Comparative analysis of the photosynthetic physiology and transcriptome of a high-yielding wheat variety and its parents
Hujie Liu, Qidi Zhu, Xinxin Pei, Guozhen Xing, Xingqi Ou,*, Hu Li,*
aCollege of Life Science, Henan Agricultural University, Zhengzhou 450002, Henan, China
bHenan Institute of Science and Technology, Xinxiang 453003, Henan, China
cHenan Collaborative Innovation Center of Modern Biological Breeding, Xinxiang 453003, Henan, China
Keywords:Photosynthetic efficiency Chlorophyll and carotenoid Lhcb1 Transcriptome Wheat
A B S T R A C T Photosynthesis is the fundamental basis of plant growth and development, and the improvement of photosynthetic efficiency can therefore promote increased crop yields.In this study,a comparative analysis of photosynthetic physiology and transcriptome was conducted between the high photosynthetic efficient variety BN207 and its parents BN64 and ZM16. The higher chlorophyll fluorescence, chlorophyll and carotenoid contents, and Lhcb1 protein accumulation in BN207 improved photosynthetic efficiency by promoting light energy absorption and conversion. Chloroplasts being distributed more closely to the cell membrane and the higher Rubisco enzyme activity of BN207 enhanced carbon assimilation, resulting in more carbohydrate accumulation in BN207. Transcriptome analysis revealed that there were several key genes mediating the high photosynthetic efficiency of BN207:TraesCS5D02G364100(chlorophyllase), BGI_novel_G006617 (lycopene ɛ-cyclase), TraesCS4A02G034800 and TraesCS4A02G035100 (Zeaxanthin epoxidase), TraesCS6B02G122500 (light-harvesting complex II chlorophyll a/b binding protein 1). These genes improved the photosynthetic efficiency of BN207 mainly by reducing chlorophyll degradation, promoting carotenoid synthesis, and increasing Lhcb1 protein accumulation. These findings provide important background information for the cultivation of wheat varieties with high photosynthetic efficiency.
1.Introduction
Wheat is widely grown throughout the world, and about 40%of the worlds population consumes wheat as a staple.Although wheat yields have been dramatically increased by scientific plant breeding and the development and use of fertilizers, the rise in demand for wheat is expected to continue [1]. To ensure food security, high and stableyielding wheat varieties are urgently needed.
The production of wheat yield is a complex process that is ultimately limited by the efficiency of the energy of sunlight being harvested through photosynthesis,and it also relies on the ability of sucrose transport from source (e.g., mature leaves)to sink(e.g.,grain)[2],with sink capacity also playing a key role[3].Several strategies for raising the yield potential of wheat have been proposed in a recent review by Reynolds et al.[4].They proposed that optimizing the partitioning to grain is important for maximizing yield potential, which can be achieve by improving spike development and grain weight through increasing the availability of assimilates.In addition,the harvest index(i.e.,the proportion of biomass that is grain)can also significantly affect grain yields. The main way for breeders to increase harvest index has traditionally been reducing plant height, while among current cultivars, the wheat harvest index approaches the maximum theoretical value of 0.6. This means that the space for further improvement in the harvest index is limited [5,6]. Grain yield is proportional to the harvest index multiplied by the total amount of biomass;therefore,to further increase grain yields,the total biomass may be increased while holding the harvest index constant [1]. Ultimately, it is clear that these processes could be achieved by improving photosynthetic capacity[7–9].
The close relationship between photosynthesis, yield, and biomass has been discussed by several reviews[7,9,10].Many CO2-enrichment studies have also shown that increasing photosynthesis can increase yield when other factors are unrestricted [11]. Accordingly, it is obvious that increasing photosynthetic capacity and efficiency are important approaches for raising potential wheat yields. Parry et al. [9]discussed several strategies to increase photosynthesis,including the promotion of the regeneration of ribulose-1,5-bisphosphate (RuBP), improving ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) activity, and increasing CO2at the Rubisco catalytic site by introducing CO2-concentrating mechanisms. Rubisco can carboxylate RuBP to 3-phosphoglycerate and also oxidize RuBP to execute photorespiration metabolism [12], while photorespiration can consume 30%of carbohydrates formed in C3photosynthesis[13].Thus,Long et al.[7]proposed ways to improve photosynthesis by reducing photorespiration.In addition,a limiting factor for photosynthesis is light-harvesting efficiency, and expanding the solar spectrum used by photosynthesis should be considered as an approach for solving this problem [14]. For example, Chl d and Chl f, which are found in oxygenic photosynthetic organisms, may be introduced into higher plants to utilize the 700–750 nm spectral region that no known eukaryotic photosynthetic organism can use. All of these strategies require improving the composition of photosynthetic organisms through genetic manipulation. Accordingly,evaluation of the photosynthetic physiology of cultivars with particularly high photosynthetic efficiency and high yields may thus illuminate the genetic manipulations required to produce such germplasm.
Bainong 207 (BN207) is high-yielding wheat variety that has been cultivated in the Huang-Huai-Hai plain of China in recent years.The current cultivated area of BN207 is as high as 1.3 million ha, and the average yields of BN207 can reach 9000 kg ha−1,which is 8%to 10%higher than that of the main local varieties [15]. Photosynthesis is the basis of yield formation, and our studies showed that BN207 had higher photosynthetic efficiency compared to its parental lines Bainong 64 (BN64) and Zhoumai 16 (ZM16). Accordingly, this study compared the photosynthetic physiology [including photosynthetic pigment content, chloroplast structure, photosystem II(PSII)activity,carbon assimilation ability,etc.]and transcriptome expression between BN207 and its parents,expecting to find key factors or genes that increase photosynthesis efficiency that can be applied to genetic manipulation of wheat.
2. Materials and methods
2.1. Plant growth
Seeds of the wheat variety Bainong 207 (BN207) and its parents, Bainong 64 (BN64) and Zhoumai 16 (ZM16), were surface sterilized, sowed into a 3:1 (v/v) vermiculite:nutrient soil mixture, and grown at 25/22 °C (light/dark) with a 14 h photoperiod under 300 μmol m−2s−1light intensity and 70%relative humidity. Two weeks later (at the three-leaf stage),the first leaf was collected for analysis.
