Sex-biased gene discovery from the gonadal transcriptomes of the large yellow croaker(Larimichthys crocea)
Junzhu Xiao,Kuan Cao,Yu Zou,Shijun Xiao,Zhiyong Wang,Mingyi Cai
Key Laboratory of Healthy Mariculture for the East China Sea,Ministry of Agriculture,Fisheries College,Jimei University,Xiamen,361021,China
Keywords:Sex differentiation Large yellow croaker Transcriptome Gonad Gene expression
A B S T R A C T The investigation of sex determination in fish is not only of great scientific value,but also has important practical significance,especially in fishes grown in aquaculture exhibiting obvious growth-related sexual dimorphism,such as the large yellow croaker(Larimichthys crocea).While growth of the female large yellow croaker is faster than that of the male,little information exists on the mechanisms that drive the differentiation of the testis and ovary in this species.In this study,the gene expression profiles of large yellow croaker testis and ovary were analysed using RNA-Seq technology.A total of 222 million reads were mapped to the reference genome(89.80%).The differential gene expression analysis identified 13 genes specifically expressed in ovary and 897 specific to testis.Among these,were candidate sex determining genes,such as Dmrt1,Gsdf,Sox9,Amh.This study explored gene expression profiles in large yellow croaker gonads at the transcriptome level by RNA-Seq technology.We found many DEGs related to the development of each specific organ,including the candidate testis differentiation genes Dmrt1,Gsdf,Sox9,Sox3,Amh and ovary differentiation gene Foxl2 by analogy with other fish species,providing essential foundation clarifying the molecular mechanism involved in early sex differentiation.
1.Introduction
Sex determination and sex differentiation are important topics to understand sexual reproduction and play crucial roles in animal evolution.Fishes exhibit a large variability in the sex-determination system XX/XY,XX/XO,WZ/ZZ,and polygenic systems(Martínez et al.,2014).The diversity of sex-determination and sex-differentiation make fish a good model for studying molecular mechanisms and evolution of sex in vertebrates.A better understanding of these processes may contribute to develop novel techniques of sex-control for improving production in aquaculture,since many fishes exhibit sexually dimorphic growth(Ashida,Ueyama,Kinoshita,&Kobayashi,2013;Mei&Gui,2015).
While the testis and ovary show morphological and functional differences they also share some common features which should be accessible through the analysis of gene expression.Early attempts to explore differential gene expression used the polymerase chain reaction(PCR)of one or a few genes(Veith et al.,2006).In a few cases,overexpression and loss of function studies have also been employed(Amrein,Gorman,&Nöthiger,1988;Matsuda et al.,2002;Salvemini et al.,2003).However,in recent years,the development of Next-Generation Sequencing(NGS)technologies has reshaped biological studies.Owing to the low background signal,good repetitiveness and no requirement of reference sequences,RNA-Sequencing(RNA-Seq)has been widely applied in large-scale transcriptome analysis in model and non-model organisms(Liu,Zhang,Liu,Quan,&Zhang,2013).Recently,RNA-Seq was used in transcriptome analysis of fish gonads,such as Xiphophorus(Zhang et al.,2011),rainbow trout(Oncorhynchus mykiss)(Salem et al.,2012),Nile tilapia(Oreochromis niloticus)(Tao et al.,2013),Channel cat fish(Ictalurus punctatus)(Sun et al.,2013),Tripterygion delaisi(Schunter,Vollmer,Macpherson,&Pascual,2014)and yellow cat fish(Pelteobagrus fulvidraco)(Chen et al.,2015).Such studies are contributed to the understanding of sex determination and sex differentiation in fish.
The large yellow croaker(Larimichthys crocea)is a member of the Sciaenidae family and is an important economic species,ranking as the top cage-cultured marine fish in China(Liu&Han,2011).The species exhibits sexually dimorphic growth and females growing significantly faster than males(Xu et al.,2006).Therefore,breeding all-female stocks through sex control is of great practical significance.However,knowledge about sex determination and sex differentiation in large yellow croaker is still limited.A male heterogametic XX/XY sex determination system was hypothesized as almost all artificially induced gynogenetic progenies were female(Lin et al.,2017;Wang et al.,2007;Wang,Wang,Liu,Xie,&Liu,2006).The histological features of the gonadal differentiation process has been characterized(Lin,Zhang,Luo,Zhen,&Shi,1992;You,Cai,Jiang,&Wang,2012)but knowledge of the underlying molecular mechanisms are still limited to a few sexrelated sequences and expression profiles of some candidate genes(Zhou,Zhang,Wang,Xie,&Zhou,2009;Zhou,Zhang,Wang,Zhon,&Xie,2010;Chen et al.,2015.).Although a survey of expressed sequence tags(ESTs)of a mixed cDNA library with gonads from both sexes is available(Zhou et al.,2010),knowledge about testis and ovary specific gene expression is still poor.Therefore,in this work,RNA-Seq was employed to investigate gene expression in the ovary and testis of large yellow croaker in early development stage with the aim of elucidating the molecular mechanisms that determine sexual differentiation and gonadal maturation.
