Functional Analysis of Dunaliella salina Calmodulin Kinase Gene
Zhenyu XING Mingfang WANG Xiangnan GAO Weiwei XU Yuting CONG Xiaojie CHAI


Abstract [Objectives] This study was conducted to investigate the function of Dunaliella salina calmodulin kinase (CaMK) gene.
[Methods] The sense and antisense gene fragments (223 bp) and spacer sequence (129 bp) of D. salina calmodulin kinase gene were cloned and inserted into the downstream part of the 35S promoter of the eukaryotic expression vector pMDCMGN-Cat. The siRNA expression system of CaMK gene was successfully constructed. The pCaMK-RNAi expression vector was transformed into D. salina cells by the LiAc/PEG-mediated method, giving transgenic D. salina. The expression of CaMK gene was then analyzed by real-time fluorescence quantitative PCR.
[Results] The expression of CaMK gene in the transgenic D. salina was significantly reduced, by 70% compared with the control group, suggesting that the expression of CaMK gene was significantly inhibited. The examination of the growth status of D. salina showed that D. salina cell division and proliferation were also affected. It is proved that CaMK gene has a positive regulation effect on the division and proliferation of D. salina cells.
[Conclusions] The study provides important information for further elucidating the function and action mechanism of D. salina calmodulin kinase gene.
Key words Dunaliella salina; CaMK; RNAi; LiAc/PEG-mediated method; Real-time fluorescence quantitative PCR
Dunaliella salina is a kind of single-cell eukaryotic green alga that is widely distributed in oceans, salt fields, and salt lakes. D. salina cells are small and diverse in shape. It has two equal-length flagella located at the front which allow it to swim freely, but has no cell wall. Due to its strong tolerance to high salt, D. salina has attracted widespread attention from biologists. Over the years, scholars at home and abroad have studied the osmotic adjustment of D. salina in high-salt environments[1-3], salt tolerance genes[4-5], D. salina proteome[6-7], etc., and obtained some research results. However, so far, the molecular mechanism of D. salina in response to salt stress signals has not been fully understood.
Calmodulin kinase (CaMK) is a protein kinase that depends on Ca2+ and calmodulin. Intracellular Ca2+·CaM activates calmodulin-dependent kinase (CaMKK), which in turn activates calmodulin kinase (CaMK), which transmits information during the cascade phosphorylation reaction of the target protein and exerts its biological effect. This protein kinase plays an important role in life activities such as gene expression, cell division, and differentiation of organisms[8-9]. Studies have shown that CaMK plays an important regulatory role in plant growth and development and stress response[10-13]. However, there are few reports in alga research. Therefore, studying the function of D. salina CaMK gene has important scientific significance for elucidating the molecular mechanism and signal transduction pathways of D. salina.
In this study, the calmodulin kinase gene fragments of D. salina were cloned by genetic engineering technology, and an efficient small interference RNA (siRNA) expression system was construct for inhibiting the expression of calmodulin kinase gene by RNAi technology, with an attempt to provide reference for further elucidating the molecular mechanism of the adaptation of D. salina to high salt environments and the molecular pathway of D. salina in response to salt stress signals.
Materials and Methods
Experimental materials
D. salina was provided by the Key Laboratory of Hydrobiology, Dalian Ocean University. The D. salina was statically cultured in Conway solution[14] at 25 ℃ in an 12 h light (1 250 lx)/12 h dark photoperiod standing culture. The Escherichia coli strain DH5α and pMDCMGN-Cat plasmids were kept by the laboratory, and pMDTM19-T simple was purchased from TaKaRa Company.
RNAiso Plus, DNAiso Reagent, DNA Marker DL-2000, restriction enzymes, Taq enzyme, T 4 DNA ligase, lysozyme, IPTG, X-gal, DEPC, DNA Marker PrimeScriptTM RT Master Mix kits, and TB Green Premix Ex TaqTM kits were purchased from TaKaRa Company. Plasmid extraction kits were purchased from Sangon Biotech. Gel recovery kits were purchased from Axygen Biotechnology Co., Ltd. Other reagents were all made in China, analytically pure.