For yield analysis experiments, different varieties of wheat were grown on the research farm in Henan Institute of Science and Technology, fertilization and watering are managed as normal.
2.2. Photosynthetic parameter measurement
The photosynthetic parameters were measured by the method of Wang et al. [16]. Assay was performed with a LI-6400XT Portable Photosynthesis System (LI-6400; LI-COR,Lincoln, NE, USA) with an integrated light source. The leaf chamber temperature was set to 25 °C, and the average CO2concentration was maintained at (382.0 ± 2.5) μL L−1. Photosynthetically active radiation (PAR) was generated with an LED-based (mixed red and blue illumination) system with 800 μmol m−2s−1. For each variety, at least five leaves were measured.
2.3. Analysis of chlorophyll fluorescence
An ultra-portable modulated chlorophyll fluorometer (MINIPAM-II, Heinz Walz GmbH, Effeltrich, Germany) was used to determine chlorophyll fluorescence. According to the methods of Li et al. [17], after leaves were dark adapted for 20 min, they were measured with an actinic light intensity of 600 μmol m−2s−1and a saturated flash intensity of 8000 μmol m−2s−1. Then, Fv/Fm, Y(II), ETR, and qPvalues were calculated using the included software according to the following equations: [Fv/Fm= (Fm− Fo) / Fm; Y(II) = (Fm′ − F) / Fm′; ETR = PAR × 0.84 × 0.5 × Y(II); qP= (Fm′ − F) / (Fm′ − Fo′)]. All measurements were replicated at least five times.
2.4. Chlorophyll and carotenoid content determination
Approximately 0.2 g leaf samples were homogenized in 95%ice-cold ethanol at 4 °C. After centrifugation at 10,000×g for 10 min, the supernatant was used for subsequent measurements. The concentrations of chlorophyll and carotenoid were determined with a UV/VIS spectrophotometer at 665,649, and 470 nm [18]. Chlorophyll a (Ca), chlorophyll b (Cb), and carotenoid (Cc) concentrations were calculated according to the following formulae: Ca= 13.95 × A665−6.88 × A649; Cb=24.96×A649−7.32×A665;Cc=(1000×A470−2.05×Ca−114.8×Cb)/245.
2.5. Western blot assay
The detection of Lhcb1 and D1 proteins by immunoblot analysis was carried out as previously described [19]. Leaf tissue was homogenized with liquid nitrogen,transferred into ice-cold extraction buffer [50 mmol L−1Hepes (pH 7.5),10 mmol L−1NaF,5 mmol L−1MgCl2,and 0.33 mol L−1sorbitol],and centrifuged at 3000 ×g for 3 min, and the precipitate was then resuspended in 50 mmol L−1Hepes (pH 7.5) including 10 mmol L−1NaF,10 mmol L−1MgCl2,and 0.1 mol L−1sorbitol.This suspension was separated by 15% SDS-PAGE. The antibodies for detection of Actin,Lhcb1,and D1 were obtained from Agrisera (http://www.agrisera.com/). Actin was used as the internal control.The signal was detected using MonProTM ECL Prime Substrate (Monad Biotech, Shanghai, China) according to the manufacturers instructions.
2.6. Rubisco activity detection
Rubisco activity detection was performed as described by Flexas et al. [20]. Briefly, samples were ground into a fine powder in liquid nitrogen and homogenized in 1 mL of extraction medium(50 mmol L−1Bicine,pH 8.0;20 mmol L−1MgCl2; 50 mmol L−1βmercaptoethanol; 2 mmol L−1phenylmethylsulfonyl fluoride;30 mg of polyvinylpolypyrrolidone; 2 mmol L−1benzamidine;2 mmol L−1ε-amino-n-caproic acid; and 1% protease inhibitor cocktail), centrifugated at 4 °C and 11,000 ×g for 2 min, and the supernatant was kept on ice prior to the assay.Rubisco activity was determined based on the method by Ward and Keys[21],and all assays were conducted at 25°C.
2.7. Paraffin sectioning and transmission electron microscopy(TEM)
Leaf tissue morphology was observed from paraffin sections.First, leaves were fixed in FAA [70% ethanol, glacial acetic acid,and formaldehyde, 18:1:1 (v:v:v)]. Then, leaves were embedded according to the method by Sun et al.[22],sectioned using a slicer(Leica RM2235;Leica Camera AG,Wetzlar,Germany),Finally,leaf samples were dewaxed,dyed,dehydrated,and observed under a microscope(Leica DMIL LED;Leica Camera AG).
Transmission electron microscopy(TEM)was conducted to analyze chloroplast structures.Leaf samples were fixed at 4°C in 2.5%glutaraldehyde for 4 h;then samples were rinsed three times with PBS buffer for 10 min each time, fixed again with 1% osmic acid at 4 °C, and rinsed another three times.Subsequently, leaf samples were dehydrated with an ethanol series of 30%, 50%,70%, 90%, and 100%ethanol,embedded in Epon812 epoxy resin, and solidified at graded temperature 37 °C, 45 °C, and 65 °C. The sections were sliced with an ultramicrotome (Ultracut, Reichert-Jung, Depew, NY, USA)and were stained with uranium peroxide acetate and lead nitrate.The processed samples were finally examined using a transmission electron microscope (JEM-1200EX, JEOL, Tokyo,Japan).
2.8. Glucose, fructose, sucrose, and starch content measurement
Soluble sugar extraction was conducted following the protocol described by Peleg et al. [23]. First, 0.2-g leaf samples were homogenized in 80% ethanol, incubated at 95 °C for 3 min, and centrifuged at 5000 ×g for 10 min, and the supernatant (soluble carbohydrate fraction) was used in the subsequent assays.Glucose was determined using a glucose oxidase–peroxidase coupled assay following Jørgensen and Andersen [24]. Sucrose and fructose contents were determined using the methods described by Turner [25] and Roe and Papadopoulos [26],respectively.
