were sown into each row to obtain at least 16 seedlings. The tested lines,including 9311 and RBPH327, were sown randomly with 2–3 repeats. About 11 days after sowing, 3-leaf seedlings were infested with second- to third-instar nymphs at a density of 8–10 insects per plant. A seedbox of the same size was used for the WBPH resistance test, with 20 seedlings of each line arranged in individual rows,but with the seedbox not divided.Each line, including 9311 and RBPH327, was sown in single rows, with a total of 16 lines per seedbox. About 7 days after sowing, 2-leaf seedlings were treated with second- to thirdinstar nymphs at a density of 8–10 insects per plant. After infestation, each seedbox was covered with a fine lighttransmitting nylon hood (44 × 34 × 44 cm) and maintained in a greenhouse under the same conditions described above for insect rearing. The degree of damage on each seedling was recorded when>90%of the 9311 plants had died(about 9 days for BPH and 15 days for WBPH). The grading criteria followed the standard evaluation system proposed by the International Rice Research Institute and the description by Qiu et al.[26]:0,no observed damage; 1, very slight damage; 3, first leaf yellowed or first and second leaves partially yellowed;5, first and second leaves yellowed, or pronounced yellowing and stunting;7,mostly wilted,but the seedling is still alive;and 9,completely wilted or dead seedling.The same resistance tests were used to evaluate the resistance level of the NILs. Both BPH and WBPH resistance tests were repeated 2–3 times, and the resistance score of each line was the mean of all tested individuals.
The seedling survival rate of the NILs was evaluated in the same manner with minor differences. The seedlings infested with BPH and WBPH were 14 and 10 days old,respectively.The surviving plants of each line were counted when >90% of the 9311 plants had died(about 15 and 20 days for BPH and WBPH,respectively). The test was repeated three times.
2.3. Conventional gene mapping
Conventional BSA was used to screen for markers linked to the resistance gene [18]. Because of the contrasting insect resistance phenotypes, two DNA bulks containing 10 extremely resistant or susceptible individuals were screened using more than 1200 markers spanning 12 rice chromosomes to identify those for which the two bulks were polymorphic.Additional polymorphic markers around the target locus were detected and then used to genotype the F2population.Finally,JoinMap 3.0 [27] was used to construct a local genetic linkage map and MapQTL 5[28]was used to identify candidate loci or genes associated with insect resistance.
2.4. Sequencing library construction and high-throughput sequencing
High-throughput sequencing was conducted using 2 DNA bulks containing 40 extremely resistant or susceptible individuals derived from BC1F2,and the phenotypes were verified by their corresponding F2lines. Together with RBPH327 and 9311, the leaves of all selected individuals were collected at the 4-leaf seedling stage and immediately frozen in liquid nitrogen for genomic DNA isolation. Equal amounts of DNA from each plant within each group were mixed to obtain the resistant and susceptible bulks at a final concentration of 40 ng μL−1. RBPH327, 9311, the resistant bulk, and the susceptible bulk were designated as R01, R02, R03, and R04,respectively.
The mixed DNA samples were sheared into approximately 350-bp fragments with a Covaris S2/E210 (Covaris Inc.,Woburn, MA, USA) ultrasonicator, followed by end-repair and the addition of single A nucleotide overhangs using Klenow fragment (3′ →5′ exo; New England Biolabs, Hitchin,Herts, UK) and dATP at 37 °C. Barcodes and Illumina sequencing adapters were ligated to the A-tailed fragments with T4 DNA ligase.The sequencing depth of the two parental lines was about 10×,whereas that of each bulk was about 50×.PCR was performed using diluted sheared-ligated DNA samples, dNTP, Q5 High-Fidelity DNA Polymerase, and primers. The PCR products were purified using Agencourt AMPure XP beads (Beckman Coulter, Herts, UK). Fragments ranging from 300 to 500 bp(with barcodes and adaptors)were excised and purified using a gel extraction kit(Qiagen,Hilden,Germany). The gel-purified products were diluted for 150 bp paired-end sequencing on an Illumina HiSeq instrument(Illumina, Inc., San Diego, CA, USA) using the manufacturer's standard protocol.
2.5. Single nucleotide polymorphism and insertion-deletion collection
Low-quality reads(with quality score<20e)were filtered,and the raw reads were sorted by barcode sequence.The barcodes were trimmed from the high-quality reads, and clean reads for each sample were mapped onto the Nipponbare genome(http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/) using the Burrows–Wheeler Aligner[29].SAMtools[30]was used for marking duplicates, and genome analysis tool kit (GATK) [31]was used for local realignment with base recalibration. The single nucleotide polymorphism (SNP) set was generated by combining GATK and SAMtools SNP calling with default parameters. SNPs between the bulks were assigned as polymorphic for association analysis. The same procedure was followed for insertion-deletion(InDel)identification.
2.6. Association analysis
The parameters SNP-index and Δ(SNP-index) were applied to identify significant differences in genotype frequency for the insect resistance region [32]. Here P and M indicate the susceptible and resistant parents and aa and ab the resistant and susceptible bulks, respectively. The Δ(SNP-index) was calculated as follows:SNP-index(aa) = Maa/(Paa + Maa),SNPindex (ab) = Mab/(Pab + Mab), and Δ(SNP-index) = SNP-index(aa)–SNP-index(ab).Maa is the depth of population aa derived from M,and Paa is the depth of population aa derived from P.Mab indicates the depth of population ab derived from M,and Pab indicates the depth of population ab derived from P.Because an F2population was used, the upper limit Δ(SNPindex) value of the trait-associated tags was expected to be 0.5. The same parameters were calculated for InDels. To identify an association threshold, LOESS regression fitting was performed following Abe et al.[32].The regions above the threshold were considered trait-related candidate regions.
2.7. Host choice test and survival rate measurement
To characterize the insect resistance gene associated with NIL-9311, a host-choice test was performed following Qiu et al. [26] and insect survival rates were measured. In the BPH host-selection test, several pre-germinated seeds of NIL-9311 and 9311 were sown into one square with 7-cm length in a bucket (29 cm diameter, 20 cm height) containing a 15-cm layer of rice field soil. At 3 weeks after sowing, only one NIL-9311 and 9311 seedling each were retained, and second- to third-instar nymphs were released in a mean quantity of 15 per plant. The number of BPH insects attached to each line was recorded every day after infestation. Six replicates were conducted each time,and the test was repeated three times.
The insect survival rate was determined by infesting one 3-week-old seedling growing in a plastic cup (10 cm diameter,16 cm height) with field soil with 10 second - to third-instar nymphs. The shoot base of the seedling was covered with a smaller plastic cup with a hole in the bottom. The surviving insects were counted 7 days after release. Six plants were tested for both NIL-9311 and 9311, and the experiment was repeated three times.
