3-乙基-2-乙酰吡嗪缩N(4)-(对甲苯)氨基硫脲Gaバ、Inバ配合物的合成、晶体结构和DNA结合性质
薛文招 赵晓雷*, 闫雪雪 张雪洁 吴伟娜 王 元 陈 忠
(1河南理工大学化学化工学院,河南省煤炭绿色转化重点实验室,焦作 454000)(2江西科技师范大学材料与机电学院,南昌 330013)
In the past few decades,thiosemicarbazones(TSCs)and their metal complexes have been a focus of chemists and biologists because of their wide range of pharmacological effects,such as antibacterial,antiviral,antifungal,and,most intriguingly,antitumor activity[1-4].It has been demonstrated that biological activities of the thiosemicarbazones are closely related to the parent aldehyde or ketone group,aminoterminal substitution and metal chelation ability[5-8].The presence of a heterocyclic ring or a bulky group at the terminal nitrogen in the synthesized TSCs plays major roles in extending their pharmacological properties[1].Considering the excellent antitumor activity of the TSCs bearing pyrazine ring against K562 cell line[9],the ligand derived from 3-ethyl-2-acetylpyrazine and N(4)-(p-tolyl)thiosemicarbazide, is selected in this work.On the other hand,extensive research in the quest for drugs bearing improved therapeutic indexes and wider activity spectra has led to metal coordination and organometallic compounds that show interesting antitumor activities[1,10-11].In particular,the interest in galliumバ and indiumバ coordination chemistry has evolved from this approach[12-14].For instance,a prototypic,highly cytotoxic galliumバcomplex with 2-acetylpyridine N,N-dimethylthiosemicarbazone has been reported[15].Further research proved that coordination of galliumバto organic ligands could improve the antimicrobial action of galliumバby increasing its lipophilicity and bioavailability.In most case of TSCs,metal-ligand synergism could occur[16].On the contrary,the indiumバcomplexes with TSCs are relatively scarce,one reason may be that the crystals suitable for X-ray diffraction study of these compounds have been difficult to obtain[14].
Taking into account this background and while searching for bioactive compounds,two complexes,namely[Ga(L)2]NO3·4CH3OH (1)and[In(L)2]NO3·1.75CH3OH(2)(HL=3-ethyl-2-acetylpyrazine N(4)-(ptolyl)thiosemicarbazone),were synthesized and characterized by X-ray diffraction methods.In addition,the biological potentiality of this series of compounds has been explored by evaluating their cytotoxicity on three human tumor cell lines.
1 Experimental
1.1 Materials and physical measurements
All starting materials were obtained commercially and used as received.Elemental analyses were carried out on an Elemental Vario EL analyzer.The IR spectra(ν=4 000 ~400 cm-1)were determined by the KBr pressed disc method on a Bruker V70 FT-IR spectrophotometer.1H NMR spectra of HL was acquired with Bruker AV400 NMR instrument using CDCl3solvent.The X-ray diffraction measurement for complexes 1 and 2 were performed on a Bruker SMART APEXⅡCCD diffractometer equipped with a graphite monochromatized Mo Kα radiation(λ=0.071 073 nm)by using φ-ω scan mode.Semi-empirical absorption correction was applied to the intensity data using the SADABSprogram[17].The structures were solved by direct methods and refined by full matrixleast-square on F2using the SHELXTL-97 program[18].C33 and O4 atoms in complex 2 were refined isotropically due to the occupancy value of 0.75 according to the elementary analysis result.All the other non-hydrogen atoms were refined anisotropically.The H atoms for C33 and O4 atoms in complex 2 were not added.All the other H atoms were positioned geometrically and refined using a riding model.
CCDC:1882572,1;1882571:2.
