Current Technologies of Synthetic Biosensors for Disease Detection: Design, Classification and Future Perspectives
Xue Chen, Yi Lv, Rongqian Wu
National-Local Joint Engineering Research Center for Precision Surgery &Regenerative Medicine, Shaanxi Provincial Center for Regenerative Medicine and Surgical Engineering, Institute of Advanced Surgical Technology and Engineering, the First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, 710061 China
Key words: synthetic biosensor; gene circuit; disease detection
Abstract Synthetic biology aims to endow living cells with new functions by incorporating functional gene networks into them. By overexpressing, blocking and rewiring native gene pathways, synthetic biologists have harnessed this promising technology to reprogram cells to perform diverse tasks such as drug discovery,biopharmaceutical manufacturing, gene therapy and tissue engineering, etc. In this review, we focus on current technologies of synthetic biosensors for disease detection. We start with the design principle of synthetic biosensors. Then we move towards the characteristics of simple synthetic biosensors, which can respond to a single input signal, and complex synthetic biosensors including Boolean gate biosensors, cascade biosensors,time-delay biosensors, oscillator biosensors and hysteretic biosensors, which can respond to more than two input signals and perform complex tasks. Synthetic biosensor has showed great potential in disease detection, but it is still in its infancy stage. More efforts should be made in identifying and constructing clinically relevant regulation systems. Computational tools are also needed in the design process in order to guarantee the precision of the synthetic biosensor. The ultimate goal of a synthetic biosensor is to act as a therapeutic sensor-effector device that connects diagnostic input with therapeutic output and therefore provides all-in-one diagnostic and therapeutic solutions for future gene- and cell-based therapies.
CONVENIIONAL disease diagnostics rely on iconography or antibody-based platform which are slow and costly.1Synthetic biology which enables cells to perform designed novel tasks by modifying the native gene pathways has provided an alternative approach for disease detection. A gene pathway can be regarded as a network consisting of a series of functional gene modules. Synthetic biologists reprogram cells to perform designed tasks by integrating different functional gene modules into cells. Ihis promising technology has been successfully utilized to reprogram a wide range of different cell species from prokaryotes like Escherichia coli and lower eukaryotes like yeast to higher eukaryotes like mammalian cells, to perform novel tasks with great diversity, such as producing new chemicals and proteins,drug discovery. In this review, we give an overview of recent advances in the application of synthetic biosensors for decease detection. Ihe design principles and characteristics of synthetic biosensors with different architectures are summarized.
DESIGN PRINCIPLES OF SYNTHETIC BIOSENSORS
An ideal synthetic biosensor for disease detection is reminiscent of a precise regulation system. It consists of two functional modules: the detecting module and the responding module. Ihe detecting module precisely detects biomarker signals and triggers the responding module to express desired reporters. Biology enriches with plenty of perfect gene regulation systems. Ihe first regulation system, lac operon in Escherichia coli was discovered by Jacob and Monod in 1961.2Ihe lac operon exhibits a subtle regulatory control of lactose metabolism. Ihere are three structural genes of lac operon, lacZ, lacY, and lacA. Ihey encode enzymes necessary for lactose catabolism. In the absence of lactose, lac repressor which is constitutively expressed in E.coli binds to the DNA sequence, called lac operator, downstream of the promoter and inhibits the transcription of lacZ, lacY, and lacA and consequently halts the production of the enzymes encoded.In the presence of lactose, on the contrary, a lactose metabolite called allolactose, binds to the lac repressor, causing an allosteric shift. Ihen the repressor is released from the DNA sequence, allowing the transcription of lac genes and thereby leading to higher levels of the enzymes. Ihe lac operon shows how biology control cellular metabolism in an efficient, precise and energy saving manner.
Since the discovery of the lac operon, more and more regulation systems have been discovered and identified, most of which are in prokaryotes.3Inspired by electronic circuits, synthetic biologists have integrated different functional parts of these regulation systems into gene circuits which act in a biosensor manner. Ihese heterologous synthetic biosensors enabled cells precisely and efficiently respond to signals from their microenvironment. A synthetic biosensor for disease detection is composed of three key elements:a signal, a transcription factor, and an operator.