2.8. Agronomic trait detection
The resistant NILs were evaluated by agronomic traits. Each line of NIL-9311,NIL-BR54,9311,and BR54 was arranged in 10 rows, with each row containing 10 plants at a density of 20 × 30 cm (20 cm between plants and 30 cm between rows).Three replicates of each line were randomly distributed in a rice field under conventional field management. When the rice matured,20 plants(excluding outer plants)were selected from each replicate and used for agronomic trait measurement. Plant height and tiller number were directly measured in the field. All the spikes of the corresponding plants were collected independently to record seed setting rate and 1000-kernel weight when the grain was dry.
2.9. Statistical analysis
Phenotypic means were compared using one-way analysis of variance with the least significant difference test at a 5%or 1%significance level.Survival rates(%)were arcsine-transformed before the analysis.
3. Results
3.1. Genetic analysis of BPH and WBPH resistance
The RBPH327 introgression line showed high BPH resistance at the tillering stage(Fig.1)as well as high WBPH resistance at the seedling stage. The mean resistance scores of RBPH327 and 9311 were 3.1 and 8.7, respectively, after treatment with BPH insects (Fig. 2a). BPH resistance scores measured for 132 F3lines ranged continuously from 2.4 to 9.0,with an apparent peak at 6.0–6.9 in the distribution curve(Fig.2a).
RBPH327 and 9311 were highly resistant and susceptible,respectively, to WBPH insects. In the seedling bulk test, the mean resistance scores were 3.3 and 8.5 for RBPH327 and 9311,respectively (Fig. 2b). The WBPH resistance scores of 112 F3lines showed a continuous distribution, like the BPH resistance scores. Moreover, the BPH and WBPH resistance scores were significantly correlated(r =0.49,P =0.02).
3.2. Rough mapping
A total of 1136 simple-sequence repeat (SSR) and InDel markers on 12 rice chromosomes were screened using the two DNA bulks and parents.Fragments polymorphic between the resistant and susceptible bulks for markers RM16563,RM16720, and RM16763 on chromosome 4 suggested that the resistance gene was located in this region.Additional markers in the target region that were polymorphic between the parental lines were used to genotype 132 F2plants. In a local linkage map spanning 47 cM of chromosome 4, the order of the markers was identical to that in the Nipponbare genome(Fig. 3a; http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/). In the 10.8 cM region between RM16563 and RM16763,interval QTL mapping revealed one locus for BPH resistance with a large LOD score of 26.7, and it was close to RM16720(Fig.3a).Given that no BPH resistance genes or QTL have been reported in this chromosomal region, this gene was designated as Bph38. Bph38 explained 62.8% of the phenotypic variance for BPH resistance in the F2population.

Fig.1– BPH-infested RBPH327 and 9311 at the seedling and tillering stages.(a) Before infestation,(b) after infestation,and(c)tillering-stage plants after infestation.
Although BPH and WBPH resistance were correlated in the mapping population, some differences were apparent in the resistance score distributions (Fig. 2). Using two DNA bulks corresponding to the extremely WBPH-resistant and-susceptible phenotypes, we screened 1182 SSR and InDel markers.The same markers identified as polymorphic in BPH resistance gene mapping—RM16563, RM16720, and RM16763—were found to be polymorphic in the WBPH analysis,indicating that the WBPH resistance gene was located in the same region as Bph38. One QTL for WBPH resistance was identified in the region between M16563 and RM16763 with a LOD score of 5.8,explaining 30.1%of the phenotypic variance(Fig. 3b). The target region was identical to that of Bph38.These independent mapping analyses indicated that the same chromosomal region confers resistance to both BPH and WBPH.
3.3. NGS-BSA
The numbers of clean reads for RBPH327(R01),9311(R02),the resistant bulk (R03), and the susceptible bulk (R04) were 48,344,523;37,938,313;72,709,208;and 73,320,363,respectively,and the corresponding Q30 percentages were 93.07, 93.03,92.74, and 93.12, respectively (Table S1). In total, 853,096 and 156,649 SNPs and 211,994 and 38,375 InDels were detected for the two parents and bulks, respectively (Fig. 4). Correlation analysis of the detected SNPs and InDels on the 12 rice chromosomes and phenotypes revealed one clear peak of abundance (indicating numerous sequence differences) on chromosome 4, and no clear peaks were detected in other chromosomes,suggesting the presence of the resistance gene on the target chromosome.For the analyzed SNPs and InDels,the peak region was 3.16–16 Mb and 3.72–15.83 Mb on chromosome 4, respectively (Fig. 5; Fig. S1). These sequence differences suggested that the same chromosomal region conferred insect resistance. Moreover, these regions encompassed the mapping regions identified using the conventional method. Thus, conventional gene mapping and NGS-BSA identified the same chromosomal region as the location of an insect resistance gene.
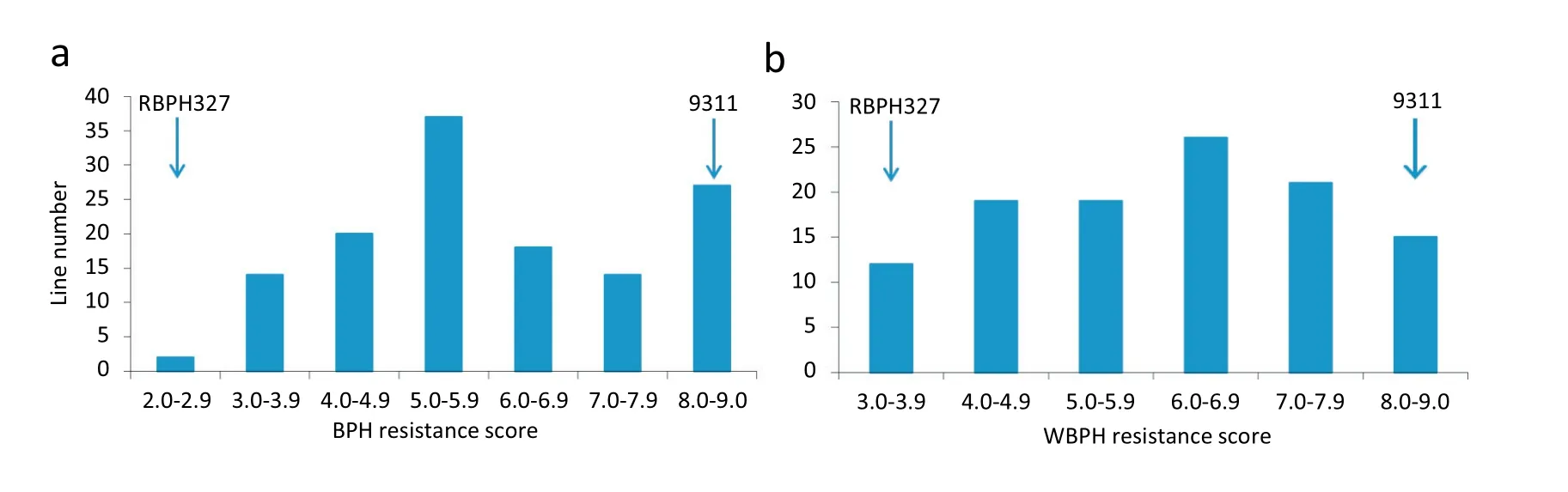
Fig.2– Frequency distribution of BPH(a)and WBPH(b) resistance scores of an F2 population derived from the cross 9311/RBPH327.Three-leaf seedlings were treated with 8 BPH insects per plant for 9–10 days.The mean resistance scores of RBPH327 and 9311 were 3.1 and 8.7, respectively.Lower scores indicate higher resistance.