2.Materials and methods
2.1.Ethics statement and sample collection
This study was approved by the Animal Care and Use Committee of the Fisheries College of Jimei University.Nine female and nine male eight-month-old large yellow croakers(13-15 cm longand 40-60 g in weight)were obtained from the breeding section of Jimei University in Ningde,Fujian,China.Gonadal tissue(ovary or testis)from each individual was taken and cut into two halves.One was stored in Bouin's solution for histology and the other part was immersed in RNA later storage solution for RNA extraction.
2.2.Histology and RNA isolation
Histological analyses were performed following the protocol of You et al.(2012).Brie fly,the gonads werefixed in Bouin's solution processed by dehydration with graded ethanol,vitrified in dimethylbenzene,and then embeddedin paraffin.Serial sections of approximately 4-7μm were stained with hematoxylin and Eosin and observed with an Olympus BX53 microscope(Olympus,Tokyo,Japan).
Total RNA was extracted with a Total RNA Kit II(Omega,USA),according to the manufacturer's instructions.RNA degradation and contamination were monitored on 1%agarose gels.RNA concentration and integrity were measured with a Qubit®RNA Assay Kit in a Qubit®2.0 Flurometer(Life Technologies,CA,USA)and 2100 Bioanalyzer(Agilent Technologies,Palo Alto,Calif.USA)with the following parameters:concentration ≥200 ng/μL,RNA integrity number(RIN)=8.9.Three samples each of ovary and testis were pooled at random with the same concentration and volume for the subsequentanalysis.
2.3.Library preparation and sequencing
Library preparation and sequencing were entrusted to Novogene(Beijing,China).Sequencing libraries were generated using the NEBNext®UltraTM RNA Library Prep Kit(New England Biolabs,USA)for Illumina following the manufacturer's instructions.Brie fly,mRNA was purified from total RNA(200 ng/μL)with NEBNext®UltraTM RNA Library Prep Kit using poly-T oligo-attached magnetic beads and fragmented using divalent cations under elevated temperature.(New England Biolabs,USA)First strand synthesis using random hexamer primers and subsequently second strand cDNA synthesis were performed using DNA Polymerase I,RNase H,dNTPs and buffer(New England Biolabs,USA).Overhangs were converted to blunt ends with exonuclease/polymerase.After connecting the adapters,cDNA fragments werefiltered and used as templates for PCR amplification.The library fragments were purified with the AMPure XP system(Beckman Coulter,Beverly,USA)to select cDNA fragments of preferentially 150-200 bp in length.Then PCR was performed with Phusion High-Fidelity DNA polymerase,universal PCR primers and Index(X)Primer.(New England Biolabs,USA).The sequence of the RNA 5′Adapter was:5′ATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCT CTTCCGATCT and the RNA 3′Adapter was(the 6bp bases with underline is the Index)5′GATCGGAAGAGCACACGTCTGAACTCCAGTCA CATCACGATCTCGTATGCCGTCTTCTGCTTG.The index-coded samples were clustered on a cBot Cluster Generation System using TruSeq SR Cluster Kit v3-cBot-HS(Illumia).The libraries with an insert length of 125 bp were sequenced on an Illumina Hiseq 2000 platform to generate paired-end reads.
2.4.Differential gene expression analysis
FastQC v1.10.1(https://github.com/s-andrews/FastQC)was applied to assess the quality of the short sequencing reads and Trimmomatic v0.36(Bolger,Lohse,&Usadel,2014)was employed to remove the reads with a high portion of low-quality bases.Clean reads were mapped to the reference genome with Hisat2 v2.1.0(Kim Langmead,&Salzberg,2015)with the default parameters.HTSeq v0.6.1(Anders,Pyl,&Huber,2015)was used to count the differentially expressed reads for each gene.The level of gene expression was estimated according to the number of reads mapped onto the reference genome.Genes were considered differentially expressed if the log2of the gene expression ratio(|FC|,fold change)>1 and the false discovery rate(FDR)≤ 0.01.The nucleotide sequence of potential differentially-expressed gene(DEGs)were used to interrogate the NCBI nonredundant(nr)database using BLASTX(Altschul et al.,1997)with an E-value threshold of 1×10-5.R was used to generate the scatter plots,and R VennDiagram was used to generate the venn diagrams(Chen&Boutros,2011),pheatmap was used for cluster analysis(https://github.com/raivokolde/pheatmap),and clusterProfiler was used for enrichment analysis(Yu,Wang,Han,&He,2012).The DEGs were further annotated against Gene Ontology(GO),and Kyoto Encyclopedia of Genes and Genomes(KEGG)database with Blast2GO(Conesa et al.,2005).
