Rhizosphere soil bacterial community composition in soybean genotypes and feedback to soil P availability
ZHOU Tao, WANG Li, DU Yong-li, LlU Ting, Ll Shu-xian, GAO Yang, LlU Wei-guo, YANG Wen-yu,
1 College of Agronomy, Sichuan Agricultural University, Chengdu 611130, P.R.China
2 Key Laboratory of Crop Ecophysiology and Farming System in Southwest, Ministry of Agriculture, Chengdu 611130, P.R.China
Abstract Soil with low phosphorus (P) availability and organic matter contents exists in large area of southwest of China, but some soybean genotypes still show well adaptations to this low yield farmland. However, to date, the underlying mechanisms of how soybean regulates soil P availability still remains unclear, like microbe-induced changes. The objective of the present study was to compare the differences of rhizosphere bacterial community composition between E311 and E109 in P-sufficiency (10.2 mg kg-1) and P-insufficiency (5.5 mg kg-1), respectively, which then feedback to soil P availability.In P-sufficiency, significant differences of the bacterial community composition were observed, with fast-growth bacterial phylum Proteobacteria, genus Dechloromonas, Pseudomonas, Massilia, and Propionibacterium that showed greater relative abundances in E311 compared to E109, while in P-insufficiency were not. A similar result was obtained that E311 and E109 were clustered together in P-insufficiency rather than in P-sufficiency by using principal component analysis and hierarchical clustering analysis. The quadratic relationships between bacterial diversity and soil P availability in rhizosphere were analyzed, confirming that bacterial diversity enhanced the soil P availability. Moreover, the high abundance of Pseudomonas and Massilia in the rhizosphere of E311 might increased the P availability. In the present study, the soybean E311 showed capability of shaping rhizosphere bacterial diversity, and subsequently, increasing soil P availability. This study provided a strategy for rhizosphere management through soybean genotype selection and breeding to increase P use efficiency, or upgrade middle or low yield farmland.
Keywords: soybean, low P, rhizosphere, bacterial community, soil P availability
1. lntroduction
Phosphorus (P) is one of the essential and key limiting nutrients for crop growth in agroecosystems (Richardson and Simpson 2011). Low soil P limits crop productivity on 30-40% of the world’s arable land (von UexkÜll and Mutert 1995). Additionally, of the applied fertilizer P each season,crop uptake is only 10-25% (Johnston et al. 2014), as P is strongly bound to soil particles (Hinsinger 2001; Shen et al. 2011).
Roots in low P soils show highly morphological and physiological plasticity to increase the capacity of P acquisition (Lambers et al. 2006; Shen et al. 2011), like increasing the root surface area contact with soil, or increasing carboxylate, proton, and phosphatase exudation for releasing soil-bounded P (Rengel and Marschner 2005;Pang et al. 2010; Wang et al. 2010; Mehra et al. 2017).These adaptations of root morphology and physiology are considered to be strategies for improving the P-mining capacity of the root system, which have been effectively elucidated. Besides, soil microorganisms are integral to the P cycle and play an important role in the mobilization of poorly soluble P and then subsequently increasing the amount of available P for the acquisition of the plant(Richardson et al. 2011; Richardson and Simpson 2011).The mechanisms of microorganisms enhancing the soil P availability can be summarized as microbial turnover that are effective in increasing net transfer of orthophosphate ions into soil solution or facilitating the mobility of organic P (Seeling and Zasoski 1993), and through induction of metabolic processes that are effective in directly solubilizing and mineralizing P from sparingly available forms of soil inorganic and organic P (Richardson et al. 2009). The soil bacterial communities are influenced by the presence of roots, in the rhizosphere, generally bacterial communities that are less diverse (Marilley et al. 1998; Marilley and Aragno 1999) and of greater size (Lynch and Whipps 1990;Semenov et al. 1999) than those associated with root-free soil. Because carbon availability often limits microbial growth in soils and the plant-derived carbon as a labile pool is preferentially utilized by microorganisms than other rhizosphere carbon pools (Dennis et al. 2010; Ramirez-Villanueva et al. 2015). Thus, it is important in developing strategies for rhizosphere management through plantmicroorganism interactions to increase P use efficiency and upgrade middle- or low-yield farmland.