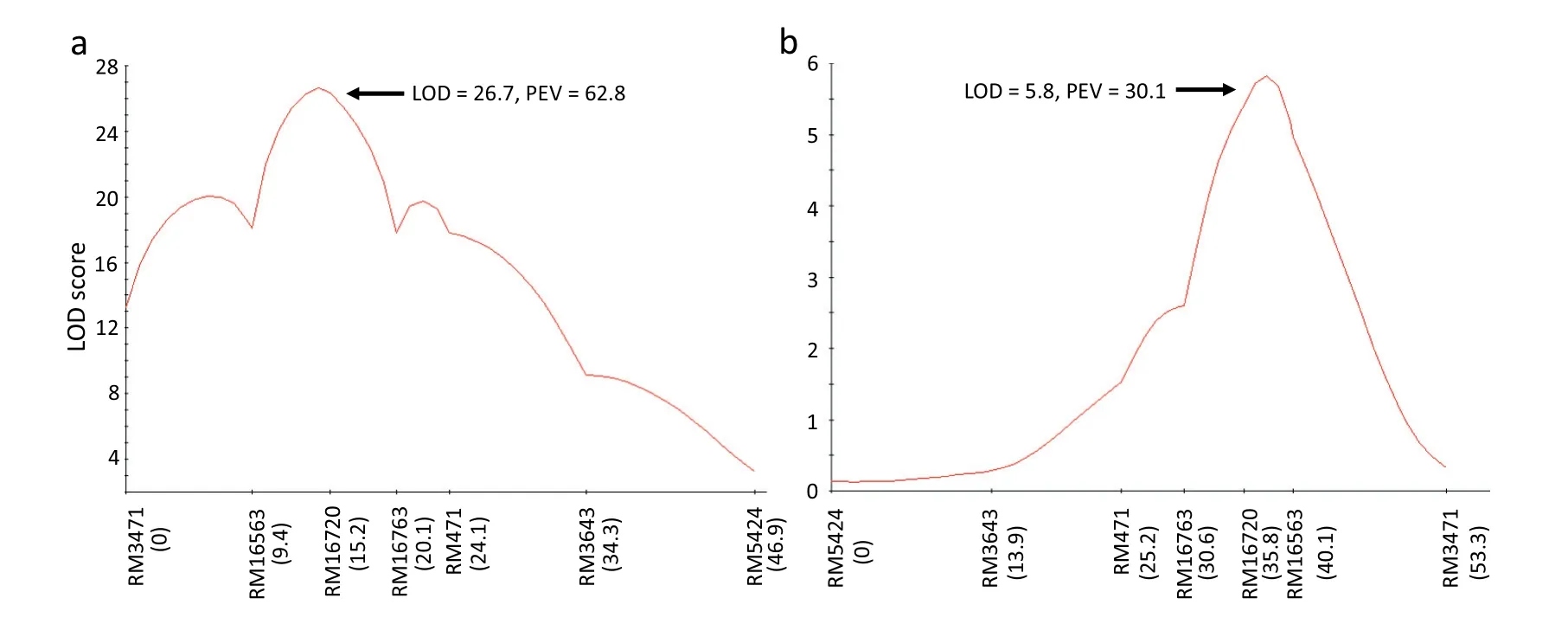
Fig. 3 – Location of the BPH (a) and WBPH (b) resistance gene on the linkage map of rice chromosome 4. Markers are indicated along the X axis with distances (in cM), and LOD scores on the Y axis. PEV, phenotypic variance explained by the locus.
3.4. Fine mapping of the resistance gene
For fine mapping of the resistance gene, we conducted three rounds of recombinant screening and polymorphic marker analysis. Because rough mapping suggested that the same region confers BPH and WBPH resistance, we used the same markers to screen the recombinants; however, we evaluated insect resistance independently. First, we identified 42 recombinants between markers RM16763 and RM16563 among 520 BC1F2seedlings, and self-pollinated them to develop BC1F2:3lines. We then tested the recombinants for BPH and WBPH resistance. In addition to RM16720, we identified polymorphic markers YM91 and YM92 and used them to genotype the recombinants (Table S2; Fig. 6a). The BPH resistance scores of recombinants 183 and 193 were 4.9 and 5.1,respectively,in the seedling test,indicating moderate BPH resistance. Recombinants 13 and 479 were highly resistant to BPH and had low resistance scores of 3.1 and 2.8,respectively, whereas recombinants 85 and 112 were highly susceptible to BPH and had high resistance scores (Fig. 6b).The recombinants showed the same phenotypes when treated with WBPH insects. Accordingly, the genotypic and phenotypic analyses of recombinants 13, 85, and 183 delimited the right margin, and those of recombinants 479,112, and 193 delimited the left margin of Bph38 (Fig. 6b).Together, they narrowed the map interval to the region between markers YM92 and RM16720, which corresponds to a physical distance of 1.2 Mb in the Nipponbare genome.
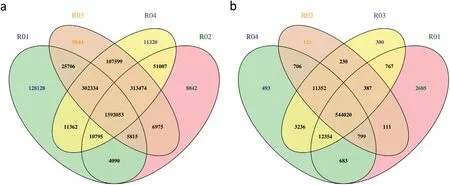
Fig.4– Venn diagram of the collected SNPs(a)and InDels(b)in four sequencing samples. R01,R02,R03,and R04 denote RBPH327,9311,the resistant bulk,and the susceptible bulk,respectively.
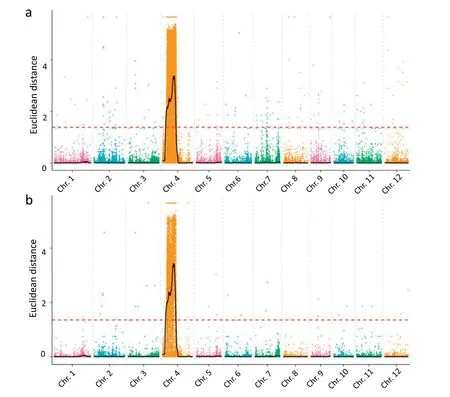
Fig. 5 – Euclidean distance-based association values of SNPs (a) and InDels (b) on each chromosome.