Experimental methods
Extraction of total RNA and genomic DNA from D. salina
Algal cells were collected from 10 ml of purified and cultured D. salina (2 parts) in the logarithmic growth phase (the culture contained 3.0 mol/L NaCl). Total RNA was then extracted according to the instruction of the RNAiso Plus kit (TaKaRa, China), followed by the detection of RNA quality by 1% agarose gel electrophoresis. Genomic DNA was extracted according to the instruction of DNAiso Reagen kit (TaKaRa, China). The DNA quality was detected by 0.6% agarose gel electrophoresis.
Cloning of sense fragment, antisense fragment and spacer sequence of D. salina CaMK gene and establishment of siRNA expression system
Primers were designed according to the CaMK gene sequence obtained from D. salina transcriptome sequencing (NCBI-SRA database: PRJNA471570): S1, S2, and A1, A2. Primers P1 and P2 were designed according to the type I intron sequence (GenBank: GCA_002284615.1) of the D. salina genome on the NCBI (Table 1). The sense and antisense fragments of the CaMK gene were amplified using the reverse transcribed cDNA as a template, and S1, S2 and A1, A2 as primers, respectively. The spacer sequence was amplified using the D. salina genomic DNA as a template and P1 and P2 as primers. The PCR reaction was started with pre-denaturation at 94 ℃ for 5 min, followed by 30 cycles of denaturation at 94 ℃ for 30 s, annealing at 53 ℃ for 30 s and extension at 72 ℃ for 1 min, and completed by extension at 72 ℃ for 10 min. The target fragments were ligated with pMDTM19-T simple vector to obtain a positive monoclone. The recombinant plasmids pMDCaMKs, pMDCaMKa, and pMDspacer were sent to Sangon Biotech for sequencing.
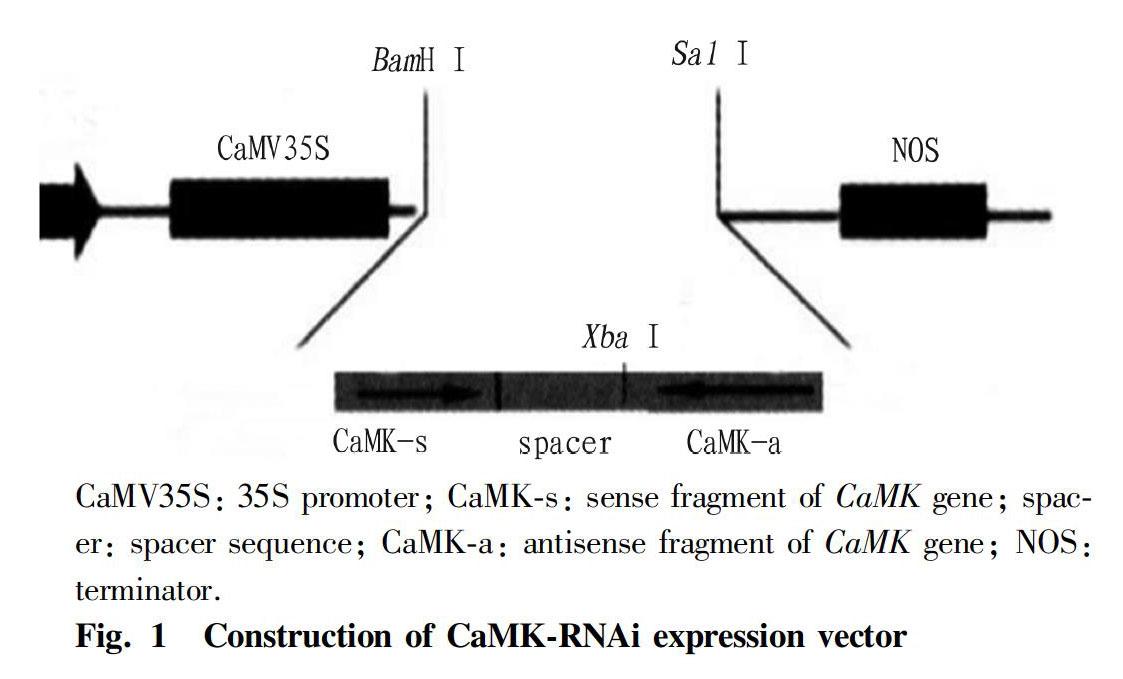
Recombinant plasmids pMDspacer and pMDCaMKs were digested with EcoR I and Xba I, and the spacer fragment and the large fragment of vector pMDCaMKs were recovered. The spacer fragment was then ligated into pMDCaMKs using T4 DNA ligase. The CaMKs-spacer fragment was obtained by double digestion with Bam H I and Xba I, and then inserted into the 35S promoter of the eukaryotic expression vector pMDCMGN-Cat by T4 DNA ligase. The plasmid pMDCaMKa was double digested with Sal I and Xba I, and CaMKs was inserted into pMDCMGN-Cat, thereby successfully constructing the siRNA expression vector of CaMK gene: pCaMK-RNAi (Fig. 1).
Transformation of D. salina cells with recombinant plasmids and screening of transformants
The LiCa/PEG-mediated method[15] was used to transfer the pCaMK-RNAi expression vector into the D. salina cells, and the transformed algal liquid was cultured for 48 h for recovery. Using the wild type algal liquid as a control, the transformed algal liquid was screened with chloramphenicol at a final concentration of 200 μg/ml to obtain transgenic D. salina.
qRT-PCR analysis of CaMK gene expression