For starch determination, the precipitate obtained after centrifugation (for the above-noted extraction of total soluble sugar)was suspended in distilled water followed by the addition of 2.5 mol L−1NaOH perchloric acid and boiling for 5 min to gelatinize the starch. After cooling, the suspension pH was adjusted to 4.5 using 2 mol L−1HCl, incubated with amylase for 15 min at room temperature, and centrifugated for 10 min at 3000 ×g, and the supernatant was used for determination of starch content according to the method described by Nelson[27].
2.9. Transcriptome analysis
Total RNA was extracted from leaves using TRIzol (Invitrogen,Carlsbad, CA, USA). mRNA was purified with Oligo(dT)-attached magnetic beads and broken into short fragments. Then, firststrand cDNA was generated using random hexamer-primed reverse transcription,followed by second-strand cDNA synthesis.Afterwards,A-Tailing Mix and RNA index adapters were added by incubating in order to conduct end repair. The cDNA fragments were amplified by PCR,and the products were purified by Ampure XP Beads(Beckman Coulter,Indianapolis,IN,USA)and validated using the Agilent Technologies 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) for quality control. The double stranded PCR products from previous step were heated denatured and circularized by the splint oligo sequence to get the final library.The final library was amplified with phi29 to make DNA nanoballs (DNBs) with more than 300 copies in each molecule;DNBs were loaded into a patterned nanoarray, and 100-base paired-end reads were sequenced using the BGIseq500 platform(BGI, Shenzhen, China). All the RNA-Seq clean data can be available at the Sequence Read Archive (SRA) (https://submit.ncbi.nlm.nih.gov/subs/sra/) under the accession number PRJNA378325.
Primary sequence data were filtered into clean reads using SOAPnuke and Trimmomatic. The clean reads were mapped against the reference wheat genome(https://urgi.versailles.inra.fr/download/iwgsc/IWGSC_ RefSeq_Annotations/v1.1/) using Bowtie2, and RSEM was used to calculate the gene expression level for each sample.Differential gene expression analysis was performed using the DESeq R package. Genes with adjusted Pvalues ≤0.001 and absolute values of log2Ratio ≥1 were identified as significantly differentially expressed genes.
2.10.RT-qPCR analysis
RT-qPCR was performed according to previously described methods[19]in order to analyze gene transcript levels.Briefly,total RNA was extracted from leaves using TRIzol reagent(Invitrogen), and 1.0 μg of RNA was used as a template for cDNA synthesis. RT-qPCR was conducted with a CFX96 Realtime PCR Detection System (Bio-Rad, Hercules, CA, USA). The relative expression level was calculated using the 2–ΔΔCTmethod,with Actin as used as an internal control[28].
2.11.Statistical analysis
All data were obtained from three independent experiments.Each result, except photosynthesis and fluorescence parameters,were shown in the figures represents the mean of three replicates. Photosynthesis and fluorescence parameters were determined using five replicates.Means were compared using a one-way analysis of variance and Duncans multiple range test at the 5%level of significance.
3. Results
3.1. Grain yield
The morphology and yield indicators of three wheat varieties were shown in Table 1. The yield of BN207 was significantly higher than that of its parents BN64 and ZM16,and the length and width of the flag leaf of BN207,that is,the leaf area,were significantly larger than those of BN64 and ZM16.
3.2. Photosynthetic rate
The net photosynthetic rate (Pn) of BN207 was obviously higher than that of its parents BN64 and ZM16, and the observed patterns in stromatal conductance (Gs) and intercellular CO2concentration(Ci)were consistent with that of Pn,with those of BN207 being higher than those of BN64 and ZM16(Fig.1).
3.3. Chlorophyll fluorescence
Chlorophyll fluorescence has been widely used to assess plant photosynthesis. It can reflect the absorption, transmission,dissipation, and distribution of light energy in photosynthetic systems.Fig.2 shows that potential photochemical efficiency(Fv/Fm),actual photochemical efficiency[Y(II)],electron transfer rate(ETR), and photochemical quenching (qP) in BN207 were all significantly higher than in BN64.The Fv/Fmvalue for BN207 was also higher than that for ZM16,though the Y(II),ETR,and qPvalues for BN207 were not significantly different from those in ZM16.
3.4. Chlorophyll and carotenoid content
Chlorophyll and carotenoids play an important role in energy harvesting and photoprotection via photosynthesis. The chlorophyll (chlorophyll a, chlorophyll b, and total chlorophyll) and carotenoid contents in BN207 were significantly higher than those of its parents ZM16 and BN64. The total chlorophyll content of BN207 was 16.4% and 41.8% higher than those of ZM16 and BN64, respectively. The carotenoid content was 8.0% and 25.3% higher than that of ZM16 and BN64,respectively(Fig.3).
3.5. Lhcb1 and D1 protein in PSII
D1 protein,a core protein in PSII,can maintain the stability of the PSII reaction center by binding various cofactors. There was no significant difference in D1 protein content between BN207 and its parents ZM16 and BN64. However, the accumulation of Lhcb1, a light-harvesting chlorophyll a/b–binding protein, was obviously more abundant in BN207 compared to ZM16 and BN64, specifically 6.82 and 3.16 times higher,respectively(Fig.4).
3.6. Leaf morphology and chloroplast structure
Leaf morphological structure is closely related to photosynthesis, so this study included paraffin section analysis of leaves. The paraffin sections revealed that the green color of BN207 leaves (corresponding to chlorophyll) was obviously more intense than that of BN64 (Fig. 5-A), which was consistent with the chlorophyll content results (Fig. 2). While there was no substantial difference between BN207 and its parents in leaf tissue structure, the leaves of BN207 were observably thicker than those of ZM16 (Fig. 5-B) and the leaf area of BN207 was obviously higher than that of its parents(Fig.5-C).
Chloroplasts are the site of photosynthesis, and their shape, distribution, and structure therefore have great influence on photosynthetic efficiency. The TEM results indicated that BN207 and its parents differ in their chloroplast distributions and thylakoid structures.The chloroplast distribution of BN207 was closer to the cell membrane than that of ZM16,and its thylakoids were stacked more tightly than were those of BN64(Fig.6).In short,the chloroplasts were distributed more closely to the cell membrane in BN207 leaves,and the tighter thylakoid structure of BN207 contributed to the high photosynthetic efficiency of the variety.