2.5.Real-time PCR
Ten DEGs were analysed by semi-quantitative RT-PCR(qRT-PCR)in ovaries and testis from 4 individuals each.The cDNAs were generated using primeScript™RT reagent Kit with gDNA Eraser(TaKaRa,Dalian,China).Real-time PCR was carried out on a StepOnePlus™Real-Time PCR System with SYBR®Premix Ex Taq™ II(Tli RNaseH Plus)using the primers indicated in Table 1 that were available in house and have previously been used(Chen et al.,2015;Zhou,Zhang,Wang,Xie,&Zhou,2010)or designed de novo using Primer Premier 5.0.After incubation at 95°C for 30s,the thermal cycling protocol was 40 cycles at 95 °C for 5s and 60 °C for 30s.After amplification,a melting curve was performed according to the manufacturer's instructions of the Light-Cycler 480 real-time PCR instrument(Roche,Switzerland).Relative gene expression levels were calculated using the 2-△△CTcomparative cycle threshold(CT)method(Livak&Schmittgen,2001).Three replicates were included for each sample,and the resulting Ct values wereanalysed using a One-Way analysis of variance(ANOVA)in the SPSS 19.0 statistical program.β-actin gene was used as the reference gene.The R package ggplot2 was used to plot the log2 of qPCR expression vs log2 RNAseq expression,using 0,95 as the level of confidence.
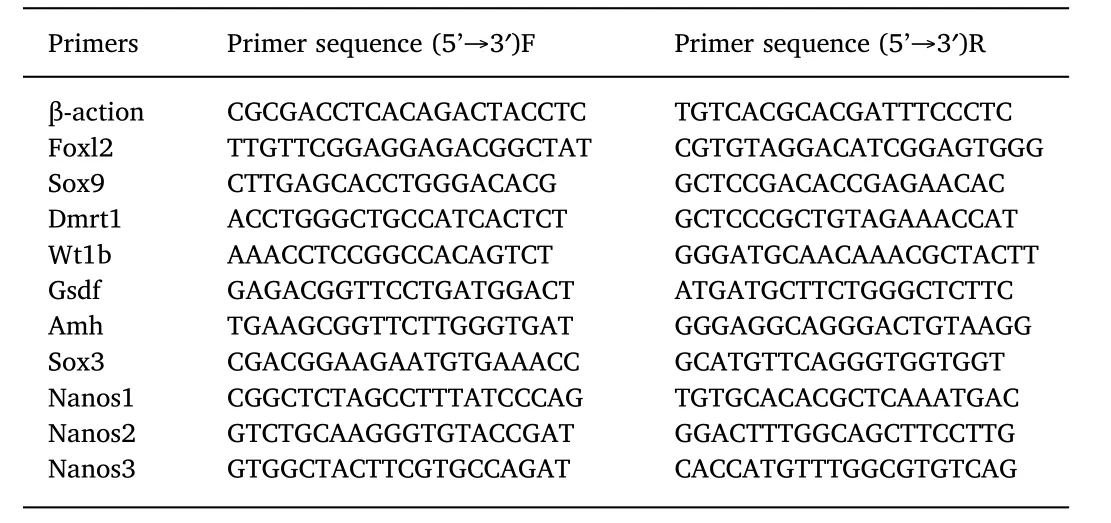
Table 1 Primer sequences used for qRT-PCR.
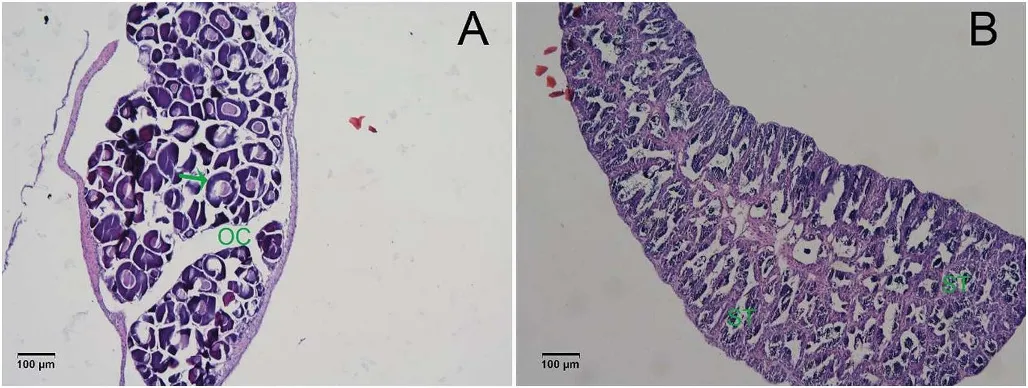
Fig.1.Sections(4-7 μm,10 times amplified)of large yellow croaker(A)Ovary and(B)Testis stained with hematoxylin and eosin.OC:ovarian cavity;ST:seminiferous tubules.(For interpretation of the references to colour in this figure legend,the reader is referred to the Web version of this article.)
3.Result
3.1.Histology
From the histology,the gonads of large yellow croaker had differentiated and their sex could be easily identified at the age of eight months(Fig.1).The ovary was well formed and the oogonia were polygonal in shape(Fig.1A).In the testis,the seminiferous tubules had many branches,forming a network,with many germ cells within the seminal lea flet(Fig.1B).
3.2.mRNA sequencing statistics
RNA-seq produced 256 million raw reads of an average size 125 bp,yielding approximately~31.83 Gb of sequence for the transcriptome analysis.The 222 million clean reads had a high average quality,with Q20 ranging from 94%to 96%(Table 2);89.80%of the reads mapped to the reference genome of the large yellow croaker and 92.33%mapped to a single loci and 7.66%mapped to multiple loci.