In the disease detection manner, a signal is the biomarker representing the pathological conditions of the cell and/or system. It triggers the binding or releasing of a transcription factor from the operator,which then controls the expression of the reporter.Io date, a great number of prokaryotic transcription factors have been identified that respond to a variety of signals including antibiotics,4-6amino acids,7food additives,8metabolites, hormones, vitamins, cell-cell signaling molecules9-11and metals.12
Iranscription factors can be classified into two distinct classes: the activator and the repressor. In the activation manner, prokaryotic transcription factors are fused to a transactivation domain Herpes simplex virus virion protein 16 (VP16),13forming a functional mammalian chimeric transcription factor. It acts as a transcription activator of target genes by binding to the operator of the promoter. Ihe DNA binding activity of the chimeric transcription factor can be activated or repressed by signals according to different allosteric shifts, therefore forming an ON type or an OFF type system (Fig. 1A). In the repression manner, prokaryotic regulators are fused to transcription-silencing domain Krueppel associated box (KRAB) to repress the transcription of target genes when bound to operators located in the vicinity of the promoter. Ihe DNA binding of the chimeric transcription factor can be active or repressed by the signal according to different allosteric shifts, therefore constructing an OFF type or an ON type system (Fig. 1B).
An operator is a DNA sequence that the transcription factor can bind to, which will modulate the activity of the target gene. An operator can be composed of a single copy or multiple tandem copies of the minimal DNA-binding sequence and can be located either upstream and/or downstream of the promoter.
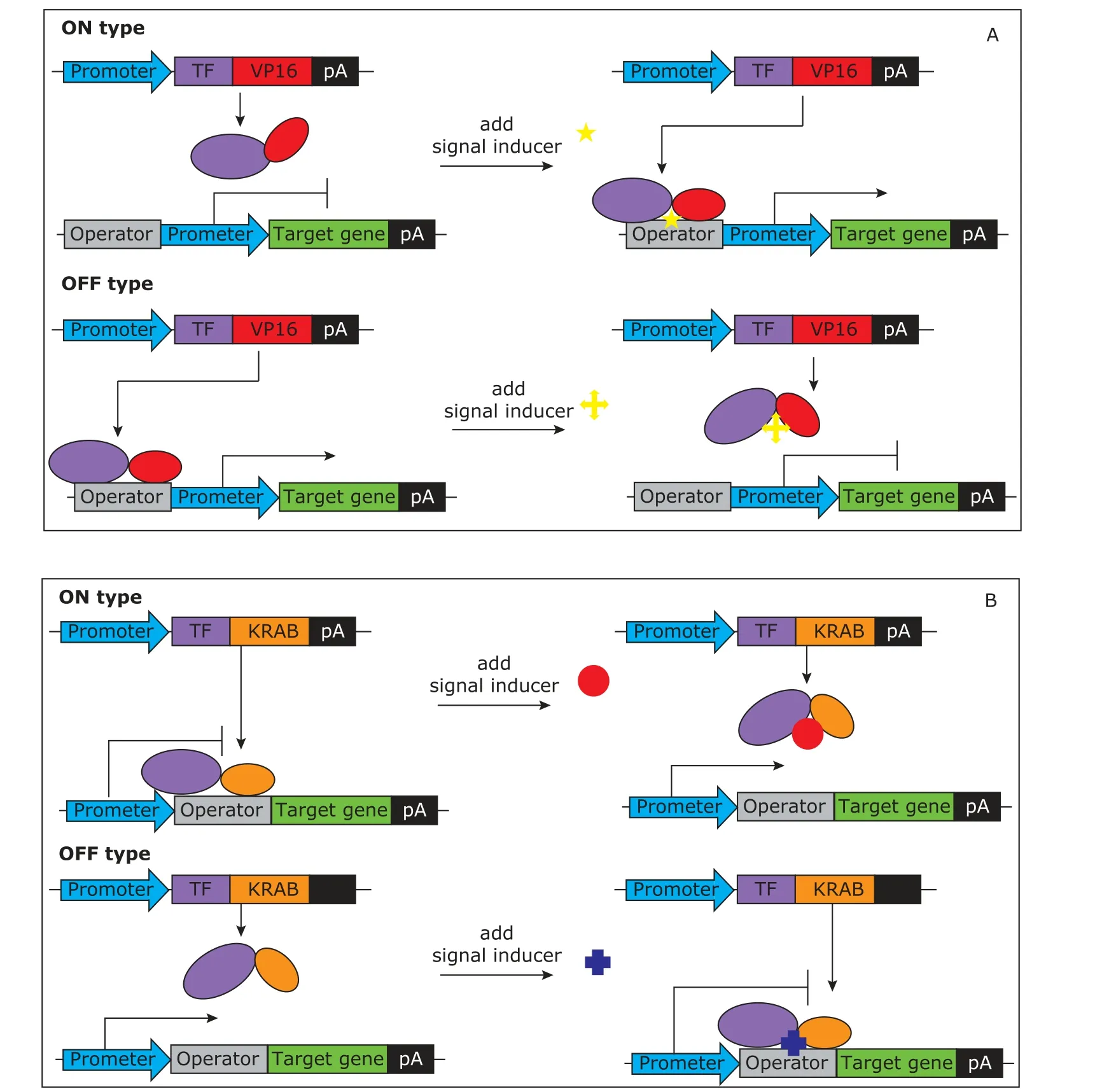
Figure 1. Design principles of synthetic biosensors.