More BC2F2individuals were genotyped for markers YM92 and RM16720, and 78 recombinants were obtained from 6130 individuals. The recombinants were self-pollinated and used for the insect resistance tests. Four additional polymorphic markers—YM68-2, RM16686, RM1236, and RM16717—were identified in the target region and used for genotyping (Table S2; Fig. 6c). Recombinants F580 and C175 were moderately resistant to BPH and had resistance scores of 5.4 and 4.4,respectively.The highly resistant recombinants A189 and B8-3 had low resistance scores of 2.8 and 2.5,respectively.Similar results were obtained when these recombinants were tested for WBPH resistance. Recombinants A189, E439-2, and F580 delimited the right margin and recombinants B8-3,C266,and C175 delimited the left margin of Bph38. These analyses narrowed the region of the resistance gene to the segment flanked by markers YM16686 and YM1236(Fig.6d).
Finally,46 additional recombinants were identified among 4200 BC2F2seedlings,and three more polymorphic markers—YM112, YM190, and YM68-1—were obtained between RM16686 and RM1236 (Fig. 6e). The recombinants were analyzed as described above for the first two rounds of gene mapping.In this manner,the resistance gene was mapped to a region between markers YM112 and YM190, corresponding to a physical distance of 79 kb in the Nipponbare genome.Several important recombinants are listed in Fig. 6f. For instance, the highly resistant recombinants 168-6, B282-2,62-9,and 123-5 had resistance scores of 2.2,2.9,2.8,and 3.2 for BPH and 3.0, 3.3, 3.4, and 3.6 for WBPH, respectively. The highly susceptible recombinant F9-3 had resistance scores of 8.6 and 8.2 for BPH and WBPH, respectively. Thus, the recombinants F9-3, 168-6, and B282-2 determined the right margin of the resistance gene and recombinants 62-9, 123-5,and 1866 determined the left margin (Fig. 6f). These findings provide further evidence that the same region controls BPH and WBPH resistance, and the resistance gene Bph38 was localized to the region between markers YM112 and YM190(Fig.6f).
3.5. Candidate gene analysis
The 12.06-Mb(3.72–15.83 Mb)region identified using NGS-BSA harbored 1883 candidate genes in the Nipponbare genome.The 79-kb region between markers YM112 and YM190 identified by fine mapping contained 12 candidate genes,including the MADS-box family genes (Os04g25870,Os04g25920, and Os04g25930), retrotransposon protein genes(Os04g25880, Os04g25890, Os04g25940, and Os04g25950), an NBS-LRR gene (Os04g25900), a hypothetical protein gene(Os04g25910), an expressed protein gene (Os04g25960), and cytokinin glucosyltransferase genes (Os04g25970 and Os04g25980). Of these genes, Os04g25870, Os04g25900,Os04g25920, Os04g25930, Os04g25970, and Os04g25980 were identified using NGS-BSA (Table 1). In particular, Os04g25870 and Os04g25900 were found to contain 87 and 122 total SNPs and InDels, respectively, between the resistant (R) and susceptible (S) bulks. Among the SNPs and InDels in the candidate gene Os04g25900, 16 were non-synonymous coding-region variants, and 6 were in the 3′ UTR. The next highest numbers of SNPs and InDels were found in Os04g25920 and Os04g25930(43 and 54,respectively).
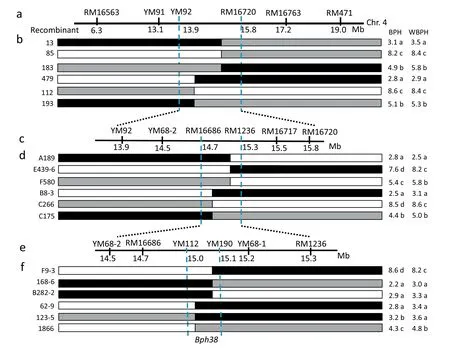
Fig.6–Fine mapping of Bph38.(a,c,e)Local genetic map with the associated markers.(b,d,f)Diagram indicating the genotypes and phenotypes of part of recombinant individuals.Black,white,and gray bars denote the marker genotypes of RBPH327 homozygotes,9311 homozygotes,and heterozygotes,respectively.Numbers below the lines denote physical distances in the Nipponbare genome.The values on the right-hand side are BPH and WBPH resistance scores obtained in the seedling bulk test.The codes listed on the left-hand side are that names of recombinants.Means labeled with the same letter within one column are not significantly different at P= 0.05.
3.6. NIL development and BPH resistance evaluation of NILs
To develop NILs in the 9311 or BR54 genetic backgrounds, we selected more than 10 BC4F1plants for each background using MAS and insect resistance evaluation. The resistant plants were self-pollinated and their agronomic traits were recorded.Plants whose appearance was the most similar to that of a recurrent parent were selected for further study(Fig.S2).
In seedling bulk tests, RBPH327 and homozygous NIL-9311R were highly resistant to BPH, with mean resistance scores of 3.2 and 3.4, respectively. Heterozygous NIL-9311H was moderately resistant to BPH and had a resistance score of 5.1. Homozygous NIL-9311S and 9311 were highly susceptible to the insects,with high scores of 8.6 and 8.8,respectively(Fig.7a).Similar results were obtained for NIL-BR54.The resistancescores of homozygous NIL-BR54R, homozygous NIL-BR54S,and heterozygous NIL-BR54H were 4.4, 8.7, and 6.9, respectively. Interestingly, NIL-9311 with Bph38 showed stronger resistance to BPH in the seedling bulk test than NIL-BR54 with Bph38 (Fig. 7b). This difference was detectable in both the resistance test and resistance scores, with significant differences between NIL-9311R and NIL-BR54R (F = 9.1, P =0.009) as well as between NIL-9311H and NIL-BR54H (F = 4.8,P = 0.04).

Table 1–Distribution of single-nucleotide polymorphisms and insertion–deletions in the candidate genes detected using next-generation sequencing–bulked-segregant analysis(NGS-BSA)and conventional gene mapping.
