ldentification of SNPs and expression patterns of FZD3 gene and its effect on wool traits in Chinese Merino sheep (Xinjiang Type)
ZHAO Bing-ru , FU Xue-feng, TlAN Ke-chuan, HUANG Xi-xia Dl Jiang, BAl Yan, XU Xin-ming,TlAN Yue-zhen, WU Wei-wei, ABLAT Sulayman ZENG Wei-dan HANlKEZl Tulafu
1 College of Animal Science, Xinjiang Agricultural University, Urumqi 830052, P.R.China
2 Key Laboratory of Genetics Breeding and Reproduction of Xinjiang Cashmere and Wool Sheep, Institute of Animal Science,Xinjiang Academy of Animal Sciences, Urumqi 830011, P.R.China
Abstract As a member of the Frizzled family, Frizzled3 (FZD3) is a receptor of the canonical Wnt signaling pathway and plays a vital role in mammalian hair follicle developmental processes. However, its effects on wool traits are not clear. The objectives of this study were to identify the single nucleotide polymorphisms (SNPs) and the expression patterns of FZD3 gene, and then to determine whether it affected wool traits of Chinese Merino sheep (Xinjiang Type) or not. PCR-single stranded conformational polymorphism (PCR-SSCP) and sequencing were used to identify mutation loci, and general linear model(GLM) with SAS 9.1 was used for the association analysis between wool traits and SNPs. Quantitative real-time PCR (qRTPCR) was used to investigate FZD3 gene expression levels. The results showed that six exons of FZD3 gene were amplified and two mutation loci were identified in exon 1 (NC_019459.2: g.101771685 T>C (SNP1)) and exon 3 (NC_019459.2:g.101810848, A>C (SNP2)), respectively. Association analysis showed that SNP1 was significantly associated with mean fiber diameter (MFD) (P=0.04) and live weight (LW) (P=0.0004), SNP2 was significantly associated with greasy fleece weight(GFW) (P=0.04). The expression level of FZD3 gene in skin tissues of the superfine wool (SF) group was significantly lower (P<0.05) than that of the fine wool (F) group. Moreover, it had a higher expression level (P<0.01) in skin tissues than in other tissues of Chinese Merino ewes. While, its expression level had a fluctuant expression in skin tissues at different developmental stages of embryos and born lambs, with the highest expression levels (P<0.01) at the 65th day of embryos.Our study revealed the genetic relationship between FZD3 variants and wool traits and two identified SNPs might serve as potential and valuable genetic markers for sheep breeding and lay a molecular genetic foundation for sheep markerassisted selection (MAS).
Keywords: Chinese Merino sheep (Xinjiang Type), FZD3, single nucleotide polymorphism (SNP), expression pattern,association analysis, wool traits
1. lntroduction
Merino sheep are renowned for producing the finest and softest wool that plays an important role in the agriculture economy. The domestication and breeding of Merino sheep would bring about high natural fiber wool production.And wool generally underpins 21% of commercial return for wool producers (Mu et al. 2017). It is well known that wool productivity and quality are the two main profitinfluencing factors in clothing and textile industries, and its value depends on intrinsic qualities and production traits,especially on mean fiber diameter and fleece weight (Chai et al. 2017; Li W R et al. 2017).
Chinese Merino sheep (Xinjiang Type) was breed through the crossbreeding among Australian Merino rams, Polwarth sheep and Xinjiang fine wool sheep. Since bred in 1985,it has become the main breed of fine wool sheep in China(Di et al. 2011). At present, Chinese Merino sheep are mainly distributed in Xinjiang Uyghur Autonomous Region,China, where a state sheep breeding farm (Xinjiang Gonaisi Fine Wool Sheep Breeding Farm) is located in. Because Chinese Merino sheep has many advantages, such as better spinning quality, relatively larger physique, finer wool and stronger hairy plexus structure, it has improved the incomes of Xinjiang herd.
