Distribution of extended-spectrum β-lactamase genes in antibioticresistant strains of Pseudomonas aeruginosa obtained from burn patients in Ahvaz, Iran
Saeed Khoshnood, Azar Dokht Khosravi, Nabi Jomehzadeh,3, Effat Abbasi Montazeri,3#,Moloudsadat Motahar,3, Fatemeh Shahi,3✉#, Morteza Saki,3, Sakineh Seyed-Mohammadi,3
1Infectious and Tropical Diseases Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
2Student Research Committee, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
3Department of Microbiology, Faculty of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
Keywords:Pseudomonas aeruginosa Burn patients Drug sensitivity Extended-spectrum beta-lactamase
ABSTRACT Objective: To evaluate the drug susceptibility profiles and the frequency of beta-lactamase encoding genes in Pseudomonas aeruginosa (P. aeruginosa) obtained from burn patients.Methods: Totally 93 non-duplicate clinical isolates of P. aeruginosa were recovered from burn patients of Taleghani Burn Hospital of Ahvaz. Antibiotic susceptibility testing was conducted by disk diffusion method according to the CLSI 2017 recommendations. PCR assay was performed by to find beta-lactamase encoding genes. Results: In this study, most clinical specimen was obtained via wound swabs [65 (69.9%)], followed by blood [14 (15.1%)] and biopsy [7 (7.5%)]. Forty-two (45.16%) patients were male and 51(54.84%) were female. High resistance was observed for most of antibiotics especially for gentamicin and ciprofloxacin(Up to 85%), whereas the highest susceptibility was reported for colistin (100.0%), followed by ceftazidime (66.7%). According to PCR results, 16.1% (15), 9.7% (9) and 14.0% (13) of isolates carried blaDHA, blaVEB and blaGES genes, respectively. It also revealed that the blaVEB gene was found to coexist within 2 isolates (2.2%). Conclusions: Antibacterial resistance is high among P. aeruginosa isolates. Colistin is highly active against multi-drug resistant P. aeruginosa isolates. Antimicrobial susceptibility testing can confine indiscriminate uses of antibiotics and resistance increase, and can improve management of treatment.
1. Introduction
Pseudomonas aeruginosa(P. aeruginosa) is one of the most important bacteria in nosocomial infections, particularly in burn units. Infections of this pathogen, especially multidrug-resistant(MDR) isolates, in burn patients, are difficult to treat and regarded as a common problem[1,2]. The increasing prevalence of MDR strains is associated with prolonged hospitalization and a significant rise in patients’ morbidity and mortality[3]. Beta-lactamases can be on the chromosome or on plasmids and may be classified as extendedspectrum β-lactamases (ESBLs), AmpCs, and carbapenemases,among other types. Nowadays, the increasing β-lactam resistance in high among P. aeruginosa isolates illustrates the significance of β-lactamases-coding genes: ESBLs, AmpC β-lactamases as well as metalo β-lactamases, which are commonly related to transmissible genetic elements promoting the resistance development[4].
ESBLs are a rapidly growing group of β-lactamases that hydrolyze penicillins and expanded-spectrum cephalosporins, and belong to class A of Ambler molecular classification system. Most predominant ESBL genes reported in P. aeruginosa include sulfydryl variable (SHV), cefotaximase (CTX-M) and temoneira (TEM) types.Other less common ESBLs, such as Guiana extended spectrum(GES), Vietnamese extended-spectrum beta-lactamase (VEB) and Pseudomonas extended-spectrum beta-lactamase (PER) types, have now been isolated on several continents[5,6].
P. aeruginosa possesses an inducible AmpC β-lactamase that hydrolyzes almost all beta-lactam antibiotics including penicillins,cephalosporins, and monobactams and may be chromosomallyor plasmid-encoded. Although plasmid-mediated AmpC β-lactamases are less prevalent than ESBLs, they’ve been found in several areas of the globe. There are different types of plasmidmediated AmpC β-lactamases: Ambler class C (ACC), AmpC type (ACT), MIR (Miriam hospital in Providence), cephamycins(CMY), moxalactam (MOX), cefoxitin (FOX) and DHA (Dhahran hospital in Saudi Arabia (DHA)[7,8]. For the first time, the DHA enzyme was identified in 1992 in the city of Dhahran, Saudi Arabia,in Salmonella enterica isolate from a stool sample of a patient with lung cancer. Consequently, blaDHA-1 producers pathogens have been found worldwide[9,10].