Table 1–The morphology and yield indicators of three wheat varieties.
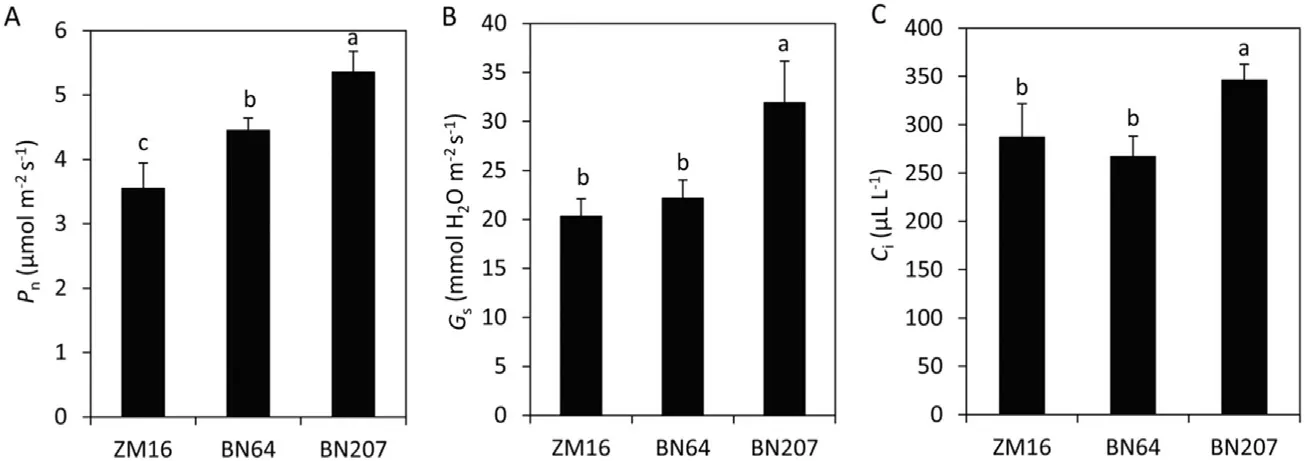
Fig.1–Comparative analysis of net photosynthetic rate(Pn),stromatal conductance(Gs),and intercellular CO2 concentration(Ci)in ZM16,BN64,and BN207.Values are the means±standard deviation(SD)(n=5).Different letters in the same column indicate statistically significant difference at P< 0.05.
3.7. Carbon assimilation
Rubisco is a key enzyme involved in CO2fixation in C3plants,and its activity in BN207 was higher than that of its parents,especially BN64 (Fig. 7-A). Rubisco plays dual roles, RuBP carboxylation and oxidization, and the latter will enter the photorespiratory process and consume carbohydrates formed in C3photosynthesis, does the increased Rubisco activity benefit the carbon assimilation process? Therefore, the accumulation of several carbon assimilation products was detected. Results showed that the glucose, fructose, and starch contents of BN207 were higher than that of its parents,but the sucrose content was lower than those of its parents(Fig.7-B–E).

Fig.2–The analysis of photochemical efficiency(Fv/Fm),actual photochemical efficiency(Y(II)),electron transfer rate(ETR),and photochemical quenching(qP)in ZM16,BN64, and BN207.Values are the means± standard deviation(SD)(n =5). Different letters in the same column indicate statistically significant difference at P< 0.05.
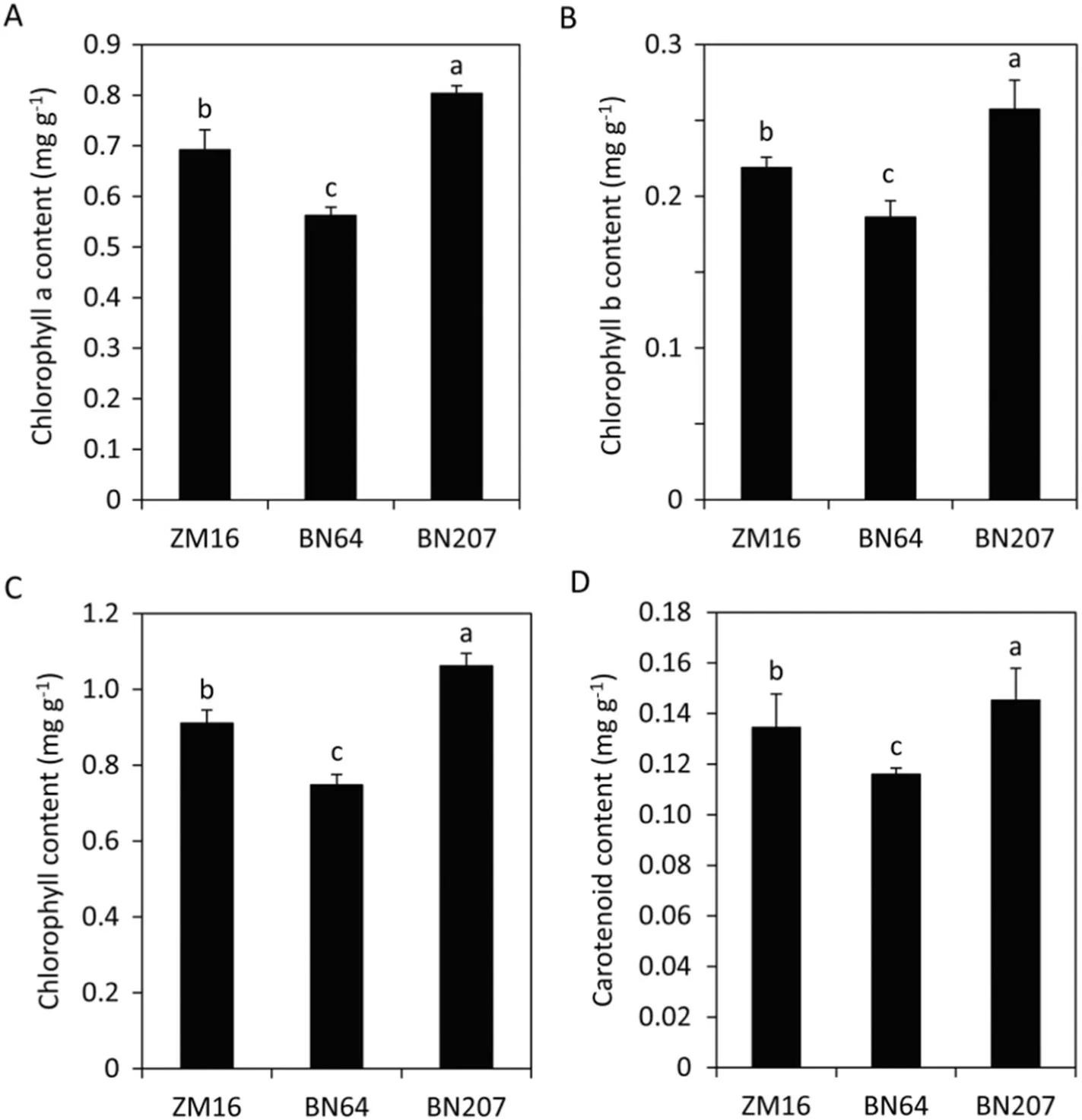
Fig.3– The content of chlorophyll a(A),chlorophyll b (B),total chlorophyll (C),and carotenoid(D)in ZM16,BN64,and BN207.Values are the means±standard deviation(SD)(n =3). Different letters in the same column indicate statistically significant difference at P< 0.05.