Total RNA was extracted from wild and transgenic D. salina, followed by the detection of purity and concentration of the RNA samples by UV spectrophotometry. Then, 500 ng of each RNA sample was taken for reverse transcription. Fluorescence quantitative PCR was performed on ABI 7300 Real-Time PCR System using target gene primers and 18S internal reference gene primers (Table 1). The reaction system included SYBR Premix Ex TaqTM II 10 μl, upstream and downstream primers (10 μmol/L) 0.8 μL each, ROX Reference Dye (50x) 0.4 μl, cDNA template 2 μl and ddH2O 6 μl. The PCR started with pre-denaturation at 95 ℃ for 30 s, followed by 40 cycles of denaturation at 95 ℃ for 5 s and annealing at 60 ℃ for 30 s.. After the reaction, the amplification curve and melting curve were confirmed. The relative expression of CaMK gene was calculated by the 2-ΔΔCt method. Statistical analysis was performed using SPSS software.
Determination of the growth curve of D. salina
Wild and transgenic D. salina were statically cultured under the same conditions (25 ℃, light intensity 1 250 lx, 12 h light/12 h dark). Based on the linear relationship between D. salina cell density and the optical density value OD682 in a certain range[16], the OD682 values of the algal liquid were used to draw the growth curve of D. salina.
Results and Analysis
Cloning of the target fragments of D. salina CaMK gene and establishment of siRNA expression system
PCR was performed using the total RNA of D. salina as a template according to the method in "Experimental methods", and the PCR product was analyzed by 1% agarose gel electrophoresis (Fig. 1-A). As shown in Fig. 1-A, there was a clear band around 200 bp, which was consistent with the expected result. After the amplification product was recovered, it was ligated with pMD19-T Simpie, followed by the transformation into E. coli DH5α competent cells. Positive clones were selected and sent to Sangon Biotech for sequencing, and the sense and antisense fragments of 223 bp were obtained, suggesting that the recombinant plasmids pMDCaMKs and pMDCaMKa were successfully constructed. PCR was performed using the D. salina genomic DNA as a template, and the PCR product was analyzed by 1% agarose gel electrophoresis (Fig. 1-B). As shown in Fig. 1-B, there was a clear band between 100-250 bp, which was consistent with the expected result. The recombinant plasmid pMDspace was sent to Sangon Biotech for sequencing, and a 129 bp spacer sequence was obtained. According to "Experimental methods", the "sense fragment-spacer sequence-antisense fragment" was ligated to the eukaryotic expression vector pMDCMGN-Cat, and the CaMK gene siRNA expression vector pCaMK-RNAi was constructed. After double digestion with BamH I and Sal I, there was a clear band between 500 bp and 750 bp, which was consistent with the expected result. The 575 bp target fragment was obtained by sequencing, which proved that the eukaryotic expression vector pCaMK-RNAi was successfully constructed.