1.2 Syntheses of the TSC ligand and title complexes 1 and 2
A mixture of 3-ethyl-2-acetylpyrazine(1.50 g,10 mmol)and N(4)-(p-tolyl)thiosemicarbazide(1.81 g,10 mmol)in ethanol(30 mL)were stirred for 4 h at room temperature.The white solid HL as a precipitate was filtered and washed three times by cold ethanol.Yield:2.35 g(75%).Elemental analysis Calcd.for C16H19N5S(%):C,61.31;H,6.11;N,22.34.Found(%):C,61.27;H,6.32;N,22.24.1H NMR(400 MHz,CDCl3):δ9.19(1H,s,NH),8.92 (1H,s,NH),8.50~8.53 (2H,t,pyrazine-H),7.50~7.52 (2H,d)/7.20~7.22 (2H,d)for phenyl-H,3.14~3.20 (4H,q,CH2),2.46 (3H,s,CH3),2.36(3H,s,CH3),1.39~1.42(3H,t,CH3-CH2).FT-IR(cm-1):ν(N=C)1 588,ν(N=C,pyrazine)1 529,ν(S=C)859.
Complex 1 was synthesized by reacting HL(0.5 mmol)with Ga(NO3)2·6H2O(nligand∶nmetal=2∶1)in methanol(20 mL)solution at room temperature.The block crystals suitable for X-ray diffraction analysis were obtained by evaporating the reaction solutions at room temperature.Complex 2 is prepared by the same procedure,while using In(NO3)2·6H2O instead of Ga(NO3)2·6H2O.
1:Red crystals.Yield:47%.Anal.Calcd.for C36H52N11O7S2Ga(%):C,48.87;H,5.92;N,17.41.Found(%):C,49.00;H,5.73;N,17.52.FT-IR(cm-1):ν(N=C)1 549,ν(N=C,pyrazine)1 509,ν(NO3-)1 384,ν(S=C)823.
2:Red crystals.Yield:58%.Anal.Calcd.for C33.75H43N11O4.75S2In(%):C,47.26;H,5.05;N,17.96.Found(%):C,47.44;H,4.83;N,18.03.FT-IR(cm-1):ν(N=C)1 579,ν(N=C,pyrazine)1 511,ν(NO3-)1 384,ν(S=C)835.
2 Results and discussion
Both crystals are easy to weather when exposed in air,and are insoluble in chloroform,diethyl ether and benzene,but fairly soluble in methanol,ethanol,DMF and DMSO.
2.1 Crystal structures of complexes 1 and 2
Crystal data and refinement results are summarized in Table 1.A diamond drawing of complexes 1 and 2 is shown in Fig.1,and bond distances and angles around the metal ions are in Table 2.Hydrogen bonds information in complexes 1 is listed in Table 3.The enolization of C=S in both complexes can be confirmed by the bond lengths of C-S in a range of 0.172 3(6)~0.175 1(5)nm,which are in excellent agreement with other previously known thiosemicarbazone complexes in the literature[19].
Complexes 1 and 2 are isostructural and crystallize in the monoclinic system,space group P21/c with different crystal methanol molecules.Thus the structure of complex 1 is discussed in detail for anexample.As shown in Fig.1a,1 is build of one mononuclear cation,one free nitrate anion and four crystal methanol molecules.The galliumバcenter is surrounded by two independent monoanionic TSC ligands with [N2S]donor set,thus forming a distorted octahedral geometry.The sulfur atoms are in a cis arrangement,with a S1-Ga1-S2 angle of 100.56(8)°and trans to the pyridinyl nitrogen atoms.Distortion from an ideal octahedral arrangement mainly arises from the small N-Ga-N and N-Ga-S chelate bite angles,which are very close to 80°.There is a slight shortening of the bond distance between galliumバand the imine nitrogen(0.200 0(5)and 0.201 6(5)nm)compared to the distance between galliumバand the pyrazine nitrogen(0.208 3(5)and 0.211 9(5)nm).Similar feature was observed in the case of complex 2(Fig.1b),with In-Npybond distances (0.229 2(4)and 0.230 7(4)nm)longer than InNiminebond distances(0.226 7(4)nm).In the crystal of 1,intermolecular NH…O hydrogen bonds between the coordination cations and crystal methanol molecules,together with N-H…O hydrogen bonds involving free nitrate anions and crystal methanol molecules link the cations into zig-zag chains along b axis(Fig.1c).