3.8. Transcriptome analysis
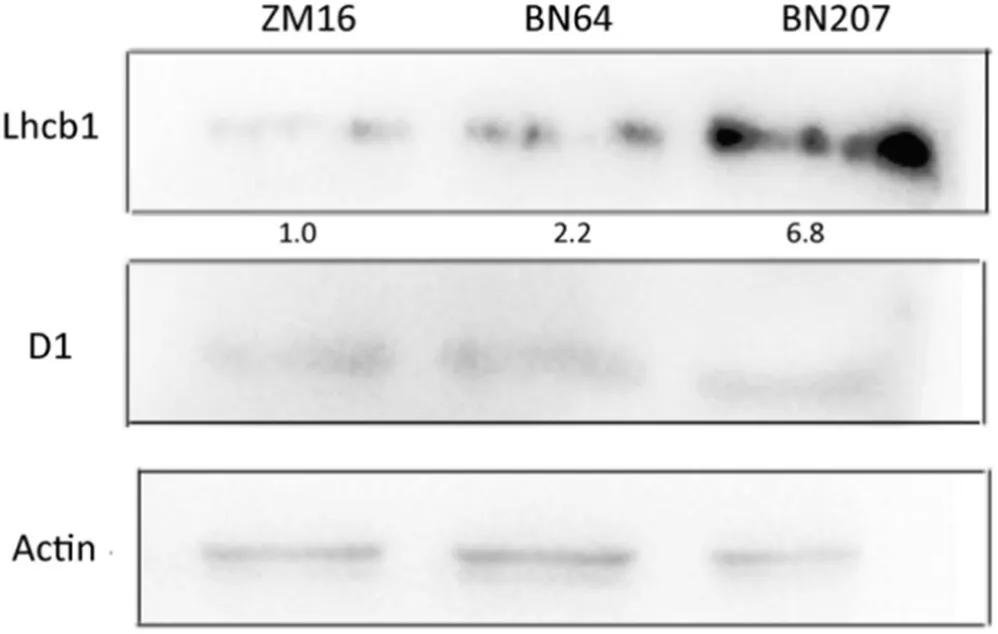
Fig.4–The western blotting of Lhcb1 protein and D1 protein in leaves of ZM16,BN64, and BN207.The bottom numbers indicated the relative protein content of Lhcb.
The above results showed that compared with its two parents,BN207 had factors enhancing its photosynthetic efficiency,including photosynthetic pigment content, photosystem activity, and chloroplast structure. In order to identify the key genes that mediated these factors in BN207, a comparative analysis of the transcriptomes of BN207 and its parents was conducted. We thus identified 7215 differentially expressed transcripts in BN64 compared with BN207 and 20,643 differentially expressed transcripts in ZM16 compared with BN207.This indicates that more BN207 alleles are derived from BN64,consistent with the results of our previous research on the genetic background of BN207(unpublished data).
We focused on photosynthesis-related Gene Ontology (GO)categories and related metabolic pathways.Accordingly,15 and 24 differentially expressed genes within the GO category‘photosynthesis’were found among the differentially expressed BN64 versus BN207 genes and ZM16 versus BN207 genes.Additionally, 16 and 39 genes involved in ‘porphyrin and chlorophyll metabolism’ were differentially expressed among BN64 versus BN207 and ZM16 versus BN207 genes.For‘carotenoid biosynthesis’, 25 and 53 genes were differentially expressed among BN64 versus BN207 genes and ZM16 versus BN207 genes(Table S1).
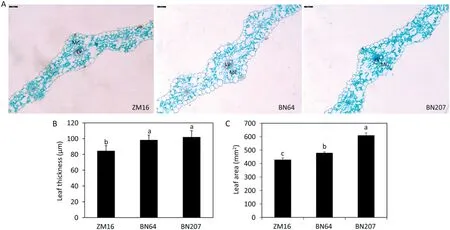
Fig.5–Leaf structure(A),leaf thickness(B)and leaf area(C)in ZM16,BN64,and BN207.Bars=40 μm.MC,mesophyll cell;LV,leaf vein.