Soybean (Glycine max L.) is widely cultivated all over the world, has at least 3 000-year cultivation history in China (Hymowitz 1970). As a historic crop, it is not only served as food for humans and animals, but also has an ecological value in agroecosystems, with roles of improving soil P availability, nitrogen fixation, and the sequestration of soil carbon (Salvagiotti et al. 2008; Liu et al. 2011; Cong et al. 2015). Commonly, the capacity of legume plants in mobilizing sparingly soluble soil P is higher than cereal crops (Houlton et al. 2008; Shen et al. 2013). However,the capacity of soybean in increasing soil P availability by root morphological and physiological adaptations is lower than most leguminous plants, such as faba bean and white lupin (Lyu et al. 2016). Interestingly, soybean also shows good adaptations to P-deficient soil, with the values of Olsen-P above 8.8 mg kg-1didn’t lead any yield increasing in southwest of China (Zhou et al. 2015). The critical value of soil Olsen-P for soybean growth is lower than most crops, with the Olsen-P values of 20 mg kg-1considered as threshold for optimal plant growth in China (Li et al. 2011).Furthermore, southwest is one of the three major soybean production regions in China, where the low P and organic matter soils exist in a large area (He 2003). A number of studies have demonstrated that soybean strongly influences the composition of the bacterial community structure in the rhizosphere soil with deficient carbon and P (Liu et al. 2011).Thus, we assumed that there are other strategies besides the adaptations of soybean root morphology and physiology to P deficiency, such as changing the composition of bacterial community in the rhizosphere to improve the soil P availability and then subsequently increase the capacity of P acquisition in soybean. Most of the studies investigated the microbial community composition in the rhizosphere of soybean responded to soil type (Xu et al. 2009), soybean genotype (Wang et al. 2014), growth stage, and soil P status(Liu et al. 2011), but soybean root drives bacterial community composition change in rhizosphere then feedbacks to soil P availability is still unclear.
The main purpose of this study described here was to investigate the bacterial community composition in the rhizosphere of E311 in low P and organic matter soil condition, which then feedbacks to soil P availability.The insights gained from this study will contribute to our understanding of soybean adapting to P-deficiency through plant-microbial interactions in agricultural soils with deficient P and organic matter.
2. Materials and methods
2.1. Plant materials and soil preparation
In the past years, we collected hundreds of soybean genotypes for evaluating the genotypic differences in P efficiency in southwest of China, fortunately a few genotypes (like E311) showed well low P adaptations (Zhou et al. 2016). In the subsequent hydroponic experiments,the results showed that P acquisition and root carbon exudation of soybean genotype E311 were higher than that of E109 (Appendix A). The rhizosphere microorganisms preferentially utilize the label carbon such as the root exudation, so the two soybean genotypes were used in this study to investigate the plant-microbial interactions, and then subsequently affected soil P availability. Moreover,E311 and E109 showed similar agronomic characters and growth period (Table 1).
Ochric Aquic Cambosol soil was obtained from the surface layer (0-20 cm) of farmland in Wenjiang, Sichuan Province, China and was air-dried and sieved through a 2-mm mesh screen. The P (KH2PO4) was applied to thesoil in prepared pots (15 L, 15 kg soil) at the rates of 10 and 100 mg P kg-1soil. In addition, 50 mg K kg-1soil (KCl)and 50 mg N kg-1soil (CH4N2O) were also added to the soil. The soil was manually mixed and homogenized for 4 weeks. The basic properties of the soil after homogenization were as follows: organic matter of 8.3 g kg-1, available N of 100 mg kg-1, available K of 102 mg kg-1, pH of 6.73, Olsen-P of 5.5 mg kg-1(P-insufficiency), and 10.2 mg kg-1(P-sufficiency),54% sand, 12% clay, and 34% silt. In our previous study,soil Olsen-P values of higher than 8.8 mg kg-1didn’t lead to any increasing in the soybean yield in southwest of China(Zhou et al. 2015), thus the Olsen-P value of 10.2 mg kg-1is sufficient for soybean growth.

Table 1 The agronomic characters and growth period of E311 and E109
2.2. Experimental design
The pot experiment was a randomized complete block design with a 2×2 factorial arrangement of treatments. The factors were two soil P levels (P-sufficient and P-insufficient)and two soybean genotypes (E311 and E109). There were six replicates for each treatment, and each plant had a separate pot.
2.3. Growth conditions
The pots were arranged in the field of the Sichuan Agricultural University at Wenjiang, Sichuan from 20 June to 28 August 2016, and the pots were covered with black plastic to prevent rainfall and direct sunlight. This experiment was designed to simulate the plant growth environmental conditions maximally as in the field, the 10-daily average temperature and sunshine duration during the experimental period are shown in Fig. 1. Similar sizes of each seed were selected before sown. Except for the fertilizer application in the soil prepared stage, none of the pots received any additional applications of fertilizer, and they were manually irrigated to the field capacity and well weeded.
2.4. Soil and plant sampling

Fig. 1 Sunshine duration and average air temperature during soybean growth period, from 20 June to 28 August 2016.