3.3.Differential gene expression
A total of 11139 DEGs were identified between the testis and the ovary(Fig.2A,Fig.2B).Of the DEGs 13 and 897 genes were specifically expressed in female and male gonads,respectively.The remaining 10229 genes were expressed in both testis and ovary(Fig.3).Based on the expression level of the genes,two major clusters were found,which corresponded to the two tissues(Fig.2C),supporting the DEG results and suggesting that the testis and ovary were very different at the gene expression level in large yellow croaker.
In the testis,897 preferentially expressed genes exhibited an FC larger than 30-fold,while in the ovary,13 preferentially expressed genes had an FC larger than 30(Table 3).Hence,the number of specifically expressed genes and the preferentially expressed genes in the testis were both higher than in the ovary.
3.4.Functional categories of sex-biased genes
The DEGs were assigned to the three GO categories(cellular component,molecular function,and biological process)and the top 30 terms with the smallest q-value were selected(Fig.4).The most significant differences of DEG number between the ovary and testis suggested there were differences in ribosome and ATP binding terms,translation,and nucleosome,for each of the categories,respectively.There were also differences in tRNA processing,actin cytoskeleton organization and regulation of Rho protein signal transduction terms in the biological process category.These differences are in line with enrichment of genes within the mitochondrion,as highlighted in the cellular component category analysis.Furthermore,differences between ovary and testis in structural constituent,actin binding,Rho guanyl-nucleotide exchange factor activity,microtubule binding and methyltransferase activity terms were also found at the level of the molecular function category(Fig.4).Among the testis up-regulated DEGs,the terms protein binding and protein kinase activity ranked as the top two in the molecular function category,while in the biological process category the terms regulation of transcription,DNA-template and protein phosphorylation ranked higher and in the cellular component the terms integrin complex and hemoglobin complex were most notable.In contrast,for the up-regulated DEGs in the ovary the most represented terms for molecular function was nucleic acid binding,structural constituent of ribosome,RNA binding and methyltransferase activity,for biological processes the most notable terms were translation and metabolic process and for cellular component were ribosome and cytoplasm.
Table 4 provides detailed GO information with the top three subterms listed for each category.The terms.related to cellular activities such as nucleosome,ribosome,mitochondrion,structural constituent of ribosome,ATP binding,cytochrome-c oxidase activity,translation,regulation of cell cycle,and positive regulation of TOR signaling are indicated in the table.
The assignment of pathways for DEGs in the gonadal transcriptome analysis were also analysed using KEGG pathways.A total of 23 terms were annotated in KEGG,of which 13 were directly related to metabolism.This category included the terms Oxidative phosphorylation,Biosynthesis of amino acids,Phenylalanine metabolism,Carbon metabolism,Arginine and proline metabolism,Valine,leucine and isoleucine degradation,Vitamin B6 metabolism,Glycerolipid metabolism,Tyrosine metabolism,Histidine metabolism,Pyrimidine metabolism,Nicotinate and nicotinamide metabolism,and beta-Alanine metabolism.The terms cytokine-cytokine receptor interaction and hematopoietic cell lineage were the top two of most significant group in the all pathways.(Table 5).
The expression levels of 30 candidate genes that might be involvedin gonadal sex differentiation of large yellow croaker were chosen from the transcriptome database for further study(Table 6).The genes specifically expressed in ovary and testis are also listed in Supplementary Table 1 and Supplementary Table 2,respectively.
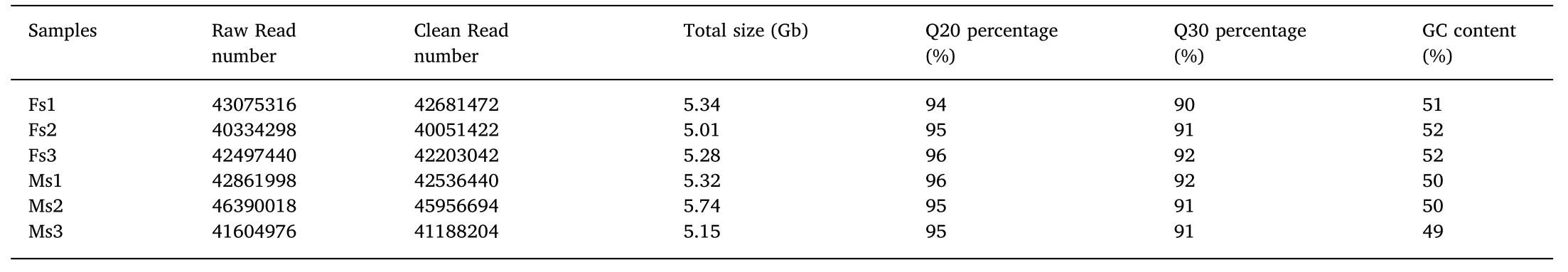
Table 2 A summary of the transcriptome sequencing data in large yellow croaker.