To find the key genes that underlie the higher photosynthetic efficiency of BN207 compared to its parents, we overlapped the differentially expressed genes related to‘photosynthesis’, ‘porphyrin and chlorophyll metabolism’,and ‘carotenoid biosynthesis’ between BN64 vs. BN207 and ZM16 vs. BN207. Accordingly, there were 5, 13, and 3 genes associated with ‘porphyrin and chlorophyll metabolism’,‘carotenoid biosynthesis’, and ‘photosynthesis’, respectively,among differentially expressed genes in both BN64 vs. BN207 and ZM16 vs. BN207 (Table 2). Among these genes,TraesCS5D02G364100 (chlorophyllase), BGI_novel_G006617(lycopene epsilon-cyclase), TraesCS4A02G034800 and TraesCS4A02G035100 (Zeaxanthin epoxidase),TraesCS6B02G122500(light-harvesting complex II chlorophyll a/b binding protein) may play key roles. They are responsible for the degradation of chlorophyll, the synthesis of carotenoid, and light energy harvesting, respectively. The up or down-regulated expression of these genes presumably reduces chlorophyll degradation, increases carotene synthesis,and promotes light energy conversion, thus yielding high photosynthetic efficiency in BN207. We randomly selected eight genes for RT-qPCR detection, and the results were basically consistent with the transcriptome data(Fig.8).
4. Discussion
Photosynthesis is the source of energy and material for plant growth and development. Breeding varieties with high photosynthesis efficiency accordingly plays an important role in improving agricultural yields.As photosynthesis occurs,plants first capture solar energy through the light-harvesting antenna complex and convert it into electrical energy at the center of the photosystem complex. Then, electrons are further transferred onto photosynthetic membranes and stored as energy in the molecules ATP and NAD(P)H. Finally, the light energy is converted into simple carbohydrates and stored as carbon-rich molecules [29]. Thus, research on high photosynthetic efficiency can improve the absorption of light energy, electron transfer and energy conversion, and carbon assimilation efficiency [30]. BN207 is the wheat variety with the largest cultivated area within the Huang-Huai-Hai plain, which is the main wheat planting area in China. It has a higher photosynthetic rate (Fig. 1)and chlorophyll fluorescence(Fig.2)than its parents BN64 and ZM16,and thus obtained higher yield(Table 1). Therefore, understanding the photosynthetic physiology of BN207 may inform the cultivation of wheat varieties with high photosynthetic efficiency and high yield.
Chlorophyll plays an extremely important role in the absorption of light energy [31]. In general, decreases in chlorophyll content and function affect the light-capturing ability of leaves, which reduces electron transfer efficiency and decreases the absorption of light energy in the photoreaction[32].Carotenoids,another key photosynthetic pigment,can also absorb light energy for photosynthesis; moreover,they play a strong role in protecting the photosystem from oxidative damage caused by stress[33].First,this means that the high chlorophyll and carotenoid content of BN207 (Fig. 3)may increase photosynthetic efficiency by enhancing the absorption of light energy and improving self-protection of photosynthetic apparatus.Second,the leaf area and thickness of BN207 (Table 1, Fig. 5-B, C) enlarged total photosynthetic area and further increased the photosynthetic pigment content per unit area, and thus it can also be accompanied by improvements in the efficiency of light energy utilization.
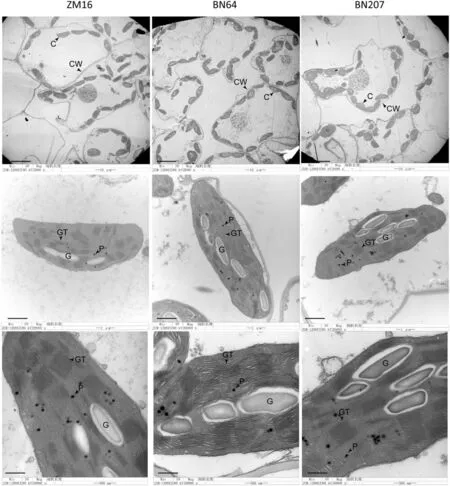
Fig.6– The leaf chloroplast ultrastructure of ZM16,BN64,and BN207.Bars= 10 μm(upper line),1 μm(middle line),500 nm(below line).P,plastoglobule; GT,grana thylakoid;G,granulose.
The light-harvesting complex of photosystem II (LHCII) in plants is the major ensemble of pigment-binding proteins,and they collect photon energy to drive solar energy conversion into chemical energy in PSII reaction centers [34,35].Lhcb1 and Lhcb2 polypeptides,two components of LHCII,can strongly bind to each PSII core unit via the Lhcb5 protein[36,37]. The Lhcb1 protein content of BN207 is significantly higher than that of its parents (Fig. 4), suggesting that BN207 has greater potential to convert photon energy into electron energy.In addition,LHCII has also been proposed to play a key role in the formation of thylakoidal grana stacks.Labate et al.[38] found that constitutive expression of pea Lhcb1-2 in tobacco increased the number of thylakoid membranes per chloroplast, grana stacking, and chloroplast number per palisade cell. The grana were markedly less tightly stacked in LHCII knock-down mutant Stm3LR3 [39]. This research confirmed that Lhcb1 plays an important role in the development of chloroplasts and thylakoids. Fig. 6 showed that the thylakoid grana stack in BN207 was more compact than that in BN64, which might also be related to the higher Lhcb1 content of BN207.