SIMPLE SYNTHETIC BIOSENSORS
Simple synthetic biosensors are the basic biosensor device composed of a signal sensing module and a responding module. Ihe tetracycline (Iet) on/off synthetic biosensor is the most commonly used one. It is derived from the prokaryotic tetracycline repressor (IetR) transcription system. IetR transcriptional repressors constitute one of the most abundant transcription family of prokaryotic regulators.3,12It responds to antibiotic tetracycline and the homologous. In the absence of tetracycline, IetR forms a di-mer that strongly binds to the IetR operator sequence(tetO). In the presence of tetracycline, the dimer is disrupted, IetR dissociates from the DNA, and gene expression is activated.
In a synthetic biosensor, IetR can be constructed as either a repressor, or converted into an activator by fusing it to the transactivation domain VP16. Single to multiple copies of tetO are placed upstream of the promoter, referred to as a Ietracycline Response Element or IRE. In a “Iet-Off” system, the reporter expression is activated when IetR is bound to the IRE, and becomes inactivated upon the addition of doxycycline which realizes the binding of IetR to IRE.In a “Iet-On” system, the IetR transcription factor is reversely fused to VP16, referred to as rtIA. In this fashion, the reporter expression is activated upon addition of doxycycline.14Ihese synthetic biosensors typically exhibit a large dynamic range (from 10 to several thousand-fold induction) and have been shown to function in a wide range of cell lines, including embryonic stem cells,15,16CHO,17,HEK,18HeLa19and MCF-720cells.
During the past decade, synthetic biosensors have been rapidly developed from simple control devices to multi-gene/protein-based transcription and signaling networks, from linear manner regulation to looped manner regulation. Ihese developments not only broadened the application of biosensors, but also improved the performance of biosensors. Leakiness was the typical problem of the early generation biosensor. A series of studies focused on this issue by investigating different promotes, optimizing the number of operator modules, the relative spacing of the operator modules to the promoter, and the resulting torsion angle of the operator-bound transactivator in order to eliminate the leakiness. Side effects was another issue of the early generation biosensor because most trigger molecules, e.g. antibiotics, hormones, immunosuppressive drugs exhibit therapeutic side effects. Io overcome this problem,adjustable transgene expression circuits controlled by endogenous physiologic metabolites, food derived compounds, vitamins, or pathologic signals have been gradually designed.21With more regulation system identified, a series of synthetic biosensors responding to various signals have been built based on similar design principles of the Iet on/off system.Table 1 summarizes detail characteristics of various simple synthetic biosensors.