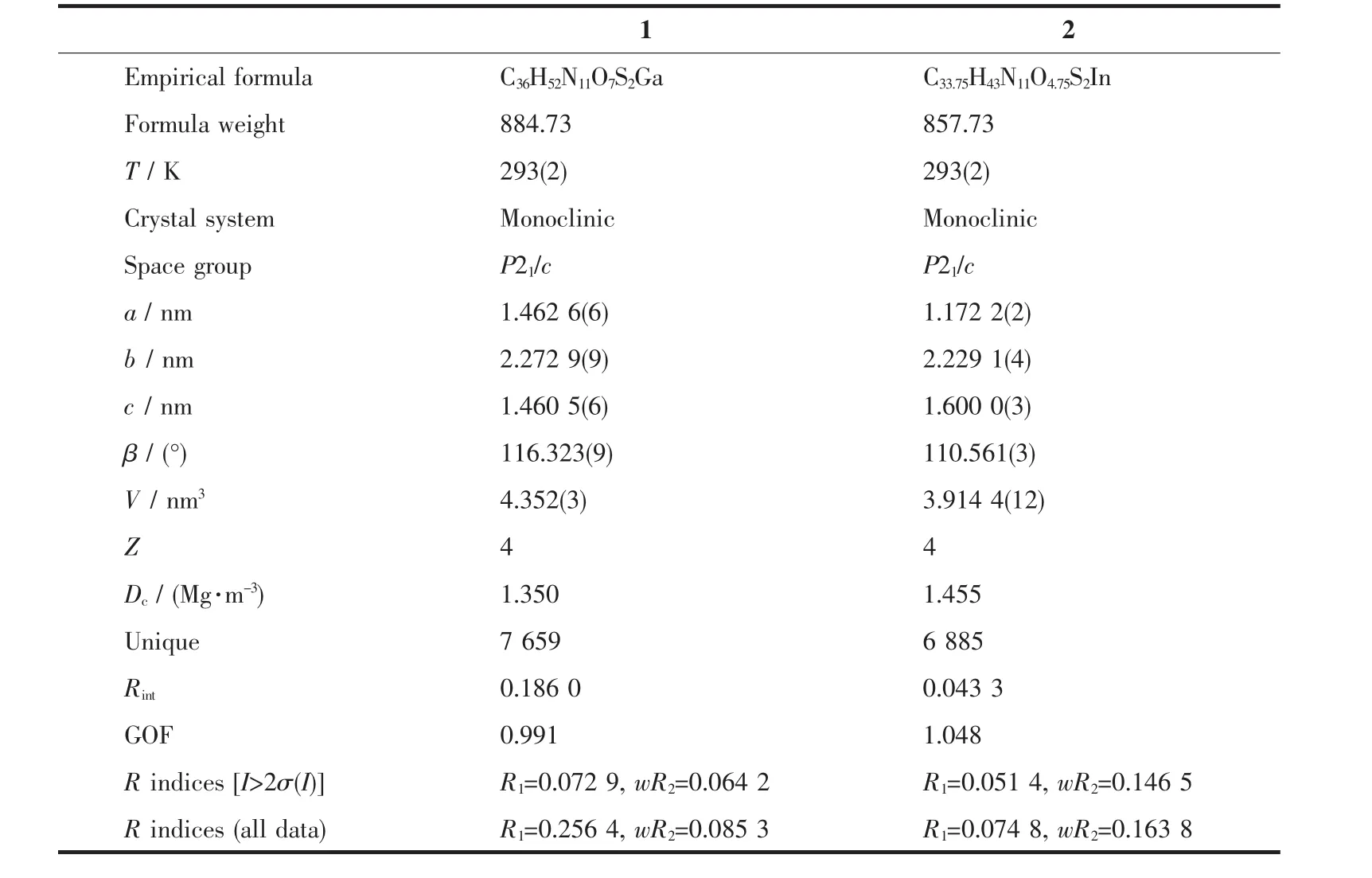
Table 1 Crystal data and structure refinement parameters for 1 and 2

Fig.1 ORTEPdrawing of complexes 1(a)and 2(b)with 10%thermal ellipsoids;(c)Extended zig-zag chain along b axis in the crystal of 1 formed by hydrogen bonds shown in dashed line

Table 2 Selected bond lengths(nm)and angles(°)in complexes 1 and 2

Continued Table 2

Table 3 Hydrogen bonds information for complex 1
2.2 IR spectra
The infrared spectral bands most useful for determining the mode of coordination of the ligands are the ν(N=C), ν(N=C,pyrazine) and ν(S=C)vibrations.Theν(N=C)band of TSC ligand was found at 1 688 cm-1,while it shifted to 1 549 and 1 523 cm-1in complexes 1 and 2,respectively.The decrease in frequency of this band is an evidence for the coordination via the imine nitrogen atom.In the TSC ligand,a band at 1 529 cm-1is assigned toν(N=C,pyrazine),whereas this band was found to shift to about 1 510 cm-1in complexes 1 and 2,confirming that pyrazine nitrogen atom takes part in coordination[4-7].Theν(S=C)vibration at 859 cm-1in the TSC ligand shifted to lower frequency (823 and 835 cm-1for 1 and 2,respectively)in the complexes,in agreement with coordination of a thiolate sulfur.In addition,the intense absorption bands at 1 384 cm-1in both complexes establish the existence of free NO3-groups[20].It is in accordance with the result of the crystal structure study.
2.3 UV spectra
The UV spectra of HL,complexes 1 and 2 in CH3OH solution (c=10 μmol·L-1)were measured at room temperature (Fig.2).The spectra of HL featured main band located around 303 nm(ε=6 567 L·mol-1·cm-1),which could be assigned to characteristic π-π*transition of pyrazine unit[21].Both bands had no shift while with absorption intensity change in the spectra of complexes 1 and 2,they were observed at 302 nm(ε=19 475 L·mol-1·cm-1)and 319 nm (ε=12 723 L·mol-1·cm-1),respectively.However,the new bands at 426 nm(ε=3675 L·mol-1·cm-1,ε=11487L·mol-1·cm-1)could be observed in spectra of complexes 1 and 2,respectively,probably due to the ligand-to-metal charge transfer(LMCT)[22].This fact supports the coordination of the ligand HL in two complexes.