After 78-day growth, three replications soybean plants were harvested at the flowering stage. Plants were gently removed from the soil to collect their roots. The shoots were oven-dried at 105°C for 30 min and at 80°C for 3 days to measure the dry weight and concentrations of P. The bulk soil was collected near the roots, and the rhizosphere soil that tightly adhered to the roots was collected by brushing(Tang et al. 2014). Rhizosphere soil samples were frozen for molecular analyses and were stored at -20°C for 6 months until the DNA was extracted. The subsamples were air-dried for the Olsen-P analysis. After the soil was collected, the whole root system was oven-dried at 105°C for 30 min and at 80°C for 3 days to measure the dry weight and concentrations of P. In another three replications, the whole root systems were gently removed from the soil,and then thoroughly washed with running water followed by distilled water immediately. The roots were separately placed in 250-mL beakers filled with deionized water and covered with black plastics to prevent light degradation of exudations. The plants were placed in a greenhouse and the details of the controlled environment chambers reference Zhou et al. (2016). After 6 h collection, 50-mL subsamples were immediately freeze-dried at -80°C, the residue white powder was dissolved in 1 mL of deionized water for plant exudation carbon analysis.
2.5. Chemical and biochemical analyses
Air-dried soil samples were passed through a 2.0-mm mesh sieve. A 2.5-g soil sample was shaken with 25 mL of 0.5 mol L-1NaHCO3at 25°C for 30 min, and after the suspension was filtered, the P concentration of the filtrate was determined according to the molybdate ascorbic acid method (Murphy and Riley 1962).
All of the plant samples were ground through a 0.149-mm mesh sieve. A 0.3 g-sample was wet-digested with concentrated H2SO4and H2O2(30%), and the P was determined by the vanadomolybdate method (Page 1982).
2.6. Determination of carbon in root exudation
The 0.5-mL concentrated sample was diluted to 5 mL,which was analysed for plant exudation carbon by the TOC Analyser (vario TOC, elementar, Inc., Germany).
2.7. DNA extraction and lllumina sequencing
Total soil genomic DNA from 0.5 g of soil samples was directly extracted using the FastDNA®Spin Kit (MP Biomedicals, Santa Ana, USA), according to the manufacturer’s instructions.Partial bacterial 16S rRNA genes were amplified using the primer set 338F (5´-ACTCCTACGGGAGGCAGCAG-3´)and 806R (5´-GGACTACHVGGGTWTCTAAT-3´) (Xu et al.2016). The PCR reactions were carried out using TransGen AP221-02 (TransStart Fast Pfu DNA Polymerase, China)and an ABI GeneAmp®9700. Amplification was assessed by analysing 10 μL of product on a 2% agarose gel. Samples with an intense major band between 300 and 500 bp were chosen for further experiments. Sequencing libraries were generated using NEB Next®Ultra™ DNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s instructions, and index codes were added. The library quality was assessed using the QuantiFluor™-ST Blue Fluorescence Quantitative System (Promega, USA), and the library was sequenced using the Illumina MiSeq platform at Majorbio Bio-Pharm Technology Co., Ltd., Shanghai,China, according to standard protocols. Specifically, Raw fastq files were demultiplexed, quality-filtered using FLASH with the following criteria: (i) The reads were truncated at any site receiving an average quality score <20 over a 50-bp sliding window. (ii) Primers were exactly matched allowing a nucleotide mismatching, and reads containing ambiguous bases were removed. (iii) Sequences whose overlap longer than 10 bp were merged according to their overlap sequence.
2.8. OTU cluster and diversity analysis
Operational taxonomic unit (OTU) clustering, species annotation, and diversity analysis were conducted as described by Wang et al. (2012). OTUs were assigned using Usearch (version 7.1 http://drive5.com/uparse)with a threshold of 97% pairwise identity and refer to the database of Silva (http://www.arb-silva.de). The normalized OTU abundance information was utilized for alpha (single sample analysis) and beta (multivariate sample community)diversity analyses. Metrics of community richness included the Chao1 and ACE estimators. Community diversity was assessed using the Shannon and Simpson indices.Coverage, indicating sequencing depth, was evaluated using Mothur version v.1.30.1 (Schloss et al. 2011).
2.9. Statistical analysis
For beta diversity, principal component analysis (PCA) was applied for the multivariate examination of OUT (Wang et al.2012), redundancy analysis (RDA) (Sheik et al. 2012) was performed in R (http://cran.r-project.org/) using the vegan package with normalized OTU abundance and environmental chemical data (DOC). The Bray-Curtis dissimilarity(Srinivasan et al. 2012) was calculated using the R package(Jami et al. 2013). Data from the three replicates were sorted by the Excel (Microsoft) Software packages. Analysis of variance (ANOVA) was conducted using SPSS 19.0 Software (SPSS Institute Inc., USA). Plant growth and soil P concentration were subjected to a one-way ANOVA to assess the effects of soybean genotypes and soil P conditions. The difference of relative abundance of bacterial phylum and genus between E311 and E109 were subjected to Tukey’s test by SPSS 19.0 Software (SPSS Institute Inc., USA).