The current study was performed to identify the occurrence of selective genotypes of β-lactamase (VEB, GES, and DHA) genes,as well as antibiotic resistance profiles in P. aeruginosa isolates obtained from hospitalized burn patients in this geographic region of Iran.
2. Materials and methods
2.1. Ethics
This research was approved by the ethics committee of Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran (Ethic of Number: IR.AJUMS.REC.1396.1011).
2.2. Bacterial isolates
In this cross-sectional study, between March and August 2017, 93 non-duplicate clinical isolates of P. aeruginosa were collected from patients attending Taleghani Burn Hospital in Ahvaz. The specimens yielded these isolates included wound, blood, biopsy, urine, ear,stool and, catheter. These isolates related to different wards of hospitals, including ICU, Gynaecology, Andrology, Pediatrics, and Plastic surgery departemt. Bacterial identification was performed by using standard culture and biochemical methods as described previously[11].
2.2. Antimicrobial susceptibility testing
Antibiotic susceptibility was carried out using the Kirby-Bauer disk diffusion test on Mueller-Hinton agar (Merck, Germany)plates according to the Clinical and Laboratory Standards Institute recommendations[12]. The antimicrobial agents were ciprofloxacin(5 μg), pipracillin-tazobactam (100/10 μg), meropenem (10 μg),gentamicin (10 μg), amikacin (30 μg), ceftriaxone (30 μg),ceftazidime (30 μg), imipenem (10 μg), and colistin-sulfate (10 μg)(Mast Co., UK). MDR P. aeruginosa was defined as isolate resistance to three or more classes of antimicrobial agents. P.aeruginosa ATCC 27 853 was used as a control strain.
2.3. DNA extraction and detection of blaVEB, blaGES and blaDHA genes by PCR
DNA extraction was performed by boiling method as described previously[13]. The DNA quantity and quality were assessed using Nano Drop Spectrophotometer PROMO (Thermo Scientific,USA) and electrophoresis on 1.5% gel agarose, respectively. PCR amplification was conducted for the detection of blaVEB, blaGESand blaDHAgenes using the set of primers described previously (Table 1)[14]. PCR mix was prepared in a final volume of 50 μL, containing 10 × buffer 5 μL, dNTPs mixture (2.5 mmol/L) 4 μL, primer (25 μmol/L) 1 μL, template 1 μL, Taq enzyme 0.5 μL, ddH2O 37.5 μL. Amplification involved an initial denaturation at 93 ℃ for 2 min, followed by 35 cycles of denaturation at 94 ℃ for 1 min,annealing at 55 ℃ for 1 min and extension at 72 ℃ for 1 min, with a final extension step at 72 ℃ for 5 min. PCR products (5 μL) were separated by electrophoresis (80 V, 40 min) using a 1% agarose gel (Sinaclon, Iran) in TBE buffer 1 × and then visualized using an ultraviolet light after staining with DNA safe stain (CinnaGen Co.,Tehran, Iran). Klebsiella pneumoniae ORI-1 containing blaGESand P. aeruginosa containing blaVEBwere used as controls.
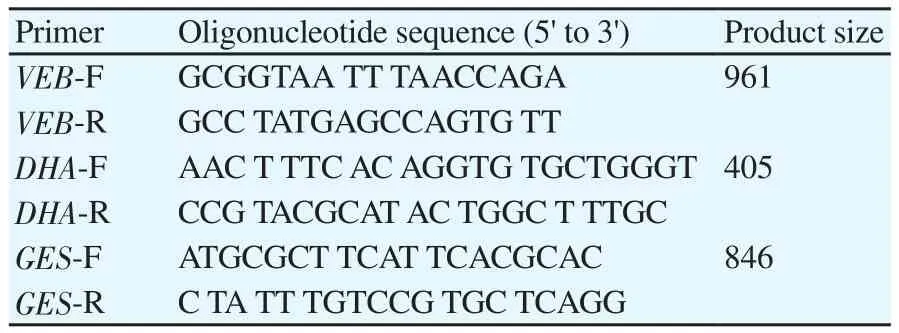
Table 1 Primers used in this study.
3. Results
Out of 93 samples, the most common clinical specimen received in the microbiology laboratory was wound swabs [65 (69.9%)]followed by blood [14 (15.1%)] and biopsy [7 (7.5%)]. Forty-two(45.16%) patients were male and 51 (54.84%) were female. The hospital wards involved in the P. aeruginosa infection was ICU [48(51.6%)], Gynaecology Department [22 (23.7%)], Plastic Surgery Department [12 (12.9%)], Andrology Department 8 (8.6%) and Pediatrics Department 3 (3.2%).