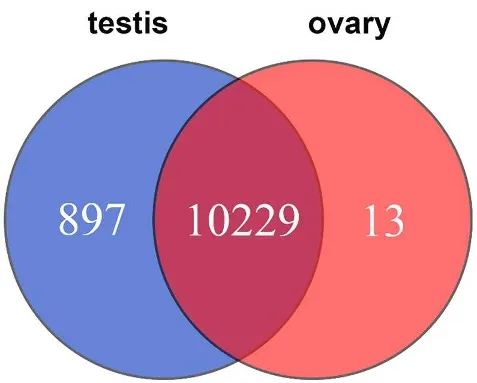
Fig.3.Venn diagram representing the genes that are common to the testis and ovary and the differentially expressed genes found only in the testis and only in the ovary.
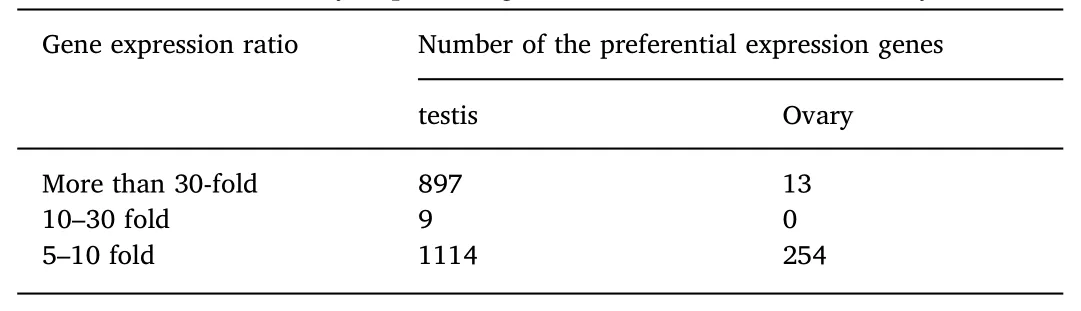
Table 3 Statistics of differentially expressed genes in the testis and the ovary.
3.5.Differentially expressed genes analysed by RT-qPCR
In order to validate the result of RNA-Seq,real time quantitative PCR(RT-qPCR)was carried out for some candidate sex differentiationrelated genes.The expression levels of the examined genes were coherent with those obtained in the RNA-Seq analysis.For example,Dmrt1,Sox3 and Nanos2 genes were not expressed in the ovary,while Foxl2 and Nanos3 were not expressed in the testis in both methods.The correlation between the RNA-Seq and RT-qPCR data gave a Spearman's rank correlation rho of 0.9515(Fig.5).
4.Discussion
This study used an RNA-Seq approach to identify DEGs(11139)between the ovary and testis of the large yellow croaker,and specific DEGs were identified and a large number of genes were preferentially(i.e.more highly)expressed in testis relative to the ovary.Although the molecular mechanism of sex determination of the large yellow croaker is still unknown,the identification of these DEGs between the testis and ovary is a useful tool for the identification of candidate sex-determining/sex-differentiation genes in this species.
The functional and pathway enrichment analysis further indicated fundamental differences existed between the testis and ovary.For example,the results of KEGG indicated differences in specific pathways between the ovary and testis,such as metabolism.The enrichment analysis combined with the GO analysis(Table 4)indicates that many DEGs are implicated in cell activities,cell cycle and metabolism.
Several transcription factors of the Sox gene family are involved in sex differentiation,as well as in neural crest development and neurogenesis(Betancur,Bronner-Fraser,&Sauka-Spengler,2010;Laudet,Stehelin,&Clevers,1993;Turner,Ely,Prokop,&Milsted,2011).Within the Sox family,Sox3,and Sox9 are the genes that are strongly associated with sex differentiation in several different fish species.For example,Sox3 was considered to be important in the process of testis development in the black sea bream(Acanthopagrus schlegeli)(Shin,An,Park,Jeong,&Choi,2009).In large yellow croaker,Sox3 was not expressed in the ovary,and had a very low expression in the testis,suggesting it may be involved in the differentiation of the testis.Simultaneously,Sox9a was expressed at low levels in the ovary,but had a higher expression in the testis.In some fish,such as the Nile tilapia,Sox9a is involved in the formation of the testis(Ijiri et al.,2008),these observations may indicate that Sox9a may be more important for testis differentiation than ovarian differentiation.
Dmrt1 is a gene that has conserved functions in testicular differentiation across vertebrates.In the Nile tilapia and blunt snout bream(Megalobrama amblycephala),Dmrt1 is involved in testis differentiation(Ijiri et al.,2008;Su et al.,2015).Furthermore,Dmrt1 was exclusively expressed during testis differentiation in rainbow trout and black porgy(Acanthopagrus schlegeli)(Baron,Houlgatte,Fostier,Fostier,&Guiguen,2005;Shin et al.,2009).In contrast,Dmrt1 exhibited sexual dimorphism in other fish species,such as in zebra fish(Danio rerio)(Guo et al.,2004)and European sea bass(Dicentrarchus labrax)(Deloffre,Martins,Mylonas,&Canario,2009),although the expression in testis was still higher than in ovary,.In the large yellow croaker,Dmrt1 wasspecifically expressed in malesat 8-months age,suggesting that it has an important role in testis differentiation and function.Other DMR family members were also expressed in the developing gonads but no significant differences existed between the ovary and testis.