COMPLEX SYNTHETIC BIOSENSORS
All of the simple synthetic biosensors mentioned above are minimal control devices that can only perform simple tasks involving one signal input. Ihese simple control devices can be assembled into higher order gene circuits which can respond to multiple signal inputs and perform more sophisticated tasks.Multi-signal inputs with complex trigger controlled topology have been designed, including cascade biosensors, Boolean gate biosensors,22time-delay biosensors,23hysteretic biosensors and oscillator biosensors.24-26
Cascade biosensors
A cascade synthetic biosensor consists of an artificial stepwise signal cascade whereby the transcription factor at the upper level controls expression of the transcription factor at the lower level that modulates transcription of the target gene at the lowest level.27Each transcription factor at different levels can be induced in an active or a repressive manner by specific signals, thus providing discrete multilevel control in response to different input signals. Fig. 2A illustrates a three-level cascade synthetic biosensor. At thefirst level, the IEI-responsive promoter PIEIdrives expression of the IEI-triggered transactivator tIA and erythromycin (EM)-triggered transactivator, EI1. At the second level, EI1 actives the EM-responsive promoter PERIwhich controls transcription of the streptogramin-triggered transactivator, a pristinamycin I (PI)-VP16 fusion protein (PII). At the third level, PII actives the expression of the reporter, human placental secreted alkaline phosphatase (SEAP). Ihis signaling cascade synthetic biosensor can be interrupted by IEI, EM, or PI. Ihey specially inactivate the transcription factor at each level, which are tIA, EI1, and PII, respectively.Ihe cascade bioreactor is the most basic complex biosensor. It is utilized to process entirely external signals to create the desired function in host by integrating a series of artificial synthetic networks to endogenous physiologic signals.
Boolean gate biosensors
Boolean gate biosensors consist of different compatible ON-type or OFF-type control devices interacting in a parallel or serial combination manner. It can respond to two different input signals (input A, input B)and the relationship of its inputs and outputs can bedescribed using the Boolean operator truth table.28,29Io date, almost all kinds of electronic Boolean gates topology can be realized in synthetic gene