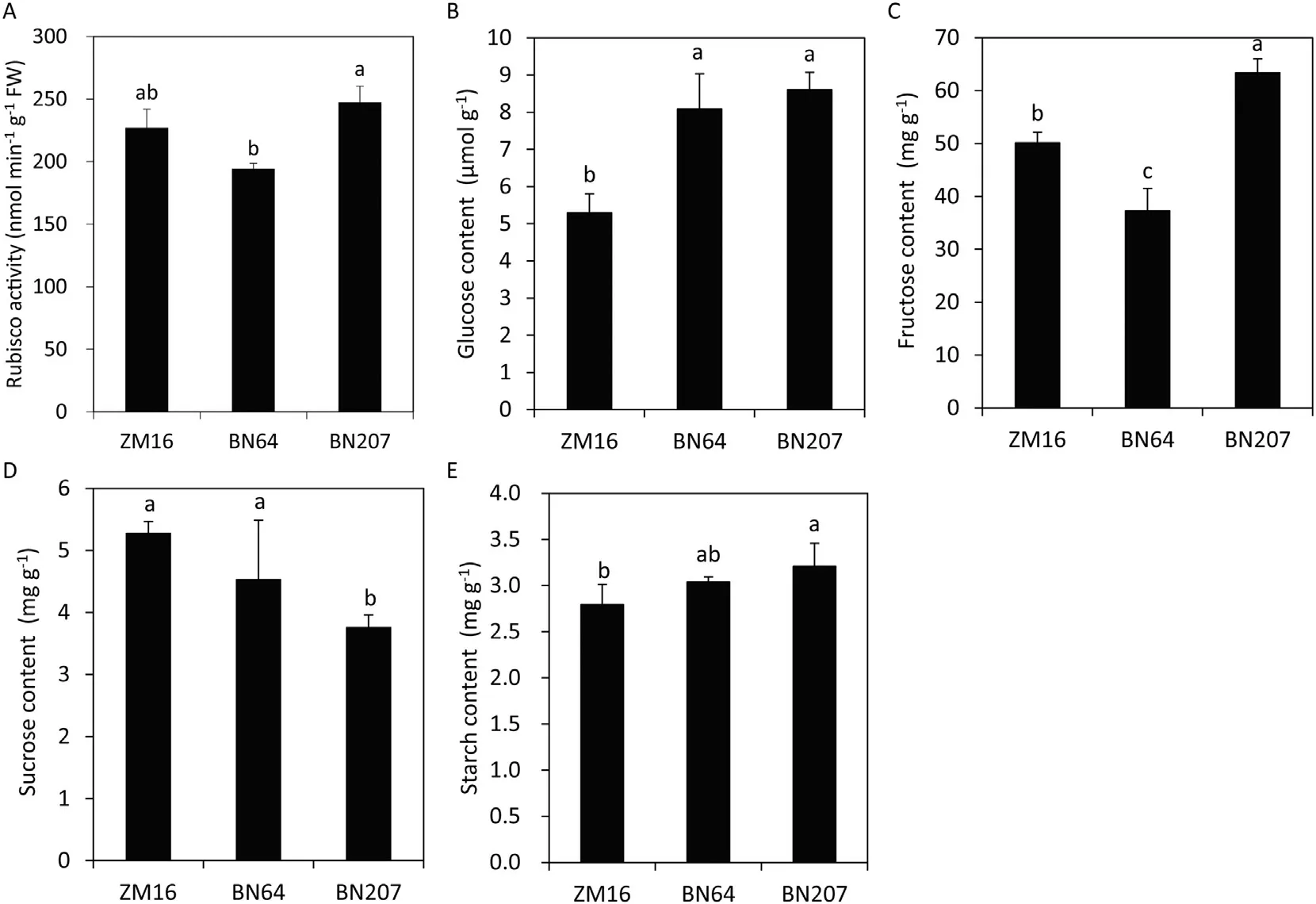
Fig.7– The Rubisco activity(A)and content of glucose(B),fructose(C),sucrose(D),and starch(E) in ZM16,BN64,and BN207.Values are the means±standard deviation(SD)(n =3). Different letters in the same column indicate statistically significant difference at P< 0.05.
Carbon assimilation converted the active chemical energy[ATP and NAD(P)H] produced during the photoreaction phase into carbohydrates for storage. Rubisco is a key enzyme involved in carbon assimilation [40]. To overcome the limitations of Rubisco enzymes on photosynthesis, scientists often explore three means of enhancing Rubisco enzyme activity[9,41].First,Rubisco content can be increased,but this method requires the increased use of nitrogen fertilizer,making it unsustainable. Second, Rubisco enzymes activity can be improved by introducing “high quality” Rubisco enzymes. For example, wheat Rubisco can be replaced with that from Limonium gibertii, providing significant increases in assimilation at concentrations of CO2up to the current ambient concentration [9]. Third, increasing the ratio of CO2to O2concentrations can enhance carboxylation efficiency.Rubisco has dual catalytic activity, that is, Rubisco can catalyze the carboxylation of RuBP and the oxygenation of RuBP.These two catalytic reactions depend on the ratio of CO2to O2concentrations, with a high ratio favoring the carboxylation reaction to fix CO2;in contrast,a low ratio promotes the oxygenation reaction via photorespiration. Accordingly, an attractive approach to improving carbon assimilation would be to introduce CO2-concentrating mechanisms into C3crops,thereby favoring the carboxylation reaction and reducing photorespiratory losses [9,42,43]. The Rubisco activity assay(Fig. 7-A) showed that the photosynthetic efficiency of BN207 also partly depends on its higher Rubisco activity. Additionally, BN207 had high stomatal conductance (Gs) and intercellular CO2concentration (Ci) (Fig. 1-B, C), and its chloroplasts were located close to the cell membrane (Fig. 6). These features promote the entry of CO2into cells and facilitate the carboxylation reaction of Rubisco, thus improving carbon assimilation and the synthesis of more carbohydrates (e.g.,starch, glucose, fructose) (Fig. 7). Sucrose is the main carbohydrate form transported from source to sink, so there is a guess that the sucrose in BN207 leaves was less than that of its parents, which may have caused the rapid export of sucrose from mature leaves to roots(sink).Unexpectedly,the root sucrose content of BN207 was lower than that of BN64,but its root weight was higher than BN64(Fig.S1). This result was consistent with the results in the leaves, that is,compared to its parents, the sucrose content in the leaves of BN207 was lower,while the glucose and fructose content was higher (Fig. 7), and the leaf area was also significantly larger than that of its parent(Fig.5). Therefore, it is believe that the reason for the lower sucrose content in BN207 leaves should be that more sucrose was utilized to promote leaf and root growth rather than sucrose transfer.
