In-vitro Synthesis and Activity Identification of Antimicrobial Peptide from Aspergillus granifolium
Huihui CAO Lei WANG Yan WANG Yanhua YAN Xiaonan SONG Yu ZHOU


Abstract [Objectives] This study was conducted to investigate the control effect of antimicrobial peptides to peanut aflatoxin contamination and achieve the purpose of biological control of peanut aflatoxin.
[Methods] Bioinformatics methods were applied to screen antimicrobial peptides with strong antifungal activity in the antimicrobial peptide database and analyze their secondary structures and bacteriostatic characteristics, and the candidated peptide was then artificially synthesized, expressed and purified and detected for in-vitro activity.
[Results] The antimicrobial peptide (AFP1) which was screened from Aspergillus granifolium exhibited strong antimicrobial activity against Aspergillus flavus. The inhibition rate of crude AFP1 at 3 mg/ml reached 58% for gram-positive bacteria, 52% for gram-negative bacteria, and 62% for A. flavus.
[Conclusions] This study provides a new approach for the prevention and control of various plant diseases including peanut disease, and a new idea for the biological prevention and control of plant diseases.
Key words Peanut; Antimicrobial peptide; Aspergillus granifolium; Anti-fungal activity; Bacteriostatic activity detection
Aspergillus flavus is an important phytopathogenic fungus that can contaminate crops such as peanut, corn and soybean. Among them, peanut is one of the crops most susceptible to A. flavus. Aflatoxin produced by A. flavus is a high-strength carcinogen. The detection rate of aflatoxin in peanuts in China exceeds 50%, which seriously affects the quality and safety of peanuts and processed products, endangers consumer health and restricts industrial development. At present, the main measure to control aflatoxin contamination in peanuts is to breed and select disease-resistant varieties, which can be supplemented by chemical control[1-2]. The effects of agrochemicals are not obvious, and the long-term, large-scale and repeated use of agrochemicals has caused serious environmental pollution and other problems. Therefore, biological and green prevention and control methods for the control of A. flavus have attracted extensive attention.
Antimicrobial peptides (AMPs), also known as host defense peptides, refer to a class of small molecules with innate immunity-related antimicrobial activity that are induced and produced in vivo. The earliest antimicrobial peptides were found in insects, and later found in various organisms, including bacteria, plants, animals and even humans. Antimicrobial peptides have the advantages of inhibiting the production of pathogens and not prone to generation drug resistance. They have good applications in biological control, breeding feed, food additives and medicine, and are also ideal substitutes for future antibiotics[3-6].
Materials and Methods
Materials
The APD database[7] (http://aps.unmc.edu/AP/) screened an antimicrobial peptide gene derived from A. granifolium with the number AP00148. The antimicrobial peptide length of A. granifolium is 159 bp and encodes 51 amino acids. Based on the nucleotide sequence of the antimicrobial peptide gene of A. granifolium, Shanghai Invitrogen was commissioned to synthesize the above gene and provide us plasmids.
Main strains and reagents: Escherichia coli XL-10-Gold, E. coli ATCC2592 and E. coli BL21 (DE3); Micrococo luteus ACCC11001; Pichia pastoris GS1115; A. flavus; pET30α-EDDIE-GFP vector, provided by Functional Genomics Laboratory, Oil Crops Research Institute, Chinese Academy of Agricultural Sciences.
Main reagents: DNA marker, tricine, DNA polymerase, urea, DTT, IPTG, plasmid extraction kit and gel recovery kit, all purchased from TaKaRa; low-molecular-weight protein marker, purchased from Invitrogen.
Methods
Prediction and synthesis of antimicrobial peptide genes
Antimicrobial peptide sequences, annotation information, antimicrobial peptide classification, secondary structure and other information were acquired using existing antimicrobial peptide databases such as ANTIMIC[8], APD[5], APPDB[9] and PhyAMP[10], to screen out antimicrobial peptides of A. granifolium with strong antimicrobial effects against bacteria, fungi and plant pathogenic fungi.
After the codons were optimized and primers were designed[11], the target gene was synthesized by PCR with the A. granifolium plasmids stored in the glycerol as a template. The primer sequences are shown in Table 1 (AFP1 and AFP2).