With continuously updating in wool and fabric processing technologies, the requirements for raw materials are becoming stricter, and then the wool products are developing lighter and softer. So the demands for superfine wool are gradually increasing. In recent years, with breeders paying more attention to the wool quality in Xinjiang, the breeding of merino sheep has been developed from fine to superfine wool strain. The mean fiber diameter of fine wool sheep wool is from 18.1 to 21.0 μm, the mean staple length is more than 9 cm, while the mean fiber diameter of superfine wool sheep is from 16.0 to 18.0 μm, the mean staple length is more than 8.5 cm, both of clean fleece weight (CFW) exceed 3 kg. They are high-quality raw materials for producing the high-grade worsted fabric. The successful cultivation of two types of strains has enriched varieties of Chinese Merino sheep, met for the different needs of sheep improvement in Xinjiang, and laid the basis for higher levels of crossbreeding in the future.
In mammals, Frizzled genes widely expressed in many tissues and organs, such as kidney epithelial induction,feather bud formation, teeth and hair follicle development(van Genderen et al. 1994; Liu et al. 2008; Gao et al. 2009).Especially, it served as receptors of Wnt ligand family through a conserved cysteine-rich Wnt binding domain and encoded seven-pass transmembrane proteins (Yang-Snyder et al. 1996; Sala et al. 2000; Schulte and Bryja 2007). Experimental perturbation of Wnt signaling resulted in aberrant hair formation and structure, thus it was critical in a variety of developmental processes (Millar et al. 1999; Andl et al. 2002; Xing et al. 2018). In addition, several Frizzled genes were specifically expressed at the sites related with Wnt/β-catenin signaling pathway, suggesting that the signaling through alternate Wnt pathways may contribute to the development and function of skin and hair (Romanowska et al. 2009; Choi et al. 2013). Meanwhile, Frizzled genes have been involved in a variety of developmental processes of many organs, such as integument, skeletal system,nervous system and gut (Drewes 2014).
Frizzled3 (FZD3) was firstly identified from Drosophila in 1999 (Sato et al. 1999). The ovine FZD3 gene is located on chromosome 2, spanning around 82.35 kb region on genome. Its coding sequence distributes in six exons intercalated by five introns and encodes one protein isoforms due to the alternative splicing: FZD3-201. Genetic and functional studies have been reported that FZD3 might regulate hair follicle morphogenesis and growth in mouse and human and expressed at several stages during hair follicle development (Hung et al. 2001). Further study showed that FZD3 and FZD6 both controlled proper midbrain morphogenesis in the mouse, but the mechanism was unknown (Stuebner et al. 2010). A study of gene knockout in mice demonstrated that FZD3 also played a crucial role in directing cell migration (Wang et al. 2006b). Multiple lines of evidence indicated that FZD3 was a key gene of hair follicle formation and maintenance, while there was no published report about the FZD3 effect on wool traits in sheep at present. Therefore, we hypothesized that FZD3 gene might exert direct or indirect effects on wool development and growth. To verify this hypothesis, we herein investigate the SNP1 and SNP2 and expression patterns of FZD3 gene and its effects on wool traits in Chinese Merino sheep(Xinjiang Type).
2. Materials and methods
2.1. Ethics statement
All animals used in this study were with the approval of the Ethics Committee of Xinjiang Academy of Animal Science,China. All experiments were conducted strictly according to the guidelines and regulations established by the Ministry of Science and Technology of the People’s Republic of China(Approval No., 2006-398, 30 September, 2006).