3. Results
3.1. lllumina sequencing and rhizosphere soil bacteria community structure analysis
After denoising and chimera, we obtained 446 359 sequences, which were then clustered into 3 635 OTUs with 97% sequence similarity. The numbers of OTUs detected for the tested treatments “E311 P-sufficiency”,“E311 P-insufficiency”, “E109 P-sufficiency”, and “E109 P-insufficiency” were 2 735, 2 403, 2 627, and 2 330,respectively (Table 2). The number of obtained OTUs was slightly lower than estimated based on Chao1 and ACE,but the values were close to the estimates. The values of Simpson and Shannon community diversity indexes were 0.0048-0.0136 and 6.31-6.50, respectively. The value of coverage was higher than 0.9719, indicates sufficient sequencing depth. These above results indicated that the sequencing data were reasonable and the number of analysed sequences can sufficiently represent the bacteria community diversity of all samples.
The rhizosphere soil bacteria community structures at the phylum and genus level in the different treatments were shown in Figs. 2 and 3, respectively. The bacteria in the four rhizosphere soil samples belonged to more than 16 phylum(Fig. 2). The Proteobacteria was the dominant bacterial phylum, with values higher than 31.19% (Fig. 2). The four rhizosphere soil samples belonged to more than 40 bacteria genus, including Dechloromonas and Propionibacterium(Fig. 3).

Table 2 Comparison of the coverage, estimated operational taxonomic unit (OTU) richness, and diversity indexes in high and low soil P level at a genetic distance of 3%
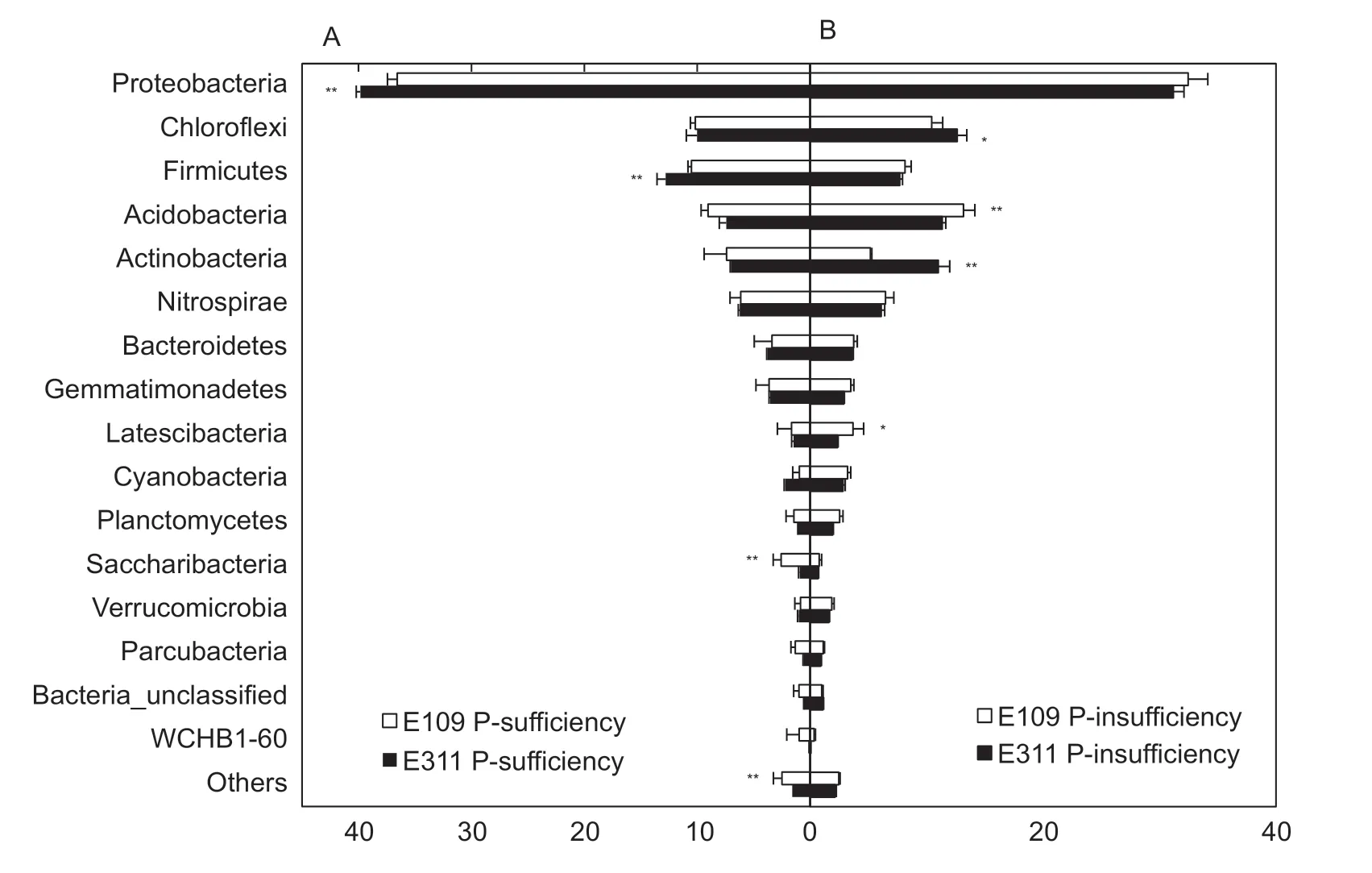
Fig. 2 Relative abundances of bacterial phyla in the rhizosphere soil with P-sufficiency (A) and P-insufficiency (B). Data are averages of three replicates and bars represent standard errors. ** and * indicate significant difference at P<0.01 and P<0.05 by Tukey’s test, respectively.