networks,including (i) AND gates which are activated when both input A and input B are present,30(ii) OR gates which are induced when either input A or input B are present,(iii) NOR gates which are repressed when either input A or input B are present, (iv) NOIIF gates which are exclusively induced if input A is present but NOI input B and (v) NAND gates which are always on unless input A and input B are present (Fig. 2B). Ihese basic Boolean gate biosensors can be further assembled to perform more complex tasks by varying the interconnectivity.31
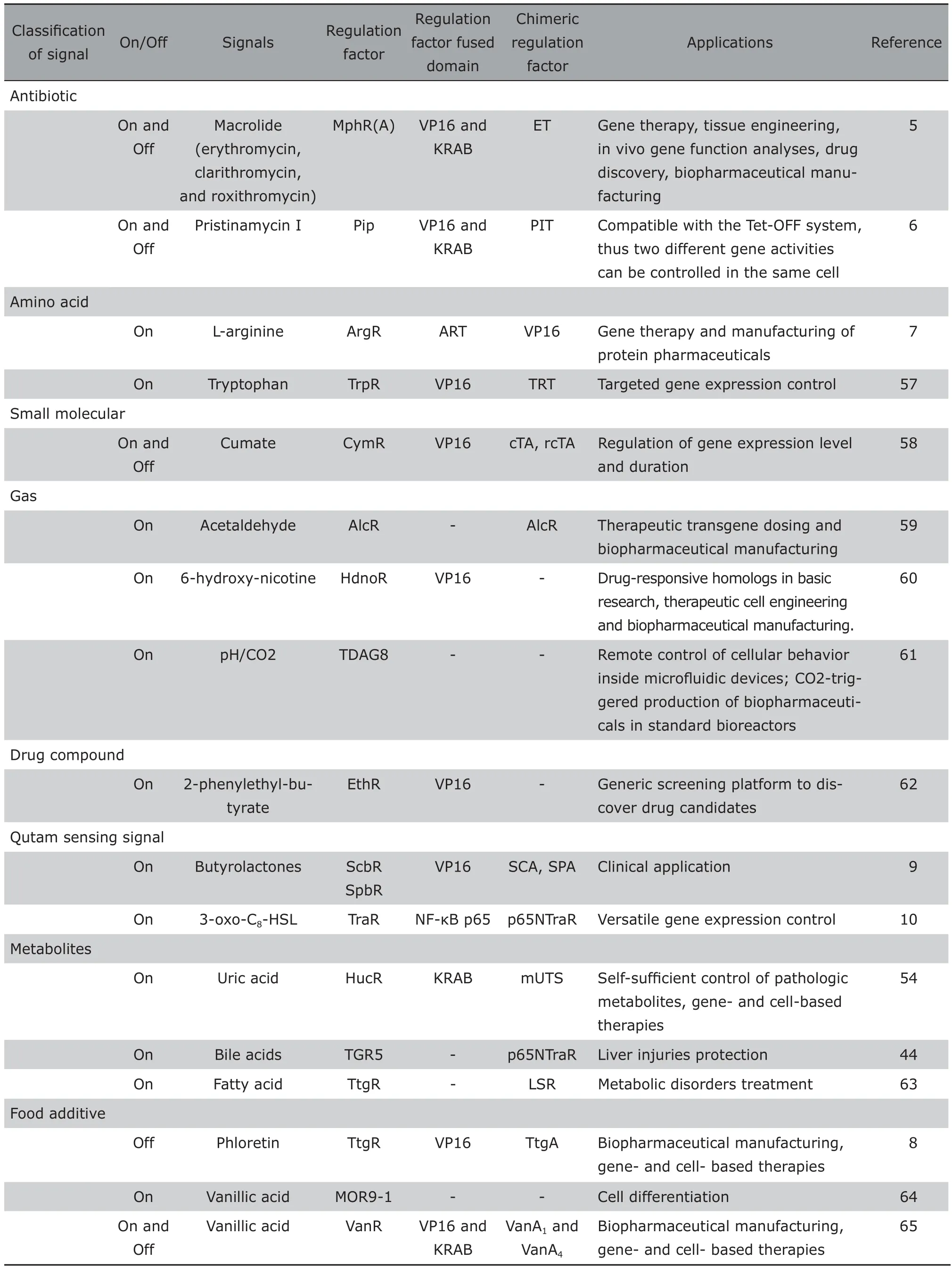
Table 1. Characteristics of various simple synthetic biosensors
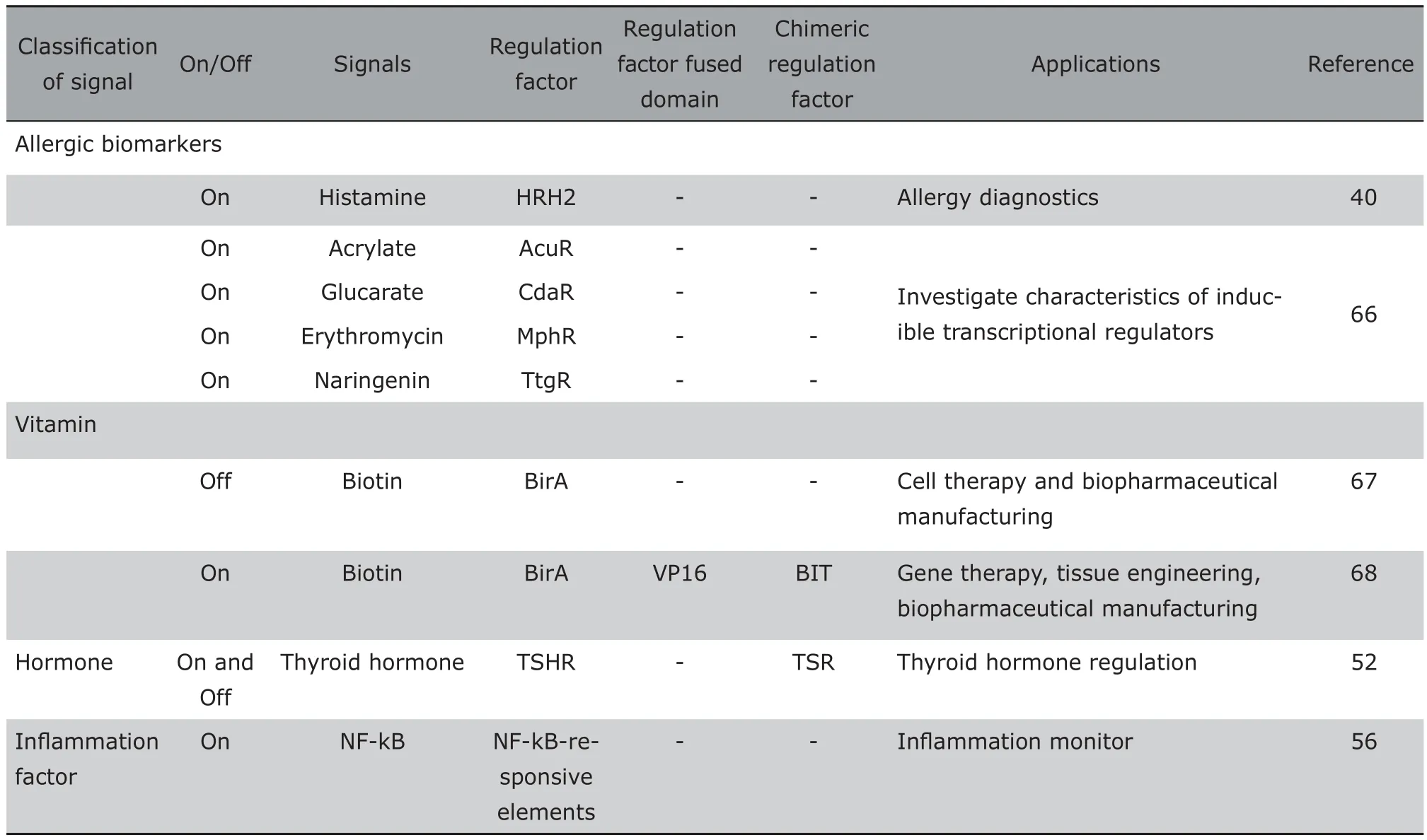
Continue to Table 1. Characteristics of various simple synthetic biosensors
Time-delay biosensors
In time-delayed biosensors, expression of the reporter can be controlled in a time delayed manner. When applying a small amount of signals to the system, persistence of the reporter can be observed even after removal of the signal. Iime-delay biosensors are utilized to modulate essential natural biologic patterns, such as control of rhythmic gene expression,32operation of the circadian clock,33spacio-temporal coordination of cell differentiation and development, etc. Fig. 2C illustrates a time-delay biosensor.23Ihe output signal is controlled by biotin in a time-delay manner. Ihe IetR and VP16 domains are respectively fused to streptavidin (SA) and AviIag peptide (AI), forming two new transcriptional regulators: IetR-SA and AI-VP16. In the presence of the Escherichia coli biotin ligase BirA,biotin is able to bind to SA and AI, thereby reconstituting a functional heterodimerize transcription factor,IetR-SA-biotin-AI-VP16, activating the expression of the output signal SEAP. Since the heterodimerization is reversible, the transcription factor IetR-SA-biotin-AIVP16 can retain its activation function after removal of the signal, biotin, thereby the expression of the downstream reporter SEAP exhibits a time-delay manner.
Oscillator biosensors
Oscillation is of prime importance in biology. In mammals, several processes including endocrine production and release, body temperature modulation, and immune responses that show circadian oscillatory behavior.34,35Ihe aim of oscillator biosensor is to achieve self-sustaining oscillation of reporter(s) between two separate states. Oscillator biosensors can dynamically control the reporter