2.2. Experimental design and treatments
There were a total of five experiments in the present study. In experiment 1, there were 12 treatments in a 3 (genotype)×2(flock)×2 (age) factorial arrangement for each SNP to clarify the associations of FZD3 with wool traits. In experiment 2, there were two treatments (superfine wool (SF) group and fine wool (F) group) to clarify the difference of mean fiber diameter (MFD) between two groups. In experiment 3, there were two treatments (SF skin tissues and F skin tissues) to clarify the changes of FZD3 expression levels in skin tissues with different MFD. In experiment 4, there were seven treatments (skin, lung, heart, skeletal muscle,kidney, liver, and spleen) to clarify the changes of FZD3 expression levels among different tissues. In experiment 5, there were six treatments to elucidate the change trends of FZD3 expression in different developmental stages. In fact, the six treatments were those of different stages of hair follicle development, including embryos at the day 65, 85,105 and 135 (E65, E85, E105, and E135), and born lambs at the day 7 and 30 (B7 and B30), respectively.
2.3. Animals and feeding
In experiment 1, a total of 628 Chinese Merino ewes (388 ewes were 24 mon old, and 240 ewes were 48 mon old)were randomly selected and the detailed arrangements in 12 treatments were shown in Appendix A. In experiments 2 and 3, 20 ewes were selected according to the measured MFD values of the 388 ewes at the age of 24 mon and divided into two treatments with 10 replicates for each treatment. Among, 10 ewes with the lowest MFD values(averaged (16.21±0.15) μm) were selected as experimental SF group, and another 10 ewes with the highest MFD values(averaged (20.68±0.30) μm) were selected as experimental F group, respectively. In experiment 4, three ewes were ramdomly selected according to the average age of 12 mon and bodyweight of 40 kg, and divided into seven treatments with three replicates for each treatment. In experiment 5, 18 ewes were selected according to the different developmental stages and divided into six treatments, and three replicates were designed for each treatment. Each sheep was a replicate in all of five experiments.
All Chinese Merino sheep (Xinjiang Type) were raised in Xinjiang Gonaisi Fine Wool Sheep Breeding Farm (China).The flock was kept on feeding natural pastures from April to November and feeding manually indoors at other months.The basal diet was formulated to meet the nutriment requirements of Chinese Merino sheep, and contained about 0.5 kg mixed concentrate, 1.5 kg mixed hay, 8 g salt,and 10 g trace element additive per sheep per day during the feeding period. To eliminate environmental effects, all sheep were given the same feed and hay on the same farm.
2.4. Sample collections and preparations
Blood and wool samples (experiments 1 and 2)628 whole blood samples of all Chinese Merino ewes were taken into ethylene diamine tetraacetic acid (EDTA) tubes from the jugular vein. Genomic DNA was extracted from blood samples via the phenol-chloroform method and stored at -20˚C for genotyping. The quantity and purity were determined by NanoDrop 2000 Spectrophotometer (Thermo Scientific, Irvine, CA, USA).
Meanwhile, wool samples from 628 ewes were respectively collected from the left mid-side regions of the sheep body and measured following the standardized methods set by China Fiber Inspection Bureau (CFIB)and International Wool Textile Organization (IWTO). The investigated indexes included mean fiber diameter (MFD),fiber diameter standard deviation (FDSD), coefficient of variation of fiber diameter (CVFD), crimp number (CN per 2.5 cm) and mean staple length (MSL). Besides, live weight(LW) and greasy fleece weight (GFW) were obtained after shearing (Di et al. 2014).
Tissue samples (experiments 3, 4 and 5)In experiments 3 and 5, skin tissues were collected from sheep in vivo.On the side of the left forelimb of the sheep, about 1 palm behind the shoulder blade, scraped the wool, and collected the skin tissues about 2 cm×2 cm with the skin sampler. In experiment 4, after a 12-h fast, three ewes were killed by cutting the carotid arteries, and then were immediately bled.Their skin, lung, heart, skeletal muscle, kidney, liver, and spleen were collected.
All collected tissues were rapidly rinsed twice with PBS,and then the tissue samples were cut into small pieces with sterilized scissors and placed in the cryopreservation tube, and immediately frozen by liquid nitrogen and stored at -80°C until analyzed.