3.2. Diversity analysis among different treatments
As shown in Fig. 2, the relative abundances of bacteria phylum differed in the treatments. In P-sufficiency, E311 was shaped a relative higher abundance of Proteobacteria and Firmicutes than E109 (Fig. 2-A). However, there was a slight difference in the relative abundance of Proteobacteria and Firmicutes between E311 and E109 in P-insufficiency(Fig. 2-B). In P-sufficiency, of the total bacteria sequences were found on the E311, 9.59% belonged to Bacillus and 6.28% to Nitrospira (Fig. 3-A). Similarly, Bacillus (9.71%)and Nitrospira (6.10%) were the dominant bacteria taxa found on E109 in P-sufficiency (Fig. 3-A). On the contrary, the dominant bacteria communities on E109 and E311 in P-insufficiency differed from that in P-sufficieny:Nitrospira and Anaerolineaceae_uncultured dominant on E109 and E311 (Fig. 3-B). Furthermore, Dechloromonas,Pseudomonas, Massilia, and Propionibacterium on E311 were higher than E109 in P-sufficiency, but a slight difference in P-insufficiency (Fig. 3). The above results were confirmed by PCA (Fig. 4-A) and hierarchical clustering analysis(Fig. 4-B) based on the beta diversity of each bacteria community. As shown in Fig. 4-A, based on the first principal component, the samples in two P treatments were clearly separated, with the E311 and E109 in P-sufficiency being separated in P-insufficient condition. In addition,E311 and E109 in P-sufficiency were separated along the second principal component, whereas E311 and E109 in P-insufficiency were not (Fig. 4-A). A similar result was obtained by using hierarchical clustering analysis, with E311 and E109 were grouped together in P-insufficiency but independently in P-sufficiency (Fig. 4-B). These results indicated that the bacteria diversity in rhizosphere soil was highly soil P and soybean genotypes specific.

Fig. 3 Relative abundances of bacterial genus in the rhizosphere soil with P-sufficiency (A) and P-insufficiency (B). Data are averages of three replicates and bars represent standard errors. ** and * indicate significant difference at P<0.01 and P<0.05 by Tukey’s test, respectively.
To further investigate the common relationship among bacteria communities of soybean genotypes in different soil P conditions, the OTU distribution was statistically analysed, and these results are presented as a Venn diagram in Fig. 5. For E311 and E109 in P-sufficiency,2 949 OTUs (141+2 506+48+254) were amplified; 2 678 OTUs (119+2 506+48+105) were amplified for E311 and E109 in P-insufficiency, indicating more abundant bacteria community diversity in P-sufficiency.
3.3. Relationship between bacteria diversity and rhizosphere soil P availability
The present study also investigated the soil P availability in rhizosphere and bulk soil. As shown in Fig. 6, the soil P availability in the rhizosphere was higher than that in bulk soil (Fig. 6-A and B). In P-insufficiency, the soil P availability in the rhizosphere or bulk soil of E311 and E109 were not significantly different (Fig. 6-B). In contrast, the soil P availability in the rhizoshpere of E311 was higher than E109 in P-sufficiency (Fig. 6-A). To further investigate the relationships between rhizosphere soil bacteria community diversity and P availability, the regression analyses were conducted (Fig. 7). The relationship between rhizosphere soil availability value and the index of community richness(ACE, Chao1) and OTUs indicated that quadratic models fit the experimental data well (Fig. 7-A). The community richness indexes and OTUs increased with soil P availability values based on a quadratic relationship with high determination coefficients (R2>0.99).