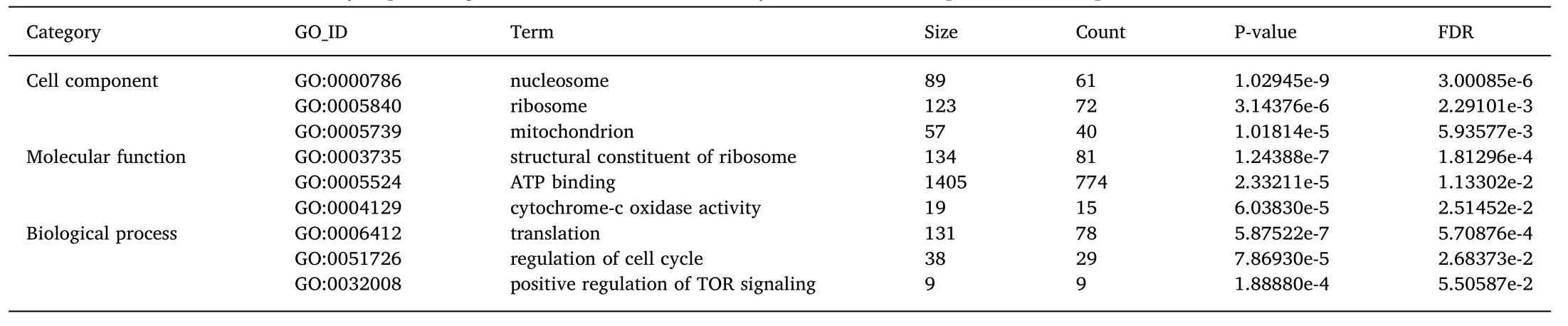
Table 4 The main GO terms for differentially expressed genes identified when the ovary and testis transcriptome was compared.
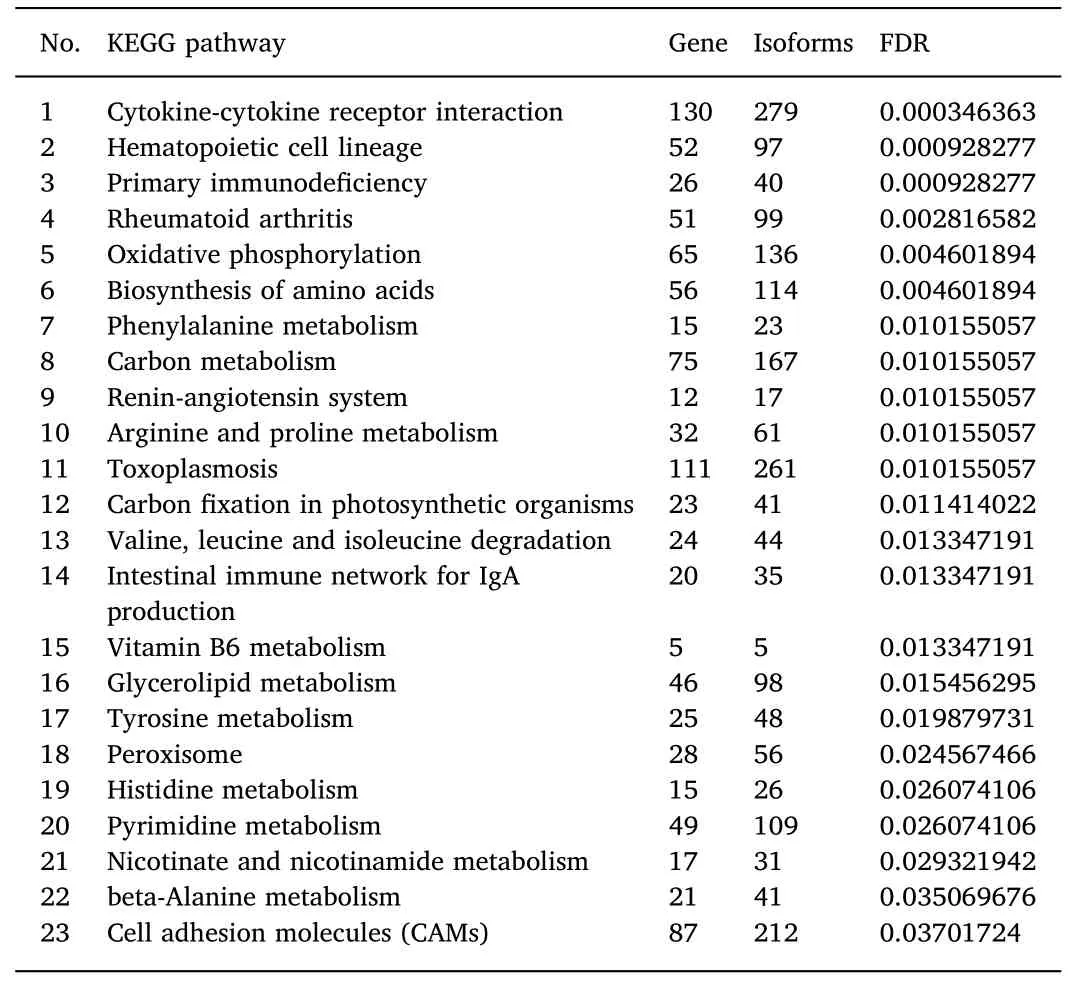
Table 5 The KEGG terms for the differentially expressed genes between the ovary and the testis in the large yellow croaker.