2.5. Sample analysis
PCR amplification (experiment 1)Based on the FZD3 sequences (GenBank accession no. NC_019459.2) in ovine genome assembly v4.0, seven pairs of primers(Table 1) were designed to amplify the six exon regions. The 20-μL PCR volume was consisted of 1 μL of genomic DNA(50 ng μL-1), 0.75 μL of each primer (10 μmol μL-1), 10 μL of 2× PCR Master Mix (1.5 mmol L-1MgCl2included) and 7.5 μL RNase-free ddH2O. PCR was conducted using thermal cycler T-gradient (Biometra, Germany) and the program was as follows: an initial denaturation at 95°C for 3 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at different temperatures for different primers (Table 1) for 30 s,and extension at 72°C for 60 s, and a final extension at 72°C for 5 min. PCR products were electrophoresed in 1.5% agarose gel, stained with nucleic acid dye (Tiangen,Beijing, China), and analyzed under the ultraviolet (UV)illuminator (Bio-Rad, USA).
Genotyping (experiment 1)Because all target fragments were less than 300 bp in length, their polymorphism was investigated using PCR-SSCP. The 7 μL PCR products were mixed with 8 μL formamide loading dye, denatured at 98°C for 10 min, then cooled rapidly on wet ice, loaded onto 12%acrylamide:bisacrylamide (37.5:1) gels and run at 180 V for16 h in 0.5×TBE buffer at 4°C. Once SNPs were determined in referenced samples, they would be included in all the subsequent gels for genotyping. The silver-stained method was used for gel analysis (Dohrmann and Tebbe 2008).

Table 1 Sequences of primers used in PCR amplification for Frizzled3 (FZD3) gene (experiment 1)
Sequencing of allelic variants and sequence analysis(experiment 1)PCR products of FZD3 gene were purified and sequenced by Sangon (Shanghai, China). To minimize the sequencing cost, each banding pattern was randomly selected in triplicate. Nucleotide sequence alignments,translations, and comparisons were carried out using the Megalign module in DNAStar Lasergene 7.1 Software(DNASTAR, Inc., Madison, Wisconsin, USA). The Align Sequences Nucleotide BLAST was used to search sequence homology at NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
RNA extraction and cDNA synthesis (experiments 3,4 and 5)Total RNA was isolated using TRIzol reagent(Invitrogen, Carlsbad, CA, USA) following the manufacturers’instructions. Subsequently, the isolated RNA was incubated with RNase-free DNase I (Qiagen, Hilden, Germany) for 30 min at 37°C to remove DNA contamination. RNA quality was assessed by 260/280 wavelength ratio and 1% agarose gel electrophoresis. A total of 1 μg RNA was reversetranscribed into cDNA using PrimerScriptTMRT Reagent Kit(TaKaRa Bio., Shiga, Japan).
Quantitative real-time PCR (experiments 3, 4 and 5)qRTPCR was performed on a CFX96TM Real-Time System (Bio-Rad, USA) using a SYBR green fluorescence kit (TaKaRa).The volume of reaction system was 25 μL, containing 12.5 μL SYBR Green Mixture, 2 μL template of cDNA, 1 μL of each primer, 8.5 μL distilled water. The PCR program was denaturized at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 60°C for 30 s, and a final cycle of melting curve analysis at 65-95°C for evaluating amplification specificity. The primers of FZD3 (GenBank accession no. XM_004004440.3)(forward primer 5´-GAAAGCCTTGCTGTTTCCTG-3´, reverse primer 5´-CAAGGCAGCAACATCGTAGA-3´) and GAPDH(GenBank accession no. NM_001190390.1) (forward primer 5´-GGTGATGCTGGTGCTGAGTA-3´ and reverse primer 5´-CAGCAGAAGGTGCAGAGATG-3´) were designed with Primer 5.0 Software (PREMIER Biosoft International, California, USA) and synthesized by Sangon.All measurements were analyzed in triplicate and the relative gene expression level was normalized by the GAPDH with 2-ΔΔCtmethod.