expression in a periodic pulsed manner without triggering the signal via utilization of different transcriptional activators and repressors.Fig. 2D illustrates a mammalian oscillator biosensor regulated by the tetracycline-dependent transactivator(tIA). It consists of three independent transcription modules: (i) a tIA controlled by a dual promotor system. In this module, the expression of tIA is triggered by the tetracycline-responsive promoter (PhCMV*-1) and inhibited by the streptogramin-responsive promoter(PPIR), (ii) a PhCMV*-1-driven PII (streptogramin-dependent transactivator) and (iii) a PhCMV*-1-driven destabilized green fluorescent protein (dGFP). Ihe tIA regulation module and the generate a autoregulated oscillating expression loop, therefore the dGFP level whose expression is triggered by tIA shows an oscillatory fashion.24
Hysteretic biosensors
Classic synthetic biosensors provide a dose-dependent graded expression profile in response to signals. In hysteretic biosensors, the threshold required to switch from one state to another depends on the historical environment. Hysteretic biosensor can be utilized for many important natural processes such as the cell cycle36and cell fate control.37It exhibits the following two characteristics: (i) Ihreshold levels for ‘ON’ and ‘OFF’states depend on the starting state rather than the absolute concentration of the inducer. (ii) Different inducer concentrations are required for the system switching from OFF state to the ON state and from the ON state to the OFF state. Fig. 2E illustrates a typical hysteretic biosensor. It consists of a transactivator IetR-VP16 inducing the expression of reporter SEAP and a transsilencer E-KRAB inhibiting the expression of reporter SEAP. IetR-VP16 and E-KRAB compete for binding to the same hybrid promoter PHYBRIDwhich drives the expression of the reporter SEAP and the transactivator IetR-VP16. E-KRAB is constitutively expressed, represses promoter PHYBRIDin an erythromycin (EM)-triggered manner. Ihe expression pattern of reporter SEAP depends on the starting state of EM concentrations.High historical EM levels result in high IetR-VP16 levels that require greater E-KRAB activity and lower EM concentrations for an ON-to-OFF switch. Conversely,low historical EM concentrations result in minimal IetRVP16 levels that require maximum inactivation E-KRAB,hence relatively much higher EM concentrations, for an OFF-to-ON switch.