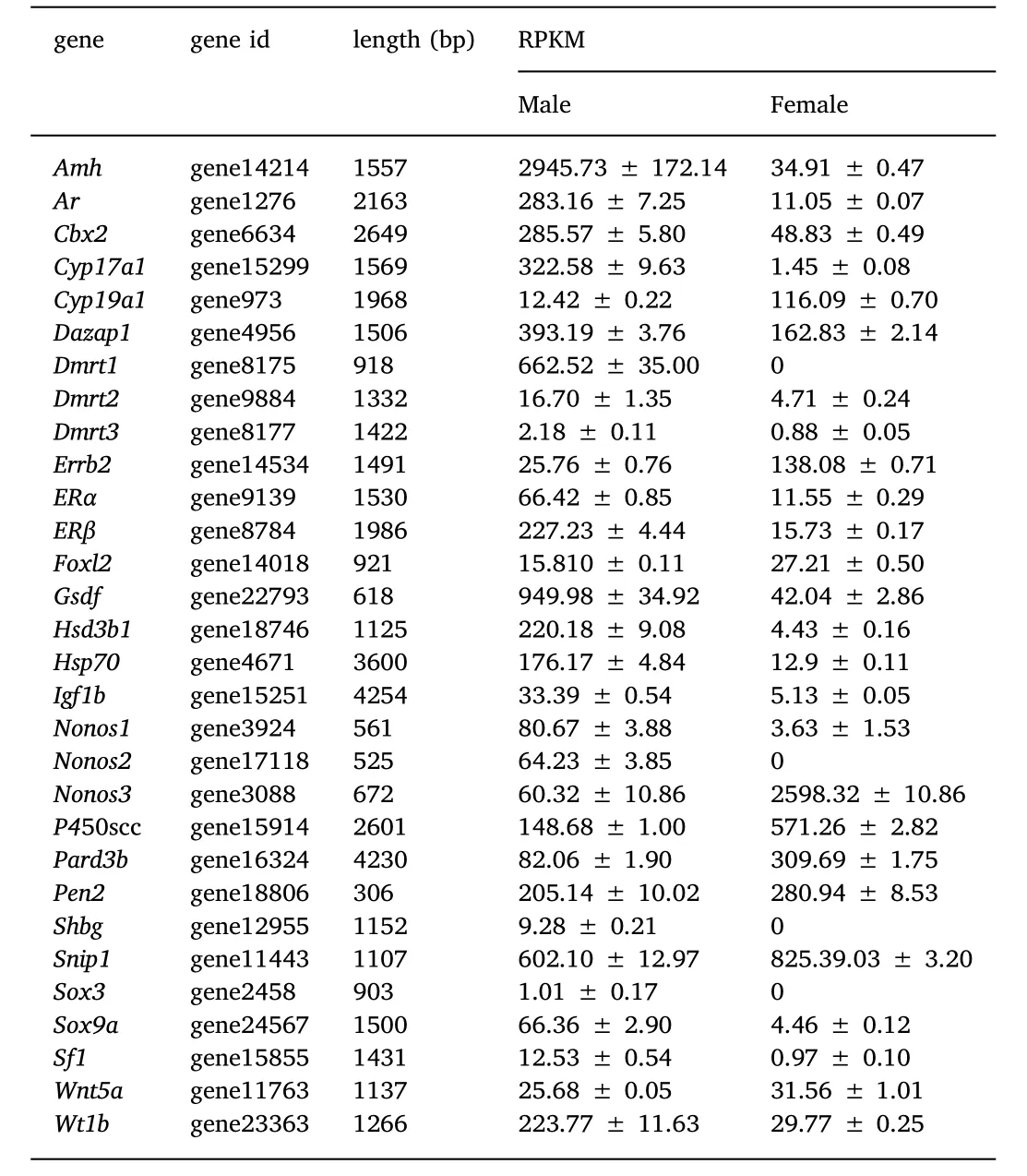
Table 6 Sex differentiation related genes and their levels of expression in the ovary and the testis.
Wilm's tumour suppressor gene 1(Wt1)(Bonetta et al.,1990)encodes a transcription factor that contains four zinc finger motifs and plays an important role in kidney and gonad development(Kreidberg et al.,1993).Wt1 has been identified in zebra fish(Perner,Englert,&Bollig,2007),Nile tilapia(Lee&Kocher,2007)and half-smooth tongue sole(Cynoglossus semilaevis)(Zhang,Chen,Liu,Wen,&Zhou,2014).In fishes,there are two paralogs,Wt1a and Wt1b and in the zebra fish,both isoforms are involved in nephrogenesis(Perner et al.,2007)but not in sex-determination as shown by knockout experiments.Likewise,Wt1b was also excluded as a sex determining candidate gene in the Nile tilapia(Lee&Kocher,2007),and only in the half-smooth tongue sole,does the expression of Wt1 appear to be significantly higher in males than in females(Zhang et al.,2014),suggesting it may have a candidate role in male differentiation.In large yellow croaker,Wt1 was expressed in both ovary and testis and as reported for the half-tongue sole,the expression was much higher in the testis than the ovary,suggesting that it might be important for the development of the male gonad.Nonetheless,further studies will be necessary to clarify the role of WT-1 in gonadal development of the large yellow croaker.