2.6. Statistical analysis
In experiment 1, the genotype, allelic frequencies, and Hardy-Weinberg equilibrium (HWE) test were calculated on each identified SNP. The data of each SNP from this experiment were analyzed by three-way ANOVA using the general linear model (GLM) procedure of the SAS 9.2 (SAS Institute Inc., Cary, NC), and treatment comparisons for significant differences were tested by the Bonferroni t-test.Linkage disequilibrium (LD) and haplotype were estimated and constructed by SHEsis platform (http://analysis.bio-x.cn/myAnalysis.php) (Shi and He 2005).
According to the characteristics of the Chinese Merino population, the statistical models were assumed to be Y=μ+G+F+A+G×F+G×A+F×A+G×F×A+e; in which, Y was the phenotype value of each trait per sheep; μ was the overall mean; genotype (G), flock (F) and age (A) were the fixed effects; G×F, G×A, F×A, G×F×A was the interaction of G by F, G by A, F by A and G by F by A, respectively;and e was the residual effect. Furthermore, the pedigree of Chinese Merino sheep in this study was incomplete,population stratification was not taken into account when testing SNP effects.
The effects of additive (a), dominance (d) and allele substitution (α) were estimated using the following equation(Yang et al. 2015): a=(AA-BB)/2, d=AB-(AA+BB)/2, and α=a+d(q-p); in which, AA, AB and BB were the least squared means of the three genotypes, p and q were the allele frequencies of corresponding locus.
The statistical analyses of experiments 2-5 were conducted using SPSS 19.0 (Chicago, IL, USA). All results were shown as mean±SE. The data from experiments 2 and 3 were analyzed by Student’s t-test. The data from experiments 4 and 5 were analyzed by one-way analysis of variance (ANOVA). Duncan test with multiple comparisons method was further used to test the differences between treatment means. Each replicate served as the experimental unit for all statistical analyses. For all tests, P<0.05 or P<0.01 was regarded as statistically significant or highly significant, respectively.
3. Results
3.1. Genetic analyses of SNPs in FZD3 gene on wool traits (experiment 1)
SNPs identification and genotypingThe six exons of ovine FZD3 were successfully amplified in the study, but polymorphisms were only detected in exon 1 (amplied by primer P1) and exon 3 (amplied by primer P4) (Fig. 1-A and B). The two mutations in exon 1 (g.101771685 T>C,SNP1) and exon 3 (g.101810848 A>C, SNP2) were detected through aligning them with the nucleotide sequences of NC_019459.2 in GenBank (https://www.ncbi.nlm.nih.gov/nuccore/NC_019459.2?from=101771243&to=101853593&report=genbank) (Fig. 2-A and B), which lead to the two amino acid changes in FZD3 protein, Phe→Ser and Asn→His. We searhced the two SNPs positions in exon 1 and exon 3 of FZD3 in the database of SNP at NCBI (https://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=780482),but no reference SNP number was found. Therefore, the two SNPs were novel for FZD3 gene. Genotype and allele frequencies of the two mutation loci were calculated and shown in Table 2. In the current analyzed population, both genotype CC and allele C frequenies were dominant at two loci. Moreover, the genotypic distributions of the two SNPs were deviated from Hardy-Weinberg equilibrium (P<0.01).
The effects of genotype, flock and age on wool traits of Chinese Merino sheep for two SNPsSNP1 genotype (G)affected MFD (P=0.04) and LW (P=0.0004), and the sheep with genotype TT had lower MFD and LW (Table 3). Flock(F) affected MFD (P=0.02), MSL (P=0.01), LW (P=0.02)and GFW (P=0.002), age (A) affects all traits except MSL(P<0.05 or P<0.01). The interaction of G×F affected LW(P=0.0013) and GFW (P=0.02), the interaction of G×A affected LW (P<0.001), the interaction of F×A affected CVFD (P=0.0029), MSL (P=0.04), LW (P<0.001) and GFW (P<0.001). The interaction of G×F×A affected LW(P=0.0043) and GFW (P=0.03).