Transformation of D. salina cells with the recombinant plasmid and screening of transformants
The pCaMK-RNAi was transformed into D. salina cells by the LiAc/PEG-mediated method. After 48 h of recovery culture, chloramphenicol was added to the final concentration of 200 μg/ml. The screening results are shown in Fig. 2. After 72 h of screening, the algal liquid of the control group turned yellow and transparent, and a white precipitate appeared at the bottom. It was found by microscopy that all the algal cells in the control group died. The experimental group was still green, and microscopic examination showed that living algal cells were successfully screened for transgenic D. salina. The screened transformants were added to 40 ml of sterile seawater to perform enlarged culture, for real-time quantitative PCR analysis and growth curve determination.
Expression analysis of D. salina CaMK gene
The expression of D. salina CaMK gene was analyzed by real-time quantitative PCR method. The results are shown in Fig. 3. The real-time quantitative PCR data was processed using the relative quantitative method. The expression of the target gene in the transformants was calculated by 2-ΔΔCT method. The CT values of the control groups CaMK gene and the internal reference gene 18S were used to calculate 2-ΔΔCT, which represented the relative change in the expression of the transformant CaMK gene. The calculation formula of ΔΔCT was ΔΔCT=(CTCaMK-CT18s)transformation group-(CTCaMK-CT18s)control group. The results of the qRT-PCR analysis showed that the expression level of the transformed CaMK gene was reduced by 70% compared with the control group, which proved that the expression of CaMK gene was significantly inhibited.
Agricultural Biotechnology2020
Determination of the growth curve of D. salina
The screened transformants were subjected to enlarged culture, and then cultured under the same conditions as the wild-type D. salina of the control group after one passage, and the OD682 value was measured during 1-8 d after passage. As shown in Fig. 4, 2 d after passage (the initial number of cells was the same), the number of cells in the transformed algal liquid was lower than that in the control group. The D. salina entered the logarithmic growth phase 5-7 d after passage, at which the difference in cell number between the two groups was more significant. Because the transformed algal fluid had been cultured for 5 d before passaging, the effects of transformation and experimental treatment on the division of algal cells could be ruled out. It could be seen that when the expression of the D. salina CaMK gene was inhibited, cell division and proliferation were also affected. It is proved that CaMK gene has a positive regulation effect on the division and proliferation of D. salina cells.
Conclusions and Discussion
Cells respond to various external signal stimuli mainly through a series of biochemical events such as phosphorylation modification of pathway proteins by protein kinases, protein interactions, and changes in downstream target gene expression, which ultimately lead to specific biological effects. Calmodulin kinase can phosphorylate many phosphorylation sites of target enzymes and activate them to achieve numerous physiological functions. It is considered to be the most important protein molecule in calcium signal transduction pathway. Studies have shown that calmodulin kinase is widely involved in the mediation of signals in various Ca2+ signaling pathways such as stress response and growth and development. As in animal studies, researchers have found that by reducing extracellular Ca2+ and intracellular calmodulin levels, cells cannot enter the mitotic phase[17]. Means[18] and Zhao et al.[19] believe that CaMK may regulate the cell cycle and the rapid growth of single-cell flagellate algae in the logarithmic phase. However, no studies have been reported on D. salina CaMK gene in this respect. Therefore, it is of great scientific significance to identify the function of D. salina calmodulin kinase gene.