Foxl2 encodes a forkhead transcription factor,the earliest recognized sexual dimorphic marker for ovarian differentiation in vertebrates(Wang,Kobayashi,Zhou,&Nagahama,2004).The expression of Foxl2 was detected specifically in developing ovarian granulosa cells in Nile tilapia(Wang Kobayashi,Zhou,&Nagahama,2004)and medaka(Oryzias latipes)(Nakamoto,Matsuda,Wang,Nagahama,&Shibate,2006).Foxl2 has also been identified and characterized in the black porgy(Acanthopagrus schlegeli)(Wu et al.,2010),wrasse(Halichoeres trimaculatus)(Kobayashi,Horiguchi,Nozu,&Nakamura,2010),cat fish(Clarias gariepinus)(Sridevi&Senthilkumaran,2011),rare minnow(Gobiocypris rarus)(Wang,Wu,Qin,Wang,&Wang,2012),Korea rock fish(Sebastes schlegeli)(Mu,Wen,Li,&He,2013),sable fish(Anoplopoma fimbria)(Smith,Guzmán,&Luckenbach,2013),Atlantic salmon(Salmo salar)(Gowen et al.,2013),willow minnow(Gnathopogon caerulescens)(Ashida et al.,2013)and rice field eel(He et al.,2014).In all of the preceding studies,Foxl2 was consistently more highly expressed in the ovary than in the testis.The present results agree with previous studies and Foxl2 is probably a key factor for yellow croaker ovarian differentiation.
Anti-Mullerian hormone(Amh)is a member of the transforming growth factor-β (TGF-β)family.In fish,Amh expression was reported to be sexually dimorphic in the gonads of the Japanese flounder(Paralichthys olivaceus) (Yoshinaga et al.,2004) and zebra fish(Rodríguez et al.,2005),with higher expression in the testis.In the protandrous black porgy,Amh prevented ectopic ovarian development in the testis and maintained male function(Wu,Li,Luo,Chen,&Chang,2015).In the large yellow croaker,the expression of Amh was low in the ovary and high in the testis,suggesting that Amh is linked to testis differentiation.
Finally,the gonadal soma-derived growth factor(Gsdf),a member of the transforming growth factor(TGF-β)superfamily,was shown to play an important role in the proliferation of primordial germ cells(PGCs)in rainbow trout(Sawatari,Shikina,Takeuchi,&Yoshizaki,2007)and to be essential for testis differentiation in medaka(Oryzias latipe)(Masuyama et al.,2012;Zhang et al.,2016).In large yellow croaker,the expression of Gsdf was also higher in the testis than in the ovary.
In conclusion,the gene expression profiles of the developing testis and ovary of the large yellow croaker were analysed by RNA-Seq technology.We found many DEGs related to the development of the testis or ovary,including the previously reported candidate testis differentiation genes Dmrt1,Gsdf,Sox9,Sox3,Amh and the ovary differentiation gene Foxl2.The mechanisms of sex differentiation in fish are numerous and complex,but the present study contributes to pinpoint genes that that may be involved in testis and ovary differentiation in the large yellow croaker.Furthermore,specific candidate genes for male differentiation were identified and further studies on their regulation during gonad development will be necessary to fully clarify their role in the large yellow croaker.
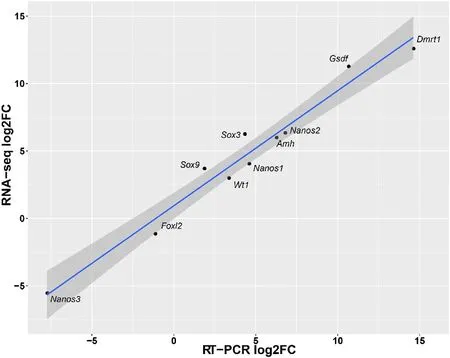
Fig.5.Comparison of the expression levels of putative sex differentiation related genes measured by qRT-PCR and RNA-seq.The RPKM(Read Per Kilobase of exon model per Million mapped reads)values are shown to 0.001.The dark grey shading around the line corresponds to the level of confidence interval(0.95).
Acknowledgments
M.C and Z.W conceived and designed the study,J.X and K.C conducted samples collection and molecular biology experiments,S.X and Y.Z performed the bioinformatics analysis,J.X and S.X wrote the manuscript.This work was funded by the National Natural Science Foundation of China(31272653,41706157),the Natural Science Foundation of Fujian Province(2017J01449).
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aaf.2019.01.001.
 Aquaculture and Fisheries2019年1期
Aquaculture and Fisheries2019年1期
- Aquaculture and Fisheries的其它文章
- Genome-wide association study identifies loci for body shape in the large yellow croaker(Larimichthys crocea)
- SuperSAGE digital expression analysis of differential growth rate in a European sea bass population
- Molecular aspects of eye development and regeneration in the Australian redclaw cray fish,Cherax quadricarinatus
- Uncovering the immunological repertoire of the carpet shell clam Ruditapes decussatus through a transcriptomic-based approach