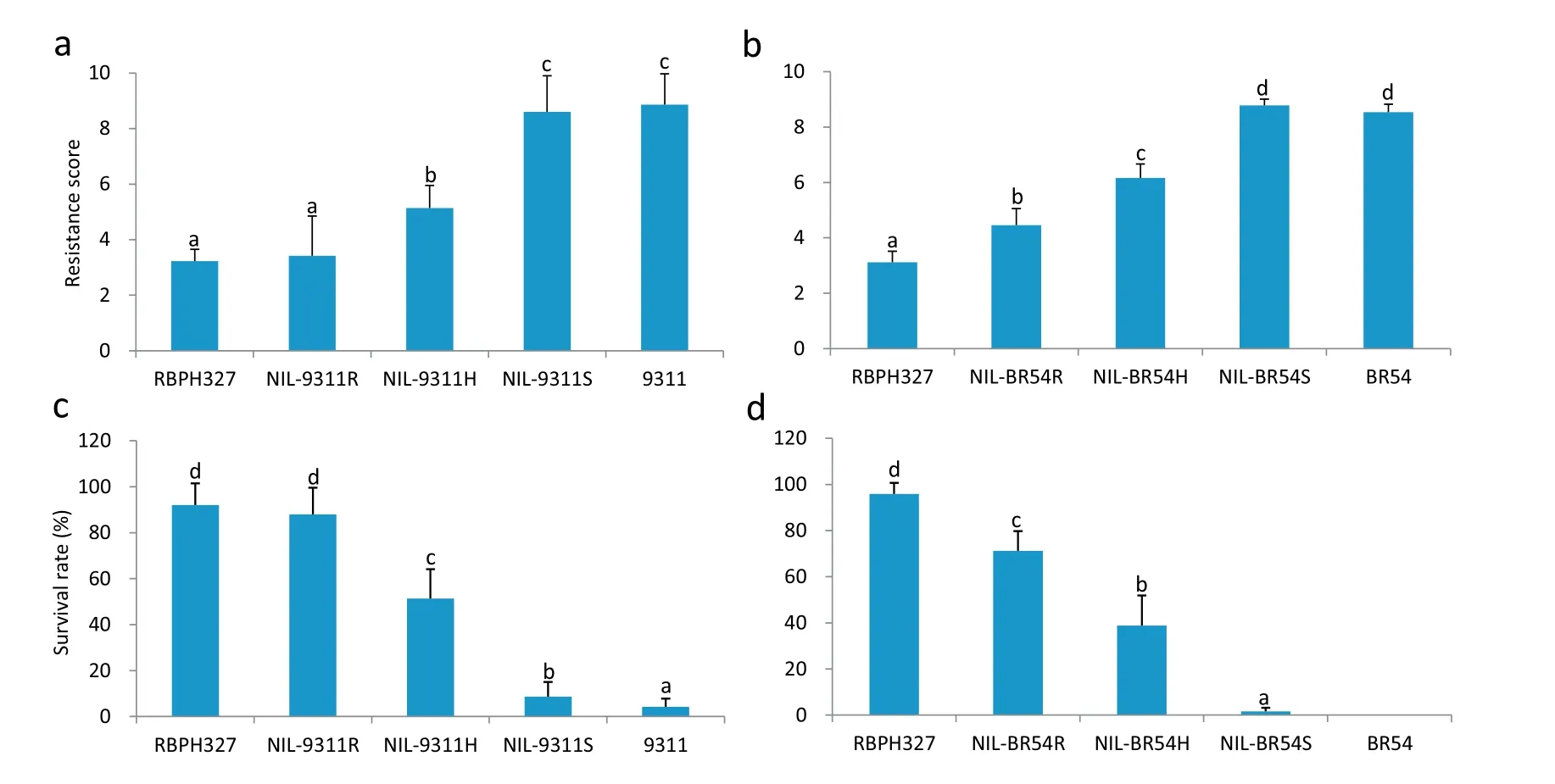
Fig.7– BPH resistance phenotype of NILs.(a,b) BPH resistance score of NILs in the 9311 or BR54 background.(c,d) Seedling survival rate(%)of NILs in the 9311 or BR54 background.Each bar represents the mean of 6–8 replicates,and error bars indicate the SEs.NIL-9311R,NIL9311S,and NIL-9311H denote BC4F2 plants homozygous for RBPH327,homozygous for 9311,and heterozygous at the Bph38 region in the 9311 genetic background,respectively.NIL-BR54R,NIL-BR54S,and NIL-BR54H denote BC4F2 plants homozygous for RBPH327,homozygous for BR54,and heterozygous at the Bph38 region in the BR54 genetic background, respectively.RBPH327,9311,and BR54 are the parental lines.Means labeled with the same letter are not significantly different at P =0.05.
Lines carrying the resistance gene showed increased seedling survival. The survival rates of RBPH327, NIL-9311R,and NIL-9311H were 92,87.9,and 51.4%,respectively,whereas those of NIL-9311S and 9311 were 8.3 and 4.3%, respectively(Fig. 7c). The same trend was observed when the NIL-BR54 lines were treated with BPH, but they showed lower BPH resistance than NIL-9311. For example, the survival rate of NIL-9311R was 87.9%, whereas that of NIL-BR54R was 71.2%(F = 6.75, P = 0.02). The survival rates of NIL-9311H and NILBR54H were 51.4 and 38.8%,respectively(F=5.34,P=0.03;Fig.7d). Thus, the NILs carrying the resistance gene showed strong BPH resistance and the resistant lines in the 9311 genetic background showed greater BPH resistance than those in the BR54 genetic background.
3.7. WBPH resistance evaluation of NILs
NILs carrying the resistance gene in either the 9311 or the BR54 genetic background showed resistance to WBPH, as indicated by the low resistance test scores and high survival rates. For instance, the resistance scores of NIL-9311R and NIL-BR54R were 3.3 and 4.1, respectively, and the corresponding survival rates were 88.2% and 77.9%, respectively. However, the lines in the 9311 genetic background showed higher resistance to WBPH than those in the BR54 genetic background. The resistance scores of NIL-9311H and NIL-BR54H were 4.8 and 5.8, respectively (F = 4.52, P = 0.043),and their survival rates were 54.9% and 45.2%, respectively(F = 5.46, P = 0.33). The same phenomenon was observed for NIL-9311R and NIL-BR54R(Fig.8).
3.8. Characterization of BPH and WBPH resistance in NILs
In host-choice tests when NIL-9311(homozygous for RBPH327 at the Bph38 region) and 9311 were planted together, significantly fewer BPH insects were observed on resistant seedlings 3 h after infestation (14 and 18 on NIL-9311 and 9311,respectively) than on susceptible seedlings, and this difference was noted throughout the test period(Fig.9a).The same phenomenon was observed for NIL-9311 and 9311 after treatment with WBPH, but insect numbers were significantly different only at 24 h after treatment (13 and 17 in NIL-9311 and 9311,respectively)and at later time points(Fig.9b).Thus,the resistance gene exerted an antixenotic effect on BPH and WBPH.
Seven days after infestation,the numbers of surviving BPH and WBPH insects were significantly lower on resistant than on susceptible plants (Fig. 9c). In particular, the BPH survival rates were 26% and 73% (F = 39.2, P < 0.001) and the WBPH survival rates were 32% and 69% (F = 18.6, P = 0.001) on NIL-9311 and 9311, respectively. These results suggest that the NIL-9311 lines with the resistance gene exerted an antibiotic effect on BPH and WBPH.