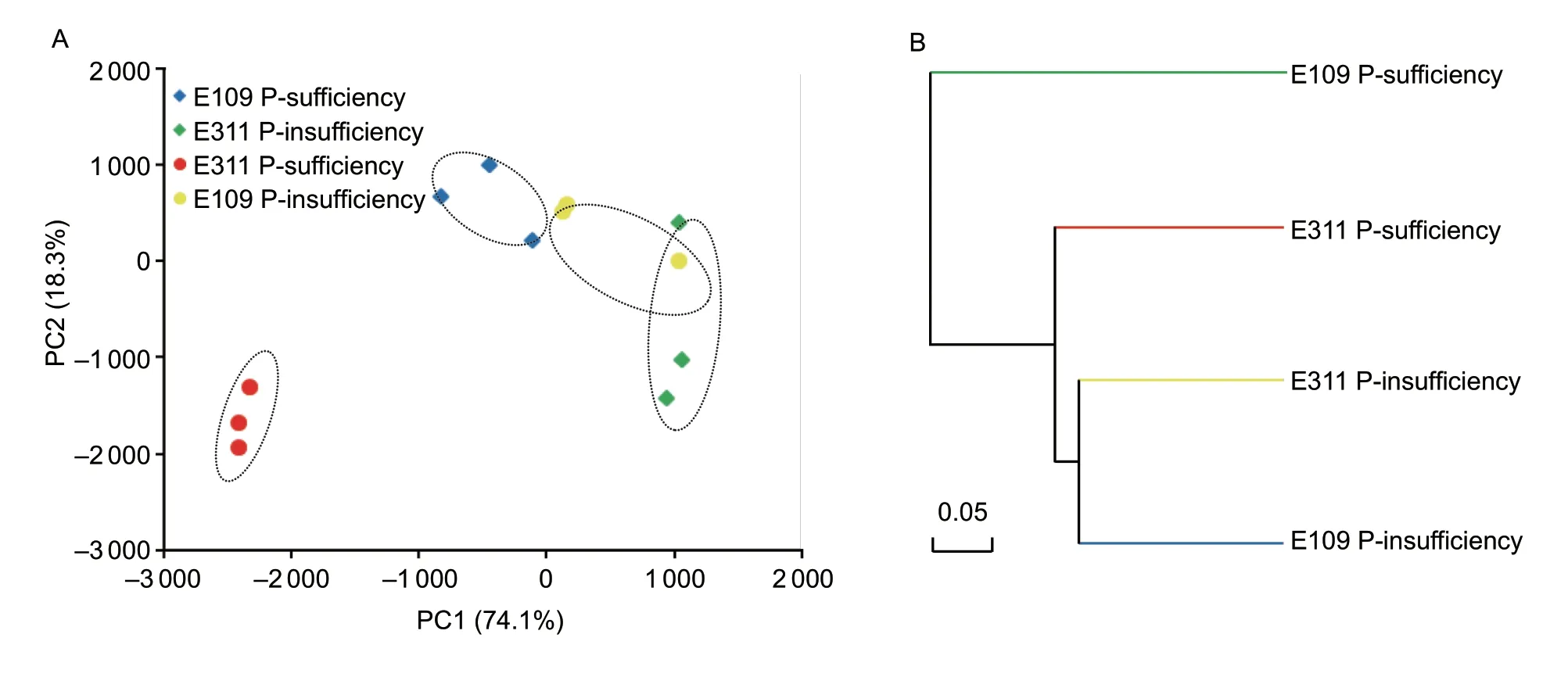
Fig. 4 Principal component analysis (A) and hierarchical clustering analysis (B) of different treatments based on their beta diversity of bacterial community composition. The dotted line frame with three points were the three replicates of the treatments.

Fig. 5 Venn diagram of OTU distribution among different treatments.
Regression analyses were also conducted between rhizosphere soil P availability and the community diversity indexes (Shannon, Simpson). The results shown in Fig. 7-B,with the increasing in rhizosphere soil P availability, the Shannon community diversity index increased based on a quadratic relationship (R2=0.9989), and the Simpson community diversity index decreased based on a quadratic relationship with high determination coefficients (R2=0.9144).
3.4. Plant growth, plant P concentration, and P uptake capacity
Shoot and root dry weights of E311were higher than E109 in P-insufficiency, but those of E311 and E109 were not significantly different in P-sufficiency (Fig. 8-A and B). The ratio of root dry weight:shoot dry weight was greater in plants grown in P-insufficiency, and that of E311 was higher than E109 irrespective of P supply (Fig. 8-C). The shoot and root of E311 and E109 had greater P concentrations in P-sufficiency than that in P-insufficiency. The shoot and root P concentrations of E311 were higher than those of E109 in P-sufficiency (Fig. 9-A and B). However, the shoot and root P concentrations of E311 and E109 were not significantly different in P-insufficiency (Fig. 9-A and B). Consequently,the plant had greater P uptake in P-sufficiency than that in P-insufficiency (Fig. 9-C). Furthermore, E311 showed higher P uptake than E109 irrespective of P supply.