Fig. 1 The PCR-single stranded conformational polymorphism(PCR-SSCP) patterns of primer P1 (A) and primer P4 (B)fragments within the Frizzled3 (FZD3) gene on silver-stained polyacrylamide gel (experiment 1).

Fig. 2 The sequencing maps of two single nucleotide polymorphisms (SNPs) found in Frizzled3 (FZD3) gene (experiment 1). A,NC_019459.2: g.101771685 T>C. B, NC_019459.2: g.101810848 A>C. The box represents the base mutation.
SNP2 genotype (G) affected GFW (P=0.04), and the sheep with genotype AA were superior on GFW compared with other genotypes (Table 4). Flock (F) affected MFD(P=0.03), FDSD (P=0.0002), CVFD (P=0.0011) and LW(P=0.0003), age (A) affects all traits except MSL (P<0.05 or P<0.01). The interaction of G×F affected CN (P=0.04)and LW (P=0.02), the interaction of F×A affected CVFD(P=0.0036), LW (P<0.001) and GFW (P=0.01). The interaction of G×F×A affected LW (P=0.03).
Genetic effects of two SNPs of FZD3 gene on wool traits in Chinese Merino sheepThe dominant effects of SNP1 on MFD (P<0.05) were identified, suggesting that the genetic values of TC individuals were inferior to those of CC and TT individuals in terms of MFD (Table 5). The additive and allele substitution effects were observed onLW at SNP1 and GFW at SNP2 as well (P<0.05 or P<0.01)(Table 5), the results suggested that additive effects were mostly consistent with those of allelic substitution effects.

Table 2 Genotype and allele frequencies and Hardy-Weinberg equilibrium test of single nucleotide polymorphisms (SNPs) in Frizzled3 (FZD3) gene (experiment 1)

Table 3 The effects of genotype, flock and age on wool traits of Chinese Merino sheep for single nucleotide polymorphism 1(SNP1) (experiment 1)1)

Table 4 The effects of genotype, flock and age on wool traits of Chinese Merino sheep for single nucleotide polymorphism 2(SNP2) (experiment 1)1)

Table 5 Genetic effects of two single nucleotide polymorphisms (SNPs) of Frizzled3 (FZD3) gene on wool traits in Chinese Merino sheep (experiment 1)1)
Haplotype analysisTo explore the linkage relationships of two SNPs, r2and D´ were used to measure the LD between them. A weak linkage between two loci (r2=0.3)was confirmed. Haplotypes generally contained more information than single SNP (Ren et al. 2014; An et al. 2018),so we further analyzed the haplotypes of the two SNPs in this study. The frequency of each of the four haplotypes(CA, CC, TA, and TC) was more than 3% (Table 6), and haplotype H2 (CC) had the highest frequency of 35.1%.
3.2. Expression patterns of FZD3 gene (experiments 2, 3, 4 and 5)
FZD3 gene expression levels in skin tissues of Chinese Merino ewes with different fiber diameters (experiments 2 and 3)SF and F groups were significantly difference(P<0.01, Fig. 3-A) on MFD. Further, our data showed that the expression level of FZD3 gene was 3.78-fold lower(P<0.05, Fig. 3-B) in SF than in F, suggesting that FZD3 gene might be contributed to the difference of MFD between the two groups.
FZD3 gene expression levels in different tissues of Chinese Merino ewes (experiment 4)FZD3 gene differentially expressed (P=0.00005) in all the investigated tissues. It highly expressed in liver and shoulder skin,moderately expressed in spleen and heart, and slightly expressed in kidney, lung and skeletal muscle (Fig. 3-C).The results indicated that FZD3 gene played an important role during wool morphogenesis and growth.
FZD3 gene expression levels in skin tissues of embryos and born lambs at different developmental stages(experiment 5)FZD3 gene differentially expressed(P=0.009) at all the six developmental stages detected in this study, with the highest expression level was found in embryos of 65 days, then showed slightly fluctuant expression levels in embryos from the 85th to 135th days,finally the lowest expression values were sustained at thetwo stages of born lambs (Fig. 3-D).