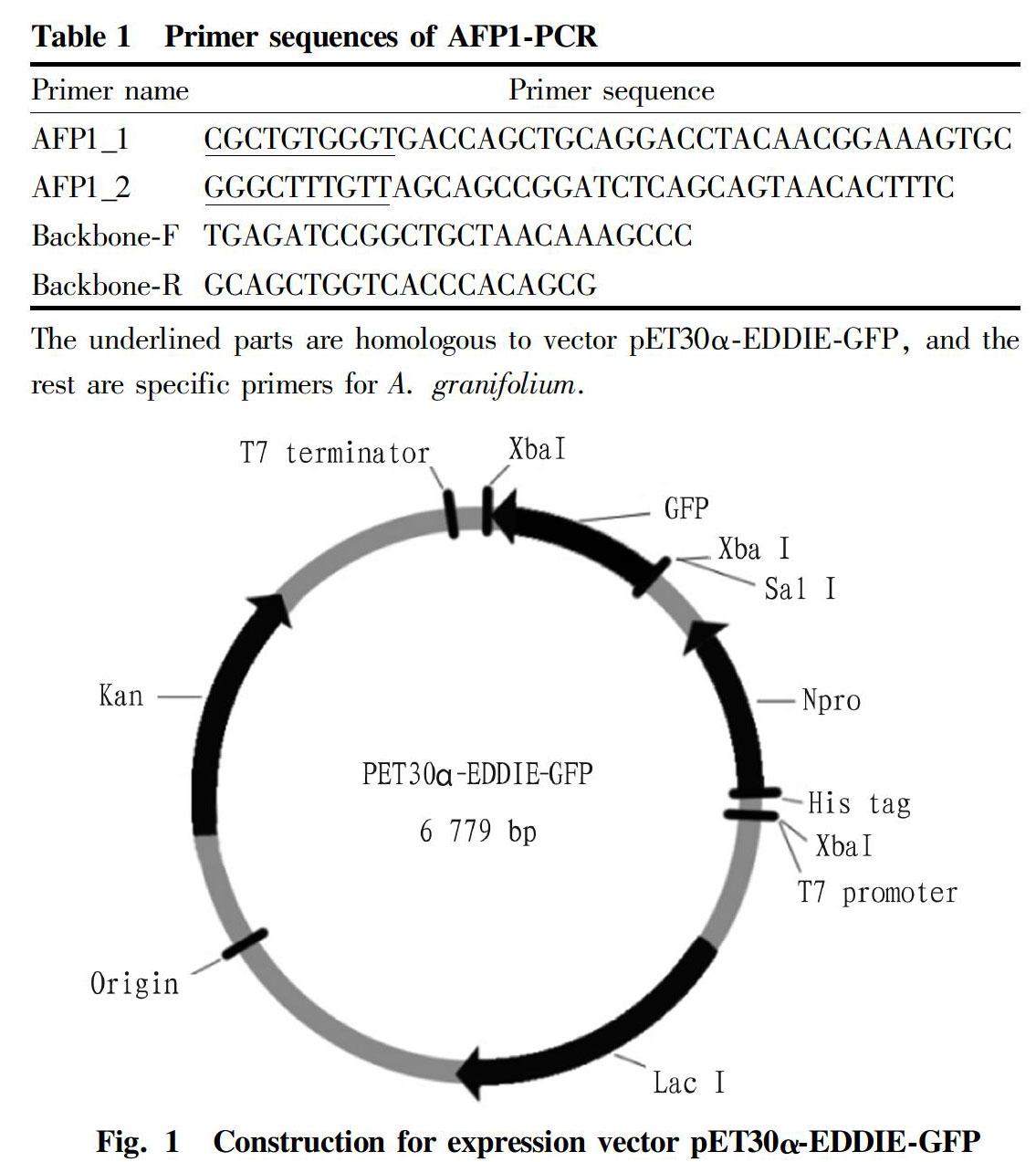
Construction of fusion expression vector pET30α-EDDIE-AFP1
The pET30α-EDDIE-GFP vector was first linearized using the linearized vectors Backbone-F and Backbone-R (Table 1) by the method reported by Ke[12]. After gel electrophoresis, 6 kb linearized vector fragments were obtained. The AFP1 gene and the linearized vector fragment were simultaneously transformed into E. coli XL10-Gold strain by in-vivo site-specific homologous recombination, to ligate the AFP1 gene to the fusion expression vector (Fig. 1).