As such,which genes were responsible for the differences in photosynthetic physiology between BN207 and its parents? To address this key question, we analyzed the transcriptome of BN207 and its parents. Accordingly, ‘porphyrin and chlorophyll metabolism’, ‘carotenoid biosynthesis’, and ‘photosynthesis’–related genes were assessed in both comparisons of BN64 vs.BN207 and ZM16 vs. BN207 (Table 2). Chlorophyllase mediates the first step in chlorophyll degradation. Lower chlorophyllase activity in eti5 mutant of Arabidopsis resulted in higher chlorophyll content than that of wild type [44]. Similarly, the expression of chlorophyllase (TraesCS5D02G364100) in BN207 was significantly lower than that of its parents, indicating that the high accumulation of chlorophyll in BN207 was attributed to the reduction of chlorophyll degradation, which is consistent with the observed strong green coloration of BN207 under cultivation. Lycopene ɛ-cyclase (LCYe) are believed to be crucial genes to lycopene cyclization and downstream carotenoid accumulation, and the transgenic tobacco expressing LCYe gene of sweetpotato accumulated significantly more βcarotene compared to the untransformed control plants [45].During the green stage of the Japanese persimmon,LCYe played an important role in carotenoid biosynthesis and was responsible for massive accumulation of lutein[46].Therefrom,the high accumulation of carotenoids in BN207 is likely related to the highexpression of lycopene ɛ-cyclase (BGI_novel_G006617). By RTqPCR detection,the expression of BGI_novel_Goo6617 in BN64 is extremely low,almost zero,but its expression can be detected in BN207 and ZM16 (Fig. 8), which suggested that BGI_novel_Goo6617 in BN207 is derived from ZM16.BGI_novel_Goo6617 is responsible for carotenoid synthesis,and it happens that the carotenoids in ZM16 and BN207 are significantly higher than that of BN64(Fig.3),which further illustrated that BGI_novel_Goo6617 has an important role in the carotenoid metabolism. Furthermore, Zeaxanthin epoxidase (ZEP) was the major contributor to carotenoid composition, with mutants lacking ZEP activity showing a remarkable 6-fold increase in total seed carotenoids relative to the wild type [47]. That means that lower expression of ZEP genes(TraesCS4A02G034800 and TraesCS4A02G035100)in BN207 compared to BN64 and ZM16 also contributed to the accumulation of carotenoid. Changes in Lhcb1 protein content mentioned above have also been verified by transcriptome analysis, with higher Lhcb1(TraesCS6B02G122500) expression occurring in BN207,thereby increasing its potential in converting light energy,and it may also dedicate to the development of more densely packed thylakoids. Together, these results suggest that TraesCS5D02G364100,BGI_novel_G006617,TraesCS4A02G034800,TraesCS4A02G035100, and TraesCS6B02G122500 contribute substantially to the high photosynthetic efficiency of BN207. Thus,these genes can be considered candidates for improving photosynthetic efficiency in wheat.

Table 2–The list of genes, involved in ‘Porphyrin and chlorophyll metabolism’, ‘Carotenoid biosynthesis’, and‘Photosynthesis’,differentially expressed in both comparison BN64 vs.BN207 and ZM16 vs.BN207.
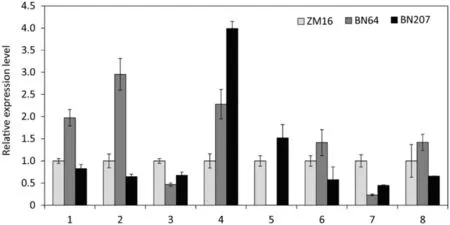
Fig.8–RT-qPCR analysis of the randomly selected eight genes differentially expressed in both comparison BN64 vs.BN207 and ZM16 vs.BN207.1,TraesCS5D02G364100; 2,TraesCS2A02G590600; 3,TraesCS7D02G137200;4,TraesCS7B02G038300;5,BGI_novel_G006617; 6,TraesCS4A02G035100;7,TraesCS2D02G125900;8,TraesCS5A02G457700.
5. Conclusions
Through photosynthetic physiology and transcriptome analyses,we identified the key factors and candidate genes associated with high photosynthetic efficiency in BN207 compared to its parents BN64 and ZM16. In brief, it appears that decreased chlorophyll degradation by reduced chlorophyllase expression, increased carotenoid synthesis by up-regulation of lycopene epsiloncyclase gene expression, promotion of Lhcb1 protein accumulation by increased Lhcb1 gene expression, which increase absorption and conversion of light energy, protect photosystem from oxidative damage in BN207 and denser thylakoid stacks.Additionally, chloroplasts were distributed more closely to the cell membranes, thereby promoting the entry of CO2into cells;this, coupled with the higher Rubisco activity in BN207, promoted carbon assimilation, enabling the accumulation of more carbohydrates, which could promote plant growth and yield formation. Accordingly, BN207 can be used as a wheat germplasm resource with high photosynthetic efficiency, and its photosynthetic characteristics and photosynthesis-related genes (e.g.,TraesCS5D02G364100, BGI_novel_G006617, TraesCS4A02G034800,TraesCS4A02G035100, and TraesCS6B02G122500) can thus be used as references breeding wheat varieties with high photosynthetic efficiency.
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2020.01.004.
Acknowledgments
This work was supported by the National Key Research and Development Program of China(2017YFD0300408).
Author contributions
HJL performed most of the experiments and data analysis.QDZ and XXP performed the plant preparation, sample collection, and RNA extraction. GZX carried out the RT-qPCR experiments. XQO helped to draft and revise the manuscript.HL participated in the experimental design, transcriptome analysis and drafted the manuscript. All authors read and approved the final manuscript.
- The Crop Journal的其它文章
- Application of moderate nitrogen levels alleviates yield loss and grain quality deterioration caused by post-silking heat stress in fresh waxy maize
- Genetic dissection of husk number and length across multiple environments and fine-mapping of a major-effect QTL for husk number in maize(Zea mays L.)
- Identification of a novel planthopper resistance gene from wild rice(Oryza rufipogon Griff.)
- Genome-wide linkage mapping of QTL for root hair length in a Chinese common wheat population
- Metabolic profiling of DREB-overexpressing transgenic wheat seeds by liquid chromatography–mass spectrometry
- Haplotype variations in QTL for salt tolerance in Chinese wheat accessions identified by markerbased and pedigree-based kinship analyses