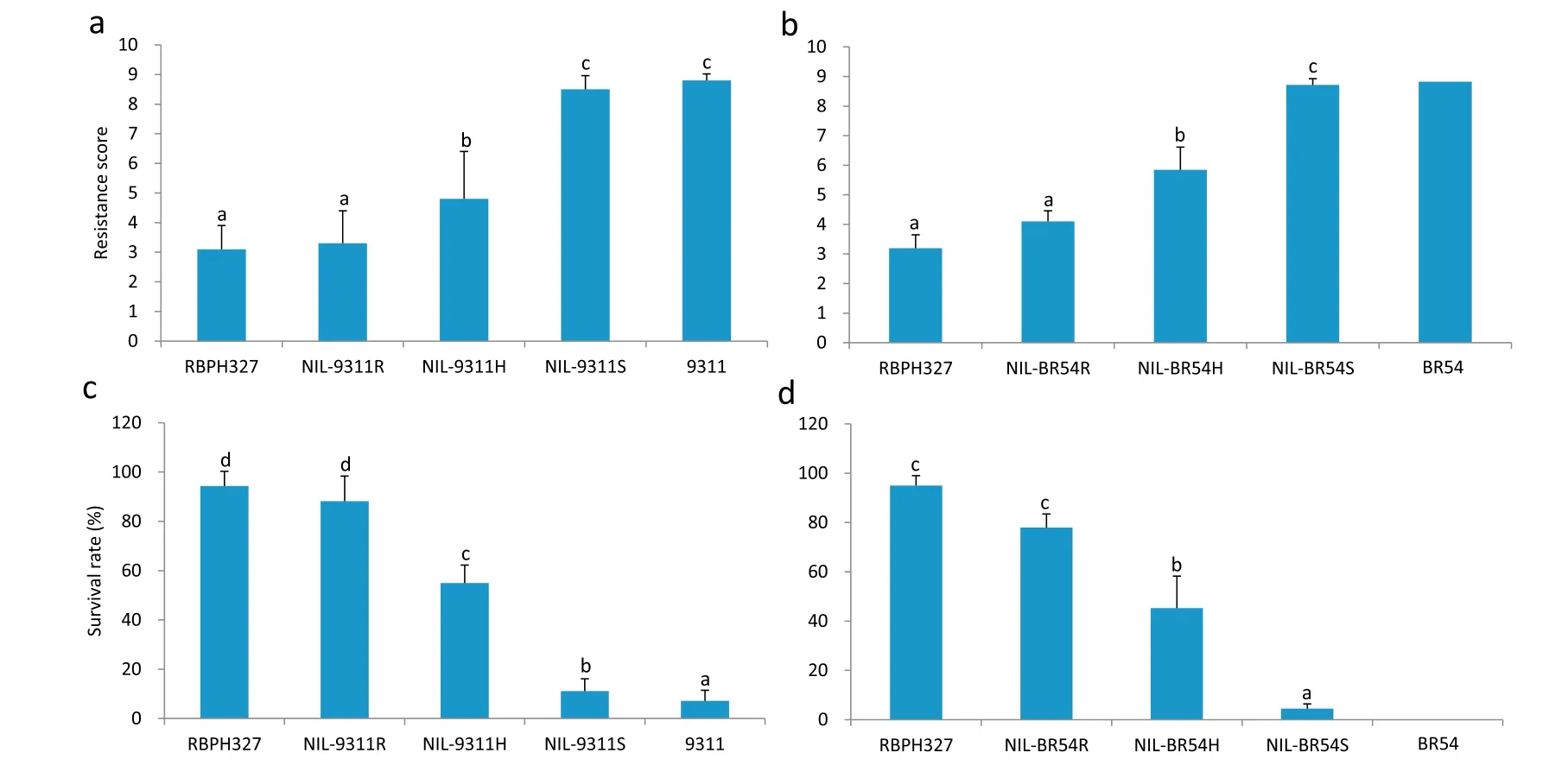
Fig.8–WBPH resistance phenotype of NILs.(a,b)WBPH resistance score of NILs in the 9311 or BR54 background.(c,d)Seedling survival rate(%)of NILs in the 9311 or BR54 background.Each bar represents the mean of 6 replicates,and error bars indicate the SEs.NIL-9311R,NIL-9311S, and NIL-9311H denote BC4F2 plants homozygous for RBPH327,homozygous for 9311,and heterozygous at the Bph38 region in the 9311 genetic background,respectively.NIL-BR54R,NIL-BR54S,and NIL-BR54H denote BC4F2 plants homozygous for RBPH327,homozygous for BR54,and heterozygous at the Bph38 region in the BR54 genetic background.RBPH327,9311,and BR54 are the parental lines.Means labeled with the same letter are not significantly different at P= 0.05.