4. Discussion
4.1. The rhizosphere soil bacterial community composition differed between soybean genotypes

Fig. 6 The value of soil Olsen-P in the rhizosphere and bulk soil of E311 and E109 (A, P-sufficiency; B, P-insufficiency). P-sufficiency means of the value of soil Olsen-P was 10.2 mg kg-1 in the initial soil, P-insufficiency means of the value of soil Olsen-P was 5.5 mg kg-1 in the initial soil. Data are averages of three replicates and bars represent standard errors. Data with different letters are significantly difference at the 5% level by LSD between two soil P levels and two soybean genotypes.

Fig. 7 Quadratic regression analysis between the soil P availability and bacterial diversity indexes. A, community richness and the values of soil Olsen-P. B, community diversity and the values of soil Olsen-P.
Studies have demonstrated that the bacterial community composition in the rhizosphere was very similar among soybean genotypes (Wang et al. 2009; Xu et al. 2009),because of the soil type and P were dominate factors in determining microbial communities (Singh et al. 2007; Xu et al. 2009). The data in the present study correspond well with the results reported before soil P determines bacterial communities. The results of principal component analysis (PCA) and hierarchical clustering analysis clearly certificated that the bacterial community composition of soybean genotypes could be categorized into P-sufficiency and P-insufficiency (Fig. 4-A and B). Except for soil P, PCA also clearly demonstrated that the bacteria community composition could be categorized into “E311 P-efficiency”and “E109 P-efficiency” (Fig. 4-A). This finding was consistent with previous reports that the soybean genotype was also the major factor in determining microbial community in low clay and organic matter content soil (Liu et al. 2011), as well as the initial soil clay (12%) and organic matter content (8.3 g kg-1) were low in the present study.The impact of plant genotype on bacterial communities varies with compounds supplied from roots, thus different compositions of root exudates were expected to select different rhizosphere communities (Benizri et al. 2002;Baudoin et al. 2003). Usually, root exudates did observe a remarkable effect on fast-growth bacterial communities(e.g., Proteobacteria) but influenced a small portion of the total bacterial communities. A number of studies have been approved that member of Proteobacteria (copiotrophic)bacteria such as Enhydrobacter, Dechloromonas,Propionibacterium, Massilia, and Burkholderia contribute to the turnover of root exudates (Lian et al. 2017). These bacteria could utilize monosaccharides, ribose, glucose,sucrose, amino acids, and fatty acids, which are components of root exudates (Benz et al. 1998; Tamura et al. 2010;Kawamura et al. 2012). In P-sufficiency, the relative abundance of Proteobacteria bacteria in the rhizoshpere of E311 was higher than that in E109 (Fig. 2), which was consistent with the results that root-exude carbon of E311 was higher than E109 (Appendix B). The capability of translocation of carbon to root in E311 plants was higher than E109, as the increasing ratio of root to shoot biomass(Fig. 8-C). Furthermore, in the RDA analysis, E311 and E109 clustered separately in P-sufficiency and the DOC was an important environmental factor which influenced the bacterial community composition (Appendix C). The increased DOC in the rhizosphere of E311 was caused by increasing of plant-exude carbon, because the DOC content was positively correlated with root-exude carbon (Appendix B). Additionally, the genus of Ropionibaterium, Massilia, and Dechloromonas in the treatment of “E311 P-sufficiency” was higher than that in “E109 P-sufficiency” (Fig. 3-A), which perhaps contributed to the turnover of the root exudates(Lian et al. 2017). The results provided the evidence of certification that E311 and E109 shaped rhizosphere soil bacterial community composition differently in the present study environment.

Fig. 8 Effects of soil P level on soybean dry matter accumulation (A, shoot; B, root) and distribution (C) between root and shoot.P-sufficiency means of the value of soil Olsen-P was 10.2 mg kg-1 in the initial soil, P-insufficiency means of the value of soil Olsen-P was 5.5 mg kg-1 in the initial soil. Data are averages of three replicates and bars represent standard errors. Data with different letters are significantly difference at the 5% level by LSD between two soil P levels and two soybean genotypes.
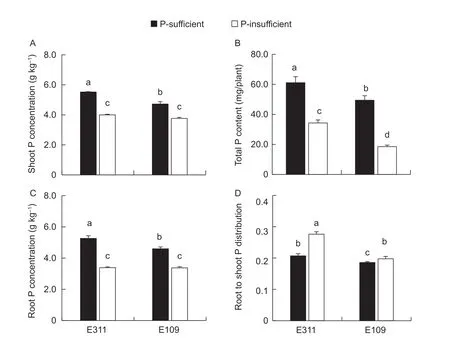
Fig. 9 Effects of soil P level on soybean P status (A, shoot P concentration; B, root P concentration; C, total P content; D, root to shoot P distribution). P-sufficiency means of the value of soil Olsen-P was 10.2 mg kg-1 in the initial soil, P-insufficiency means of the value of soil Olsen-P was 5.5 mg kg-1 in the initial soil. Data are averages of three replicates and bars represent standard errors.Data with different letters are significantly difference at the 5% level by LSD between two soil P levels and two soybean genotypes.