Patterns of antibiogram testing of P. aeruginosa isolates have been showed in detail in Table 2 and they indicated that the highest antibiotic resistance rates were recorded for gentamicin 94.6%,ciprofloxacin 93.5% and meropenem 90.3%, while all isolates(100.0%) were sensitive to colistin.
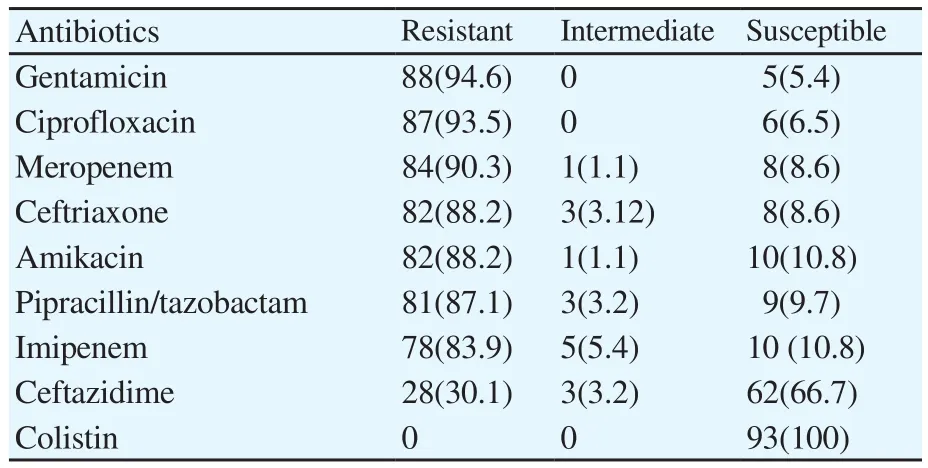
Table 2 Antibiotic susceptibility test results of P. aeruginosa isolated from different clinical specimens [n(%)].
Eighty-three (89.24%) of the P. aeruginosa isolates were resistant to at least three classes of antibiotics (β-lactams, fluoroquinolones,aminoglycosides) and classified as MDR. Moreover, there was a severe antibiotic resistance among the 48 isolates of P. aeruginosa collected from burn ICU ward in comparison with other wards isolates. According to PCR results, 16.1%(15), 9.7%(9) and 14.0%(13) of isolates carried blaDHA, blaVEBand blaGESgenes,respectively. It also revealed that the blaVEBgene was found to coexist with in 2 isolates (2.15%).
4. Discussion
A main colonizer in burn patients is P. aeruginosa, which may increase the risk of infections[15]. Higher resistance to antibacterial agents and ESBL production were observed in the MDR P. aeruginosa isolates from burn wards, where the patients were immunocompromised, and had a longer hospital stay, with a longer chemotherapy course and invasive therapeutic procedure[15,16].
The fewer incidence rates have been reported from Asian,European, and Latin America countries (16.4%-45.9%)[17-21]. In Iran, limited information is available on concerning the prevalence of MDR P. aeruginosa[22]. A recent meta-analysis and systematic review in Iran conducted by Vaez et al.[22] revealed that the prevalence of MDR P. aeruginosa was 58%. The highest and lowest prevalence of MDR P. aeruginosa reported in Tehran (100%), and Zahedan (16%), respectively. Also, the highest resistance rate was against ceftazidime (50%) and amikacin (50%).
Our findings revealed that the prevalence of MDR P. aeruginosa was greatly reduces treatment options. Farshadzadeh et al.[23] and Khosravi et al.[24] in Ahvaz reported that the MDR of P. aeruginosa isolates rapidly increased from 95.1% in 2010-2011 to 100% in 2016. In Asia, various studies have reported a significant increase in MDR P. aeruginosa rates in Pakistan[25], India[26], and Thailand[27],with resistant rates of 30%, 50%, and ≥20%, respectively, which is lower than our findings. In 2015, annual report of the European antimicrobial resistance surveillance network in thirty participated countries reported the rates of MDR P. aeruginosa isolates was< 50%[28].
In this study, highly resistance of P. aeruginosa to ciprofloxacin compared with 26.8% in Latin America[29] and 10%-32% in Europe[30,31]. de Almeida et al.[15] in a hospital-based survey in Brazil, observed the highest resistance rates of P. aeruginosa isolates against ciprofloxacin (94.3%), and gentamicin (88.6%),which is consistent with our findings. Fazeli et al.[32] showed that 21.56% and 39.21% of the P. aeruginosa strains were resistant to ciprofloxacin and gentamicin, respectively, which was lower to our results. Fluoroquinolones and aminoglycosides represent the highest resistance rate, and the increasing resistance against of P. aeruginosa isolates, respectively[15].