Fig.2 UV spectra of ligand HL,1 and 2 in CH3OH solution at room temperature
2.4 EB-DNA binding study by fluorescence spectrum
It is well known that EB can intercalate nonspecifically into DNA,which causes it to form strong fluorescence emission.Competitive binding of other drugs to DNA and EB will result in displacement of bound EB and a decrease in the fluorescence intensity[20].The effects of the ligand and complexes on the fluorescence spectra of EB-DNA system are presented in Fig.3.The fluorescence intensities of EB bound to ct-DNA at about 600 nm showed remarkable decreasing trends with the increasing concentration of the tested compounds,indicating that some EB molecules are released into solution after the exchange with the compounds.The quenching of EB bound to DNA by the compounds is in agreement with the linear Stern-Volmer equation:I0/I=1+Ksqr[20,22],where I0and I represent the fluorescence intensities in the absence and presence of quencher,respectively,Ksqis the linear Stern-Volmer quenching constant,r is the ratio of the concentration of quencher and DNA.In the quenching plots of I0/I versus r,Ksqvalues are given by the slopes.The Ksqvalues were 0.756 and 2.078 for complexes 1 and 2,respectively,while that of ligand HL was tested to be 0.554.The results indicate that the interactions of complexes with DNA are stronger than that of ligand HL,probably due to the structure rigidity and metalligand synergism effect of complexes[21].Complex 2 has higher quenching ability than 1,which indicate that the coordination environment of metal ion may be also responsible for the DNA interaction ability in some extent.

Fig.3 Emission spectra of EB-DNA system in the presence of HL(a),complexes 1(b)and 2(c)