RNA interference (RNAi) is a kind of dsRNA-mediated specific and efficient gene expression inhibition method, which has broad application prospects in gene function research. siRNA is an important intermediate effector molecule by which RNAi occurs. It has a special structure, that is, the sequence of siRNA is homologous with the target mRNA sequence; and the ends of the two single strands of siRNA are a 5′-terminal phosphate and a 3′-terminal hydroxyl group. In addition, the 3′ terminal of each single strand has 2 to 3 prominent unpaired bases[20-23]. The gene silencing efficiency of RNAi is affected by interference fragments and spacer sequences. In the selection of interference fragments in this study, all transcripts of CaMK gene in the transcriptome were comprehensively analyzed, and the interference fragments designed in the conserved region could interfere the expression of all transcripts. The successfully constructed eukaryotic expression vector pCaMK-RNAi was transformed into the D. salina cells, obtaining transgenic D. salina. The expression of CaMK gene was analyzed by real-time fluorescence quantitative PCR method. The results showed that the expression of CaMK gene in the transgenic D. salina was significantly reduced, by 70% compared with the control group, suggesting that the expression of CaMK gene was significantly inhibited. To investigate the function of the CaMK gene, we examined the growth status of D. salina. In order to exclude the effects of transformation and screening on algal cells, the algal liquid obtained after transformation and screening was subjected to recovery culture for 5 d. After detection and passage, the OD682 value of the algal liquid in the experimental group was 0.069, and the OD682 value of the algal liquid in the control group was 0.078. The initial algal cell quantities in the two groups of algal liquid were basically the same, and the influences of other factors on the growth curve results were excluded. The number of cells in the transformed algal liquid was lower than that in the control group 2 d after passage under the same initial cell number. The D. salina entered the logarithmic growth phase 5-7 d after passage, at which the difference in cell number between the two groups was more significant. Because the transformed algal fluid had been cultured for 5 d before passaging, the effects of transformation and experimental treatment on the division of the algal cells could be ruled out. It could be seen that when the expression of D. salina CaMK gene was inhibited, cell division and proliferation were also affected. It is proved that the CaMK gene has a positive regulation effect on the division and proliferation of D. salina cells. However, the preliminary functional analysis of the algal calmodulin kinase gene is just the beginning. The subsequent work will screen proteins that interact with CaMK by co-immunoprecipitation, so as to provide new information for further elucidating the action mechanism of CaMK in the signal transduction of D. salina in response to salt stress.
References
[1] ZHOU L, MENG XH, LIU CS, et al. Effects of osmotic stress on intracellular glycerol content and enzyme activity in Dunaliella salina[J]. Chinese Bulletin of Botany, 2006, 23(2): 145-151. (in Chinese)
[2] CHEN H, LAO YM, JIANG JG. Effects of salinities on the gene expression of a (NAD(+)) -dependent glycerol-3-phosphate dehydrogenase in Dunaliella salina[J]. Sci Total Environ, 2011, 409(7): 1291-1297.
[3] ZHAO LN, GONG WF, CHEN XW, et al. characterization of genes and enzymes in Dunaliella salina involved in glycerol metabolism in response to salt changes[J]. Phycological research, 2013(61): 37-45.
[4] CONG YT, MA YX, WANG Y, et al. Characterization and expression analysis of protein kinase C gene from Dunaliella salina[J]. J. Ocean Univ. China,2019,18(4):977-984.
[5] GAO XN, CONG YT, YUE JR, et al. Small RNA, transcriptome and degradome sequencing to identify salinity stress responsive miRNAs and target genes in Dunaliella salina[J]. Journal of Applied Phycology, 2019, 31(2): 1175-1183 .
[6] WANG Y, CONG YT, WANG YH, et al. Identification of early salinity stress-responsive proteins in Dunaliella salina by iTRAQ-based quantitative proteomic analysis[J]. Int. J. Mol. Sci., 2019(20): 599-612.