Table 6 Haplotypes constructed with two single nucleotide polymorphisms (SNPs) of Frizzled3 (FZD3) gene (experiment 1)
4. Discussion
Wool production and quality traits are as main economic traits of fine wool sheep, and its several influencing genes have been reported, such as DKK1 (Mu et al. 2017), FST(Ma et al. 2017), KAP 6-3 (Li S et al. 2017), and MTR (Rong et al. 2015). In a previous sheep microarray study of our laboratory (Di et al. 2013), FZD3 gene had been found to be differentially expressed in skin tissues of fine wool sheep and resulted in different fiber diameter. In the present study,we further performed FZD3 gene SNPs detection and association analysis with wool production and quality traits in Chinese Merino sheep.
We amplified the six exon regions of ovine FZD3 and identified two missense mutations, namely, g.101771685 T>C in exon 1 and g.101810848 A>C in exon 3. While no sequence polymorphism was found in the other exons,indicating that the coding sequence in ovine FZD3 might be relatively conserved or our population in this study has similar genetic backgrounds. The association analysis showed that individuals with TT genotype of SNP1 had lower MFD and LW (P<0.05 or P<0.01, Table 3). It is of great significance for artificially selecting economically advantageous traits (such as lower MFD) in Chinese Merino sheep breeding, but we also need to pay attention to the protection of the sheep against LW loss, keep breeding in balance. Dominant effects of SNP1 were observed in MFD(P<0.05, Table 5), indicating that the genetic value of TC is less than that of TT and CC. Therefore, the individuals of TC at SNP1 should be excluded in actual production in terms of MFD. The Chinese Merino sheep with AA genotype at SNP2 had significantly higher (P=0.04, Table 4) GFW than others.Meanwhile, the additive effects and allele substitution effects of SNP1 on LW and SNP2 on GFW were significant (P<0.05 or P<0.01, Table 5), which well demonstrated the individuals obtained advanced phenotype after allele substitution and provided another evidence for the accuracy of additive genetic variance based on phenotypic differences among individuals (Moghaddar and van der Werf 2017).
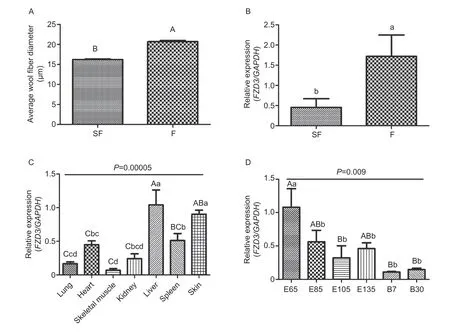
Fig. 3 The expression levels of Frizzled3 (FZD3) gene in the tissues of Chinese Merino sheep (experiments 2, 3, 4 and 5).Housekeeping gene GAPDH was used as an internal control for quantitative real-time PCR (qRT-PCR) analysis. A, the average wool fiber diameter of superfine wool (SF) (n=10) and fine wool (F) (n=10) ewes. B, FZD3 gene expression levels in skin tissues of superfine wool (SF) (n=10) and fine wool (F) (n=10) ewes. C, FZD3 gene expression levels in seven tissues (n=3). D, FZD3 gene expression levels in skin tissues of embryos and after born lambs (n=18). Each value represents mean±standard error (n=3).Bars with different lowercases or uppercases indicate they are significant difference at P<0.05 or P<0.01, respectively.
Because quantitative traits with higher additive effects could be selected in early generations during the breeding process, two SNPs could be used as a molecular marker for early selection in Chinese Merino sheep breeding. As different strains of Chinese Merino sheep had different wool characteristics, a better understanding of the genetic structural variability of candidate genes related with important economic wool traits is helpful to illustrate the genetic control of these complex traits (Gui et al. 2016).Currently, the breeding goals of Chinese Merino sheep are mainly focused on MFD and GFW, but it is necessary to consider other wool traits, such as MSL and LW, for breeding the sheep with fine wool quality, high wool yield, and stable inheritance.