3.9. Agronomic trait evaluation of the NILs
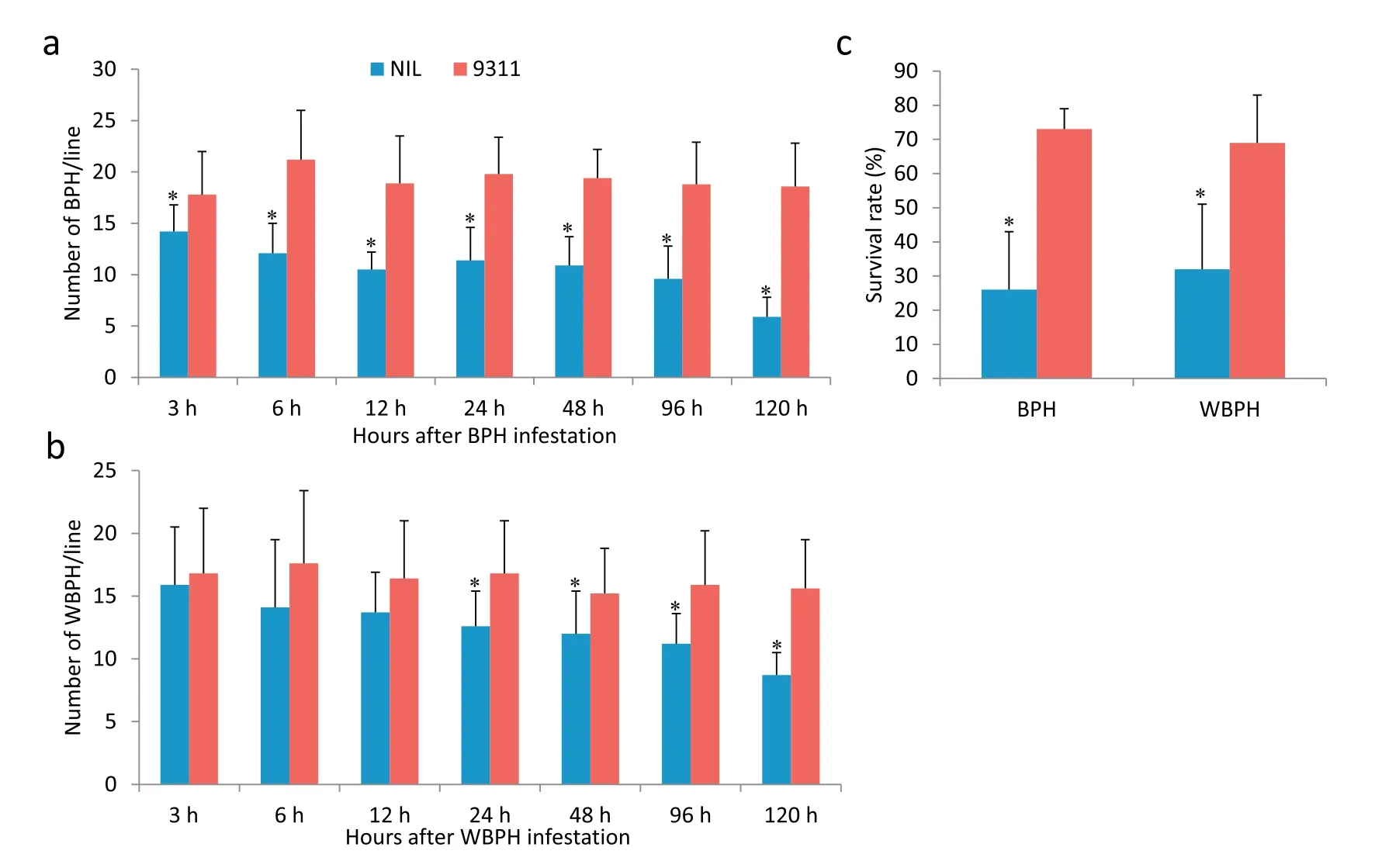
Fig.9– Characterization of BPH and WBPH resistance in NIL-9311. (a)BPH host-choice test,(b) WBPH host-choice test,and(c)BPH and WBPH survival rates.Error bars represent SEs.Means labeled with asterisks are significantly different at P <0.05.
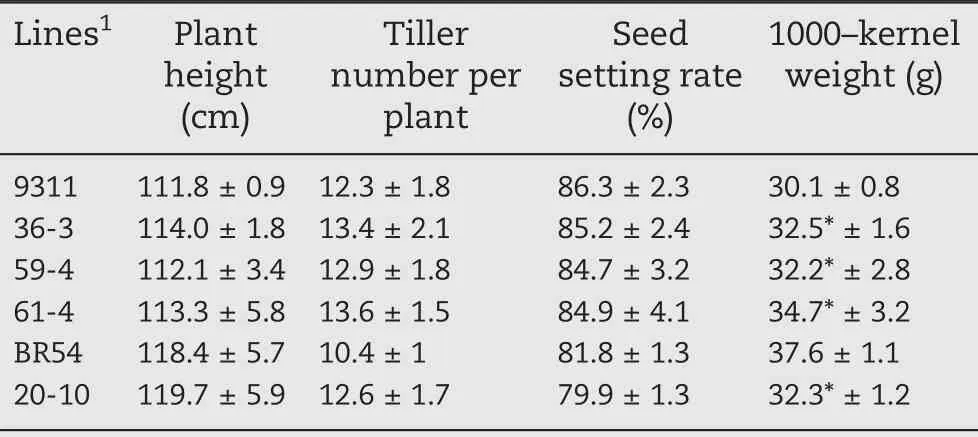
Table 2–Agronomic traits of near isogenic lines and recurrent parents.
Agronomic and yield traits of the NILs are described in Table 2.The plant heights of the NIL-9311 lines 36-3, 59-4, and 61-4 were 114, 112.1, and 113.3 cm, respectively, values slightly higher than that of 9311(111.8 cm),but the difference was not significant (P > 0.05). The height of NIL-BR54 (119.7 cm) was similar to that of BR54 (118.4 cm, P > 0.05). No significant differences in tiller number were noted between 9311 and NIL-9311, but NIL-BR54 had more tillers (12.6) than BR54(10.4;F=8.2,P=0.01).The setting rates of both NIL-9311 and NIL-BR54 were lower than those of their recurrent parents.The setting rate for NIL-BR54 decreased by 79.9%,whereas it decreased by 85.2%, 84.7%, and 84.9% for the three NIL-9311 lines of 36-3, 59-4, and 61-4, respectively. The 1000-kernel weight of 9311 (30.1 g) was significantly lower than that of the NIL-9311 lines, with weights of 32.5, 32.2, and 34.7 g for lines 36-3 (F = 34, P < 0.001), 59-4 (F = 31, P < 0.001), and 61-4(F = 151, P < 0.0001), respectively. However, the 1000-kernel weight of NIL-BR54 (32.3 g)was significantly lower than that of BR54 (37.6 g, F = 164, P < 0.0001). Notably, the developed NIL-9311 lines had greater 1000-kernel weight and more tillers than 9311.
4.Discussion
Increasing evidence suggests that host plant resistance is an effective,economical,and environmentally friendly approach for managing crop pests[8,33].Rice planthoppers,mainly BPH and WBPH,are the most damaging rice insect pests and cause severe yield losses by direct feeding and viral transmission of diseases [34,35]. Thus, the identification, mapping, and characterization of planthopper resistance genes will be valuable for establishing prolonged resistance against diverse predominant biotypes and could contribute to sustainable rice outputs.The mapping effort identified the region harbored by markers YM112 and YM190, and one major resistance locus,Bph38, conferred resistance to both BPH and WBPH. BPHresistance genes are often clustered on rice chromosomes[11,36].For example,12 BPH resistance genes(Bph6[26],Bph27[10],bph18(t)[37],Bph34[38],Bph3[39],Bph12[20,40],Bph15[13],Bph17 [41], Bph20(t) [42], Bph30 [36], and Bph33 [43]) are found on chromosome 4. Based on the marker information, the present analysis identified Bph38 as a new member of this cluster on chromosome 4, by using high-resolution mapping(Fig.6),indicating that it confers resistance to BPH and WBPH.The locus was about 4.1 Mb downstream of Bph27 [10] and 2.2 Mb upstream of Bph20(t) in the Nipponbare genome (Fig.S3)[42].
Interestingly, two independent gene mapping tests suggested that Bph38 confers resistance to both BPH and WBPH.An identical phenomenon was observed for Wbph7 and Wbph8, which were found to be in the same chromosomal regions of Bph14 and Bph15, respectively [20]. Furthermore,transgenic lines carrying Bph6 or Bph9 were resistance to BPH and WBPH[15,16].The most plausible explanation is that the two insects use the same infestation mechanism, such as physiological or molecular resistance mechanism,so that the resistance gene acts against both.