[7] KATZ A, WARIDEL P, SHEVCHENKO A, et al. Salt-induced changes in the plasma membrane proteome of the halotolerant alga Dunaliella salina as revealed by blue native gel electrophoresis and nano-LC-MS/MS analysis[J]. Mol cell proteomics, 2007, 6(7): 1459-1472.
[8] HU DW, HE ZC, SHIH M. The regulation role of calcium/calmodulin-dependent protein kinase on physiologic properties in higher plants[J]. Journal of Wuhan Botanical Research, 1999(S1): 89-98. (in Chinese)
[9] HANKS SK , QUINN AM. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members[J]. Methods Enzymol, 1991(200): 58-61.
[10] HUA W, ZHANG L, LIANG S, et al. A tobacco calcium/calmodulin-binding protein kinase functions as a negative regulator of flowering[J]. J Biol Chem.2004(279):31483-31494.
[11] LIU HT, GAO F, LI GL, et al. The calmodulin-binding protein kinase 3 is part of heat-shock signal transduction in Arabidopsis thaliana[J]. Plant J., 2008(55): 760-773.
[12] YANG T, CHAUDHURI S, YANG L, et al. A calcium/calmodulin-regulated member of the receptorlike kinase family confers cold tolerance in plants[J]. J Biol Chem., 2010(285): 7119-7126.
[13] TAKAHASHI F, MIZOGUCHI T, YOSHIDA R, et al. Calmodulin-dependent activation of MAP kinase for ROS homeostasis in Arabidopsis[J]. Mol Cell, 2011(41): 649-660.
[14] CHEN MY. Biological food culture[M]. Beijing: China Agriculture Press, 1995. (in Chinese)
[15] CHAI XJ, JIN FEI, CONG YT, et al. Establishment of a new method for genetic transformation of Dunaliella salina[J]. Agricultural Science Technology, 2017, 18(8): 1374-1377.
[16] LI KJ. Construction of a system for the transformation of foreign genes in Dunaliella salina[D]. Dalian: Dalian University of Technology, 2007. (in Chinese)
[17] LORCA T, CRUZALEGUI FH, FESQUET D, et al. Calmodulin-dependent protein kinase II mediates inactivation of MPF and CSF upon fertilization of Xenopus eggs[J]. Nature, 1993, 366(6452):270-273.
[18] MEANS AR. Calcium, calmodulin and cell cycle regulation[J]. FEBS letters, 1994(347): 1-4. PMID: 8013652.
[19] ZHAO L, LU R, YAO Z. The roles of Ca2+ calmodulin-dependent kinases in cell proliferation[J]. Chinese Journal of Cell Biology, 2007(29): 331-335.
[20] RAMASWAMY G, SLACK FJ. siRNA: A guide for RNA silencing[J]. Chem Biol, 2002, 9(10): 1053-1055 .
[21] SEO BS, KIM S, SCOTT MP, et al. Functional interaction between heterologously expressed starch branching enzymes of maize and the glycogen synthase of brewers yeast[J]. Plant Physiol, 2002(128): 1189-1199.
[22] HANNON GJ. RNA interference[J]. Nature, 2002, 418(6894): 244-251.
[23] MUCMANUS MT, SHARP PA. Gene silencing in mammals by small interfering RNAs[J]. Nature Rev Genet, 2002, 3(10): 737-746.
- 农业生物技术(英文版)的其它文章
- Enhanced Production of Natural Carotenoids from Genetically Engineered Rhodobacter sphaeroides Overexpressing CrtA
- Testis of Male Tilapia Under Methomyl Stress: Transcriptome Changes and Signal Pathway Analysis
- First Report of Phomopsis Leaf Spot on Patchouli [Pogostemon cablin (Blanco) Benth.] Caused by Diaporthe arecae in China
- Effects of Plant Growth Regulators on Tillering Ability of Ophiopogon japonicus cv
- Effect of Irrigation and Fertilization on Population Structure and Yield of Wheat
- Production and Cultivation Technology of Selenium-enriched Pueraria thomsonii Benth