In-vitro expression and purification of antimicrobial peptide AFP1
In this study, a new antimicrobial peptide expression and purification method was used. The swine fever virus protein mutant EDDIE was used as a fusion protein, and the antimicrobial peptide was expressed in the form of inclusion bodies. The subsequent renaturation process of the inclusion bodies was the process of EDDIE refolding, which can cut off the antimicrobial peptide fused at the C terminal, thereby achieving the purpose of purifying the antimicrobial peptide. The specific operation method was based on the method reported by Huang et al.[13-14]. The process of protein renaturation was the purification process of the antimicrobial peptide. The purified antimicrobial peptide AFP1 was finally tested for its protein concentration by NanoDrop-2000.
Analysis of the activity of the antimicrobial peptide AFP1 in vitro
The Oxford cup method[15] was used to detect the antimicrobial activity of the antimicrobial peptide AFP1 in vitro. The AFP1 test sample was 3 mg/ml in concentration and 200 μl in volume. Specifically, the chemically synthesized pure antimicrobial peptide AFP1 with a concentration≥98% was used as a positive control, and the renaturation liquid served as a negative control. The bacteria were cultured at 37 ℃ for 12-16 h, the yeast was cultured at 22-25 ℃ for 24 h, and A. flavus was cultured at 28 ℃ for 48-72 h, followed by the determination of the antimicrobial rate.
Results and Analysis
Gene prediction and secondary structure analysis of AFP1
The secondary structure of AFP1 was analyzed by put the amino acids in the PDB protein analysis library (http://www.rcsb.org/pdb/home/home.do) . The results showed that the secondary structures of the AFP1 were mainly β-sheets (Fig. 2).
Synthesis of AFP1 and construction of prokaryotic expression vector
pET30α-EDDIE-GFP was linearized with vector linearization primers and detected by agarose gel, and about 6K fragments were cut down, (Fig. 3). The AFP1 gene fragment was amplified by PCR and detected by 1% agarose gel electrophoresis. The size of the band was 159 bp, which was consistent with the expected size (Fig. 4, bands 1-4). The recombinant obtained in "Construction of fusion expression vector pET30α-EDDIE-AFP1" was detected, and the result met the expected band size (Fig. 4- Fig. 6).
In-vitro expression and purification of AFP1
After the induced expression of the recombinant protein pET30α-EDDIE-AFP1, the supernatant and the precipitation were tested by SDS-PAGE respectively. Fig. 5 (1-4) showed a significantly increased band at 23.6 kDa, which was consistent with the EDDIE-AFP1 fusion protein. The self-shearing results of the fusion protein are shown in Fig. 5B: lanes 1 and 2 were 5.6 KDa in size, which was in line with the size of the antimicrobial peptide AFP1. It was suggested that the EDDIE-AFP1 fusion protein was expressed in the form of inclusion bodies.
Analysis of the activity of AFP1 peptide
The in-vitro antimicrobial activity of the antimicrobial peptide was tested on the gram-positive bacteria, gram-negative bacteria, yeast, and plant pathogenic fungus, A. flavus. The results are shown in Fig. 6. The artificially purified AFP1 and the chemically synthesized AFP1 both showed inhibition zones around them, but no obvious inhibition zones were found around the control oxford cups (renaturation liquid). Three biological repeated tests showed that the AFP1 exhibited an antibacterial rate against Gram-positive bacteria reaching 58%, an antibacterial rate against gram-negative bacteria reaching 52% and an antifungal rate against phytopathogenic fungus A. flavus reaching 62%.