It is well known that follicle determines wool phenotype(Mcdowall et al. 2008). At present, there are many studies on FZD3 in humans and mice. Interestingly, a study showed that FZD3 expressed throughout the epidermis and in the outer cell layers of hair follicle, which was detected in the 7-d-old neonatal skin of mouse (Hung et al. 2001). Further,the mRNA-encoding human FZD3 in epidermal keratinocytes and HaCaT keratinocyte cell lines were identified (Hung et al.2001). FZD3 and FZD6 controlled axonal growth in the central nervous system (CNS) and hair patterning in mouse skin (Wang et al. 2002, 2006a). In our study, FZD3 showed a higher expression level in the skin tissues of Chinese Merino sheep. Therefore, FZD3 gene may play a vital role during hair follicle morphogenesis and formation.
Some previous studies indicated that the embryo growth stages of 65th, 85th, 105th, and 135th days were the important stages for primary follicle formation, secondary follicle formation, secondary-derived follicle and most primary hair follicles, and some secondary hair follicles maturing basically, respectively (Stenn and Paus 2001;Ablat S et al. 2017). To explore the potential role of FZD3 in follicle morphogenesis in Chinese Merino sheep, the expression of FZD3 in skin tissues of embryos and lambs at different growth stages were investigated in this study,and the investigated stages were the 65th, 85th, 105th and 135th days of embryos and the 7th, 30th days of born lambs.It was noteworthy that the expression levels of ovine FZD3 varied in different developmental stages of skin tissues, with peaking (P<0.01) in embryo at the 65th day (Fig. 3-D), which implied that FZD3 gene played important roles during the formation of primary hair follicle in Chinese Merino sheep.Previous studies have demonstrated that KAP8.1 and KAP1.3 exhibited developmental expression patterns in skin tissues of Chinese Merino sheep (Chen et al. 2011). Also,we found FZD3 gene had this specific expression pattern.In total, FZD3 gene might be a key gene responsible for fiber diameter in Chinese Merino sheep.
Accordingly, FZD3 gene may be a potential gene affecting wool traits or the formation of hair follicle. We further demonstrated that two SNPs of FZD3 gene showed significant associations with wool traits in Chinese Merino sheep. The identified SNP may be useful for markerassisted selection for high quality wool in Chinese Merino sheep.
5. Conclusion
In this study, FZD3 gene associations with wool traits and its expression patterns were identified in Chinese Merino sheep. These findings supported that the significant associated SNPs of FZD3 gene could be used in markerassisted selection for the breeding of desired wool traits in Chinese Merino sheep.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31360543 and 31760655), the earmarked fund for the China Agriculture Research System(CARS-39), the Open Project of Key Laboratory of Genetics Breeding and Reproduction of Xinjiang Cashmere and Wool Sheep, China (2016D03017) and the Postdoctoral Science Foundation, China (2017M623287). Sincerely thanks to all who participated in the study.
Appendixassociated with this paper can be available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
 Journal of Integrative Agriculture2019年10期
Journal of Integrative Agriculture2019年10期
- Journal of Integrative Agriculture的其它文章
- Application of virus-induced gene silencing for identification of FHB resistant genes
- Dynamic changes of root proteome reveal diverse responsive proteins in maize subjected to cadmium stress
- Strategies to enhance cottonseed oil contents and reshape fatty acid profile employing different breeding and genetic engineering approaches
- Maize/peanut intercropping increases photosynthetic characteristics,13C-photosynthate distribution, and grain yield of summer maize
- Rhizosphere soil bacterial community composition in soybean genotypes and feedback to soil P availability
- Effect of biochar on grain yield and leaf photosynthetic physiology of soybean cultivars with different phosphorus efficiencies