Numerous studies have attempted to identify resistance genes in several crops such as rice, maize, and wheat. Of the available methods, conventional positional cloning is often undertaken and has achieved considerable success. For example, map-based cloning of rice BPH resistance genes has revealed at least 8 major genes[15].However,this method is low-throughput and time consuming, with the gene discovery process involving gene mapping population production, genome-wide survey, rough mapping, and fine mapping. For quantitative traits, these steps are coupled with the subsequent validation of the most relevant candidate regions using NILs [44,45]. In the present study, we used conventional gene mapping to identify Bph38: we developed the F2mapping population for rough mapping (Figs. 2, 3);BC1F2and BC2F2populations for fine mapping(Fig.6);and NILs to verify the candidate region(Figs.4,5).As a rough estimate,at least 3–4 years are required to complete the process,assuming two successful crosses are made each year, and insect resistance evaluation is conducted efficiently. If any steps fail, more time is needed. The R and S bulks, each containing 10 extremely resistant or susceptible samples,were still very small and required accurate phenotype. Thus,despite the utility and reliability of the conventional approach, new methods are required to increase the speed and effectiveness of gene mapping and cloning in plants.
NGS technologies have accelerated the genetic analysis of traits of interest. They allow whole-genome re-sequencing and facilitate the fine mapping and identification of causal polymorphisms. Bulked-segregant techniques require more samples than conventional method and need to be combined with genome capture technology to identify candidate genes [46,47]. In the present study, two DNA bulks corresponding to 40 extremely resistant and susceptible phenotypes as well as 2 parental lines were sequenced, and numerous polymorphic SNPs and InDels were collected and compared.One region with a cluster of polymorphic markers was detected,and it was in agreement with the conventional gene mapping result (Fig. 5; Fig. S1). Although the physical position of the target region(12.06 Mb)identified by NGS-BSA was larger than that identified by rough mapping (10.8 Mb),our NGS-BSA approach required less time than conventional method and verified the mapping result. These detected polymorphic SNPs and InDels can be used to accelerate fine mapping of the gene of interest.
Plants generally show antixenosis, antibiosis, and tolerance effects against insect attack. Previous studies [10,14,26]have shown that BPH resistance genes such as Bph6, Bph14,and Bph27 exert antixenotic and antibiotic effects on insects.The resistance gene Bph7 confers mainly tolerance to BPH[48].However, the resistance mechanisms of WBPH resistance genes have not yet been reported.Based on insect host-choice tests and seedling and insect survival rates, this study indicated that the resistance of the NILs significant antixenotic and antibiotic effects on BPH and WBPH, unlike those in susceptible plants. For instance, in the host-choice test, significantly fewer BPH insects were observed on the resistant seedlings 3 h after infestation, and this result was observed 24 h after WBPH treatment(Fig.9a,b).Moreover,the BPH and WBPH insect survival rates were 26%and 32%on the resistant plants, respectively (Fig. 9c). Thus, the novel resistance gene Bph38 is a suitable candidate for breeding of insect-resistant rice.
Genetic background affects the resistance level or durability of disease or insect resistance genes or QTL [26,49,50].For example, Bph6 confers stronger resistance in the 9311 genetic background than in the Nipponbare background[26].This was also observed when Bph38 was introgressed into 9311 (indica) and BR54 (japonica). According to the seedling resistance evaluation, NIL-9311R showed higher resistance than NIL-BR54R (F = 9.1, P = 0.009), and the same result was obtained when NIL-9311H and NIL-BR54H were compared(F=4.8,P=0.04).The seedling survival rates of NIL-9311R and NIL-BR54R were 87.9% and 71.2%, respectively (F = 6.75, P =0.02),and those of NIL-9311H and NIL-BR54H were 51.4%and 38.8%, respectively (F = 5.34, P = 0.04). A similar trend was observed when these two types of NILs were infested with WBPH. The resistance scores of NIL-9311R and NIL-BR54R were 3.3 and 4.1, respectively, in the seedling bulk test, and their corresponding survival rates were 88.2% and 77.9%,respectively. Thus, the indica and japonica genetic backgrounds strongly affected the resistance level associated with Bph38. Considering the interaction between this planthopper resistance gene and genetic background, one possibility is that japonica varieties, cultivated mainly in temperate climates, are subjected to little or no selection pressure from BPH and WBPH. For this reason, planthopper resistance genes may have a relatively low effect in these cultivars during insect attack.
5. Conclusions
A novel planthopper resistance gene, Bph38, from wild rice was identified and mapped on chromosome 4 using BSA and NGS methods. Further fine mapping narrowed the resistance gene interval to a 79-kb region flanked by markers YM112 and YM190. Mandatory feeding and host-choice tests performed using developed NILs revealed that Bph38 exerted both antibiotic and antixenotic effects on both insects, with stronger effects observed in indica-background lines. The agronomic traits of the NILs were similar to those of the recurrent parents, but NILs showed high BPH and WBPH resistance.
Declaration of competing interest
Authors declare that there are no conflicts of interest.
Acknowledgments
This research was supported by National Key Research and Development Program of China (2016YFD0100600), the National Program on Research and Development of Transgenic Plants (2014ZX0800911B), the National Natural Science Foundation of China (31160276 and 31560423), the Guangxi Innovation-Driven Development Special Funding Project(Guike-AA17204070), and the State Key Laboratory of Hybrid Rice(KF201905).
Author contributions
Yongfu Qiu and Meng Yang conceived and designed the study.Meng Yang and Jiebin Lin performed most of experiments including the primary and fine mapping, insect characterization test, agronomic survey and data analysis. Ling Cheng,Hailian Zhou, and Shu Chen conducted part of primary mapping and agronomic survey. Fang Liu and Rongbai Li supervised the field tests. Meng Yang, Jiebin Lin, and Yongfu Qiu wrote and improved the manuscript.
Appendix A. Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2020.03.011.
- The Crop Journal的其它文章
- Application of moderate nitrogen levels alleviates yield loss and grain quality deterioration caused by post-silking heat stress in fresh waxy maize
- Genetic dissection of husk number and length across multiple environments and fine-mapping of a major-effect QTL for husk number in maize(Zea mays L.)
- Genome-wide linkage mapping of QTL for root hair length in a Chinese common wheat population
- Comparative analysis of the photosynthetic physiology and transcriptome of a high-yielding wheat variety and its parents
- Metabolic profiling of DREB-overexpressing transgenic wheat seeds by liquid chromatography–mass spectrometry
- Haplotype variations in QTL for salt tolerance in Chinese wheat accessions identified by markerbased and pedigree-based kinship analyses