4.2. Rhizosphere soil bacterial community composition changes feedback to soil P availability
Plant-microbe interactions improve soil P availability through microbe-induced changes in the rhizosphere involving in soil pH and the releasing of exudate that solubilize inorganic P strongly absorbed to soil (Kirk et al. 1999) or release organically bound P (Li et al. 2007). Furthermore,the microbe is a more efficient P pool for plants than a single pulse of fertilizer P in the rhizosphere (Bünemann et al. 2011), and all of the MBP are potentially available to plants over long term (Richardson and Simpson 2011).In P-sufficiency, the values of Olsen-P in the rhizosphere of E311 were higher than those in E109 (Fig. 8-A), while that was not in P-insufficiency (Fig. 8-B). Coincidentally,the rhizosphere soil bacterial community diversity of “E311 P-efficiency” was higher than “E109 P-efficiency”, whereas E311 and E109 in P-insufficiency were not (Figs. 2-4). Thus,these results imply that bacterial community composition changes maybe feedbacks to soil P availability. To confirm this hypothesis, we analysed the relationship between soil P availability and bacterial diversity indexes (Fig. 7). The results revealed a close relationship between bacterial diversity and soil P availability. In particular, there were high determination coefficients (R2>0.99) in the quadratic relationships between the community richness indexes and soil P values. With increasing in the bacterial community richness indexes, the soil P availability increased sharply.
Besides, more evidence showed that a close relationship between bacterial community composition and soil P availability was existent. It was reported that Proteobacteria was a typically representative bacteria phyla involved in soil P cycling (Wakelin et al. 2012; Tang et al. 2016), which showed a positive response to soybean genotypes in this study, with the data of relative abundance of Proteobacteria of E311 (Fig. 2-A), as well as soil P availability was higher than E109 in P-sufficiency (Fig. 6-A). Interestingly, in genus level, the relative abundance of Pseudomonas and Massilia of E311 P-sufficiency were higher than that of E109 P-efficiency (Fig. 3-A), but that in P-insufficiency were not(Fig. 3-B). These bacterial species were reported to be able to solubilize inorganic P in soil (Richardson and Simpson 2011). These results provided evidence in favour of the hypothesis that rhizosphere bacteria community (E311)changes feedback to soil P availability. Although the soil P availability and bacterial community composition showed a well relationship, but it did not automatically approve that soil P availability was only affected by bacteria. Thus more work need to do, like specials in a dominant group of bacteria,such as phosphate-solubilizing bacteria.
Increasing soil P availability through plant-drive soil microorganism is considering as an efficient strategy to improve P-mining capacity of the root system. The plant tissue (shoot and root) P concentration of E311 was higher than E109 in P-sufficient treatment, but that in P-insufficiency was not (Fig. 9-A and B). These results corresponded well with the values of rhizosphere Olsen-P of E311 and E109 in P-sufficiency and -insufficiency (Fig. 6-A and B). These results provided evidence to the certification that rhizosphere bacteria community changes resulted in increasing soil P availability, which stimulated the capability of soybean P uptake.
5. Conclusion
Our results implicated soybean genotypes E311 growing in low P (10.2 mg kg-1) and low organic matter (8.3 g kg-1) soil have capability of sharping specific soil bacterial community composition, with the data of the high relative abundance of phylum Proteobacteria, genus Dechloromonas,Pseudomonas, Massilia, and Propionibacterium. The bacterial community composition changes may be advantageous to increasing soil P availability. Additionally,ameliorative soil P availability stimulated the capability of soybean P uptake. This study provided new insights into root-rhizosphere interactions of soybean growing in the soil with low P availability and low organic matter,which is important in developing strategies for rhizosphere management through soybean genotype selection and breeding to increase P use efficiency, or upgrade middle or low yield farmland.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2016YFD0300109-3) and the National Natural Science Foundation of China(31671626, 31771728). The authors also wish to thank Gan Jianyang (Sichuan Agricultural University) who participated in the chemical analysis.
Appendicesassociated with this paper can be available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
 Journal of Integrative Agriculture2019年10期
Journal of Integrative Agriculture2019年10期
- Journal of Integrative Agriculture的其它文章
- Application of virus-induced gene silencing for identification of FHB resistant genes
- Dynamic changes of root proteome reveal diverse responsive proteins in maize subjected to cadmium stress
- Strategies to enhance cottonseed oil contents and reshape fatty acid profile employing different breeding and genetic engineering approaches
- Maize/peanut intercropping increases photosynthetic characteristics,13C-photosynthate distribution, and grain yield of summer maize
- Effect of biochar on grain yield and leaf photosynthetic physiology of soybean cultivars with different phosphorus efficiencies
- lnheritance of steroidal glycoalkaloids in potato tuber flesh