Huihui CAO et al. In-vitro Synthesis and Activity Identification of Antimicrobial Peptide from Aspergillus granifolium
Discussion
Antimicrobial peptides are a class of basic polymorphic substances with antimicrobial activity that have a small molecular weight and are generally composed of 20-60 amino acid residues. They have a broad spectrum of antimicrobial activity. Most antimicrobial peptides show hydrophobicity, cationic property and amphiphilic structures, and their secondary structures are rarely linear structures in relative terms, but mainly α-helix, β-sheet or α and β mixed structures. The bactericidal effect of antimicrobial peptides is caused by the action of antimicrobial peptide molecules on bacterial cell membranes, which causes membrane dissolution or rearrangement of the membrane structure. There are two types of action models: "carpet" and "barrel-plate"[16]. Aflatoxin contamination in China mainly occurs in agricultural products such as peanuts, corn, rice, wheat and related foods. Among them, peanut is the crop most likely to be infected with aflatoxins. According to the survey results in recent years, many provinces, including Hebei, have suffered from aflatoxin contamination to varying degrees. The detection rate of corn aflatoxin B1 in Hebei Province in 2012 reached more than 55%[1-2,17]. Ren et al.[1] tested aflatoxin B1 content in samples collected from different corn producing areas in Hebei Province, and the data showed that the aflatoxin B1 content in corn products was 2.04-10.33 μg/kg. The aflatoxin contamination in Hebei Province has been widespread and serious, and the control of the contamination has become an urgent problem in corn safety. Previous studies have found that the contamination rate of aflatoxin in peanuts is more than 20% nationwide, and the detection rate of aflatoxin B1 has reached 24.32%[2]. More than 65% of aflatoxins in agricultural products such as corns, peanuts and rice are infected in the middle and late stages of crop growth. Therefore, controlling the infestation of aflatoxins on crops during the growth period, especially in the mature period, can significantly reduce the contamination level of aflatoxins in agricultural products[18-20]. At present, A. flavus during the growth period is mainly prevented and controlled based on agricultural control through selection of disease-resistant varieties, supplemented by chemical control. The effect of agrochemicals is not obvious, and the long-term, large-scale and repeated use of agrochemicals brings serious environmental pollution and other issues[21]. Therefore, the use of biological control concepts and antimicrobial peptides to achieve the purpose of A. flavus control has received widespread attention and attention.
The antimicrobial peptide of A. granifolium is also called antifungal protein. Its 3D structure is mainly represented by Beta structures. Cysteines in the amino acid sequence can form disulfide bonds, and generally can form four pairs of disulfide bonds, in the order of Cys7-Cys33, Cys14-Cys40, Cys26-Cys28 and Cys49-Cys51[22-23]. Studies have shown that the antimicrobial peptide from A. granifolium has a good inhibitory effect on filamentous fungi such as A. niger, A. terreus, Neurospora crassa, Trichophyton rubrum, Candida albicans and other fungal pathogens, as well as fungi such as Saccharomyces cerevisiae. Studies have further shown that the antimicrobial peptide from A. granifolium has a minimum inhibitory concentration against filamentous fungi reaching 1 μM, a minimum inhibitory concentration against A. niger of 0.25 μM and a minimum inhibitory concentration against Neurospora crassais of 0.06 μM[24]. According to the good antifungal effect of the A. granifolium antimicrobial peptide on A. flavus, the gene of the A. granifolium antimicrobial peptide can be transferred into peanut plants by genetic engineering means or transgenic technology to achieve the purpose of improving the resistance of peanut to A. flavus. Existing studies have transformed the antimicrobial peptide gene of A. niger into rice by transgenic means, which has improved the resistance of rice to pathogens[25]. This study provides new ideas for the study of fungal diseases in peanut in the future.
References
[1] REN JB, ZHANG XH, GAO ZF, et al. Investigation and analysis of mycotoxins in corn of the new season of 2012[J]. Siliao Yu Xumu·Guimo Yangzhi, 2013(4): 13-15. (in Chinese)
[2] LIU X. Strengthen research work of exposure and control of mycotoxin[J]. Chinese Journal of Preventive Medicine, 2006(40): 307-308.
[3] Boman HG. Peptide antibiotics and their role in innateimmunity[J]. Annu Rev Immuno1, 1995(13): 61-92.
[4] GORDON YJ, ROMANOWSKI EG. A Review of antimicrobial peptides and their therapeutic potential as anti-infective drugs[J]. Curr Eye Res, 2005, 30(7): 505-515.
[5] WIMLEV WC, HRISTOVA K. Antimicrobial peptides: successes ,challenges and unanswered questions[J]. J Membr Biol, 2011, 239(1-2): 27-34.
[6] HANCOCK RE, CHAPPLE DS. Peptide antibiotics[J]. Antimicrob Agent Chemother, 1999, 43(6): 1317-1323.
[7] WANG Z, WANG G. APD: the antimicrobial peptide database[J]. Nucleic Acids Reasearch, 2004(32): 590-592.