Various studies have been reported P. aeruginosa carrying blaGESgene[15,23,33]. One of the most important drug of choice for treatment of MDR P. aeruginosa infections is carbapenem[34]. Our results showed a highly carbapenem-resistance in P. aeruginosa (≥87.1%),and is significantly higher than European countries[28].
In current study, resistance to ceftazidime is higher than the percentage reported from European countries (0%-6.8%)[28].However, ceftazidime-resistance (50.4%) of P. aeruginosa in different provinces of Iran is higher than the percentage reported from our study[22]. Additionally, in comparison with most European countries[28], resistance to other antibiotics is high. In current study,the prevalence of ESBL-producing P. aeruginosa strains was about 13.26%.
In several previous studies performed by Tavajjohi et al.[35] and Bokaeian et al.[36] in Kashan and Zahedan, it was 9.2% and 6.8%.Woodford et al.[37] in UK and Lim et al.[38] in Malaysia reported it as 3.7% and 4.2%, respectively, which was lower to our results. ESBL-positive P. aeruginosa strains that produce ESBLs are frequently isolated[39]. In the study conducted by Gad et al.[40] reported that 97% of P. aeruginosa isolates were beta-lactamase producers. In the human isolates, the frequency of blaDHAin P. aeruginosa is 21.56%[32].
However, in the present study, we found that blaDHAand blaGESwere most prevalent ESBLs in P. aeruginosa. The production of blaGEShas been related to expanded spectrum cephalosporin resistance[41]. An analysis performed in a hospital in Riyadh, Saudi Arabia[42] between January to April 2010 indicated that 25 (16%) were ESBL producers P. aeruginosa isolates, with 5 (20%) carrying blaGESgenes.
Another study from Riyadh conducted by Al-Agamy et al.[43] also reported blaGESin 5 (22%) of ESBL-positive P. aeruginosa isolates.The first report of blaVEBin Iran declared by Shahcheraghi et al.[44].They displayed the frequency of blaVEB, and blaGESamong the ESBL isolates (MIC ≥16 mg/L) were 24%, and 0%, respectively. Our findings showed 9/93 (9.7%) of isolates carried blaVEBgene.
Additionally, in a hospitals-based survey[36] conducted for investigation the prevalence of ESBLs -producing P. aeruginosa isolated from various clinical samples (wound, tracheal tube, urine,blood, ear discharge) of patients hospitalized in Zahedan. Their results revealed that frequency of MDR and ESBL-positive of the isolates were 19/116 (16.37%), and 8/116 (6.89%), respectively,and 4 isolates (3.4%), amplified blaVEB-1. Finally, our data indicated colistin presented an excellent activity against MDR P. aeruginosa isolates. The results of our study showed that most antibiotics used are unsuitable for the treatment of P. aeruginosa infections. Also, the frequency of ESBL producing P. aeruginosa isn’t significant in our study. Performance of antimicrobial susceptibility testing confines indiscriminately uses of antibiotics and resistance increase and improve treatment programs.
Conflict of interest statement
The authors report no conflict of interest.
Acknowlegdement
We would like to thank the Department of Microbiology, Faculty of Medicine, Ahvaz Jundishapur University of Medical Sciences, for their cooperation.
Funding
This study was supported by grants from Vice Chancellor for Research affairs, Ahvaz Jundishapur University of Medical Sciences,Ahvaz, Iran, and Tropical and Infectious Diseases Research Center of the University (Grant No. OG-96143).
 Journal of Acute Disease2019年2期
Journal of Acute Disease2019年2期
- Journal of Acute Disease的其它文章
- Tardive phenomenon presenting as isolated dysarthria: A rare entity mimicking stroke
- Shoshin syndrome: report of a treatable disaster
- Long-term diabetes-related severe complications among individuals with T2DM in Jazan, Saudi Arabia
- Polymorphism of hypoxia-inducible factor-1α gene in pediatric acute respiratory distress syndrome
- Characterization and frequency of antibiotic resistance related to membrane porin and efflux pump genes among Acinetobacter baumannii strains obtained from burn patients in Tehran, Iran
- The red cell distribution width to platelet ratio predicts 30-day mortality of acute pulmonary embolism patients