[8] BRAHMACHARY M, KRISHNAN SP, KOH JL, et al. ANTIMIC: a database of antimicrobial sequences[J]. Nucleic Acids Res, 2004(32): 586-589.
[9] HAMMANMI R, FLISS I. Current trends in antimicrobial agent research: chemo- and bioinformatics approches[J]. Drug Discovery Today,2010,15(13/14): 540-546.
[10] Hammami R, Ben Hamida J, Vergoten G, et al. Phytamp: a database dedicated to plant antimicrobial peptides[J]. Nucleic Acids Reaserch, 2009(37): 963-968.
[11] DNA Works. Automatic oligonucleotide design for PCR-based gene synthesis[OL]. htttp://mcll.ncifcrf,gov/dnaworks/.
[12] KE T, LIANG S, WU J, et al. A novel PCR-based method for high throughput prokaryotic expression of antimicrobial peptide gene[J]. BMC Biotechnology, 2012(12): 10.
[13] HUANG J, HUANG JY, KE T, et al. Cloning and bacteriostatic activity detection of a new antibacterial peptide BnPCD842895[J]. Chinese Journal of Oil Crop Sciences, 2013, 35(4): 357-363. (in Chinese)
[14] CAO HH, DONG CH, YU JY, et al. Antimicrobial activity detection of antimicrobial peptide gene BnLTP1 from Brassica napus[J]. Chinese Journal of Oil Crop Sciences, 2016, 38(1): 001-006. (in Chinese)
[15] JIANG HL. Influencing factors and control methods of cylinder-plate method for measuring antibiotic titer[J]. Chinese Journal of Veterinary Drug, 2011(8): 23-26. (in Chinese)
[16] ZHAO HX, ZHAN Y, XU ZR. Research progress of antibacterial peptide from animals[J]. Chinese Journal of Animal Science, 2003(7): 16-19. (in Chinese)
[17] YUAN YQ, ZHANG YQ, ZHANG SL, et al. Influencing factors and comprehensive control measures of peanut aflatoxin pollution[J]. Modern Agricultural Science and Technology, 2017(21): 1-2. (in Chinese)
[18] PAL K, GARDENER B. Biological control of plant pathogens[J]. The plant health instructor, 2006(17): 2.
[19] Mishra HN, Das C. A review on biological control and metabolism of aflatoxin[J]. Critical Reviews in Food Science and Nutrition, 2003, 43(3): 245-264.
[20] Dorner JW. Biological control of aflatoxin contamination of crops[J]. Toxin Reviews, 2004, 23(2): 425-450.
[21] MENDEZ-ALBORES A, DEL RIO-GARCIA JC, MORENO-MARTINEZ E. Decontamination of aflatoxin duckling feed with aqueous citric acid treatment[J]. Animal Feed science and Technology, 2007(135): 249-262.
[22] NAKAYA K, et al. Amino acid sequence and disulfide bridges of an antifungal protein isolated from Aspergillus giganteus[J]. Eur J Biochem, 1990,193(1): 31-38.
[23] Campos-Olivas R et al. NMR solution structure of the antifungal protein from Aspergillus giganteus: evidence for cysteine pairing isomerism[J]. Biochemistry, 1995(34): 3009.
[24] ANNA HUBER, DOROTTYA HAJDU, DORIS BRATSCHUN-KHAN. et al. New antimicrobial potential and structural properties of PAFB: A cationic, cysteine-rich protein from Penicillium chrysogenum Q176[J]. Sci Rep, 2018, 29, 8(1): 1751.
[25] MONTESINOS E. Antimicrobial peptides and plant disease control[J]. FEMS Microbiol Lett, 2007, 270(1): 1-11.
- 农业生物技术(英文版)的其它文章
- Enhanced Production of Natural Carotenoids from Genetically Engineered Rhodobacter sphaeroides Overexpressing CrtA
- Testis of Male Tilapia Under Methomyl Stress: Transcriptome Changes and Signal Pathway Analysis
- Functional Analysis of Dunaliella salina Calmodulin Kinase Gene
- First Report of Phomopsis Leaf Spot on Patchouli [Pogostemon cablin (Blanco) Benth.] Caused by Diaporthe arecae in China
- Effects of Plant Growth Regulators on Tillering Ability of Ophiopogon japonicus cv
- Effect of Irrigation and Fertilization on Population Structure and Yield of Wheat