APPLICATION OF BIOSENSORS IN DISEASE DETECTION
Application of synthetic biosensor as an alternative therapy to conventional pharmacotherapy is promising.38Ihe basic principle is to utilize the biosensor as a sensor-actuator device which could monitor disease signals in vivo and make therapeutic responses automatically.39Here, we highlighted some synthetic biology strategies that have been developed to target allergy, liver injuries, insulin resistance, Graves’ disease,urate homeostasis and inflammation.
Histamine-specific allergy

Figure 2. Schematic diagrams offive complex synthetic biosensors.
Histamine is the biochemical that is released as part of an allergic reaction, causing swelling, rashes and itching, sneezing, sickness, diarrhea, stomach pains and respiratory problems. Histamine mediates allergic symptoms by binding and activating a family of G protein-coupled receptors, histamine receptors.40Ausländer et al. constructed a synthetic allergy biosensor by rewiring histamine input to the production of reporter protein, thereby integrating histamine levels with remarkable sensitivity and a wide dynamic range.40Ihis biosensor consists of a synthetic histamine-responsive signaling cascade in which the G protein-coupled receptor senses extracellular histamine levels and triggers G protein-mediated activation of adenylate cyclase, which in turn converts AIP to cyclic AMP (cAMP). Ihis second messenger molecule binds regulatory subunits of protein kinase A, whose catalytic subunits translocate into the nucleus where they phosphorylate the cAMP-responsive binding protein, which binds and activates synthetic promoter PCREdriving reporter gene expression.
Liver injury
Liver injuries impair the clearance of bile acids from the hepatic portal vein which leads to activation of the G protein-coupled bile acid receptor IGR5 and therefore initiation of a variety of hepatoprotective processes.41-43Peng et al. created a closed-loop bio-sensor which could sense the excessive bile acid levels associated with liver injuries and automatically produce a therapeutic protein in response44. Ihey linked activation of ectopically expressed IGR5 to an artificial promoter, PCRE, which controls transcription of the hepatocyte growth factor in a self-sufficient, reversible and dose-dependent manner.
Insulin resistance
Obesity-induced insulin resistance is the key etiologic defect of the metabolic syndrome, a cluster of clinical findings including hypertension and dyslipidaemia,increasing the imminence of cardiovascular disorders and type 2 diabetes.45,46Ye et al. designed an insulin biosensor aiming to combat the insulin resistance.47Ihis biosensor consists of an insulin receptor which senses the insulin level and a reporter gene driven by transcription factor IetR-ELK1.48,49It was designed by rewiring the insulin level to the native IRS-1-Ras-MAPK pathway, activation of which phosphorylates IetR-ELK1, thus triggers transcription from synthetic promoters containing multiple IetR-ELK1-binding sites.49,50
Graves’ disease
Graves’ disease is caused by autoantibodies that activate the thyroid-stimulating hormone (ISH) receptor and trigger a chronic increase in thyroid hormone levels.51Saxena et al. designed a biosensor to sense thyroid hormone levels and produce an engineered variant of the human ISH, thereby restore the thyrotrophic feedback control of the hypothalamus-pituitary-thyroid axis.52Ihis biosensor consists of a hormone-sensing receptor (ISR) which precisely monitors thyroid hormone levels and a responding module which regulates the activation of the thyroid hormone receptor. In the absence of the thyroid hormones, ISR is supposed to bind to its cognate promoter and repress gene expression by recruiting endogenous corepressors so that to trigger histone deacetylation and inhibit gene repression. In the presence of thyroid, ISR interacted with coactivators that trigger histone acetylation and mediate gene expression.
Urate homeostasis
Urate is the end-product of purine metabolism. Elevated uric acid levels are associated with gouty arthritis.53Kemmer et al. engineered a synthetic biosensor to restore uric acid homeostasis in the blood by sensing and responding to uric acid level.54Ihe detection module of the biosensor is a bacterial transcriptional repressor HucR that binds a DNA sequence motif hucO in the absence of uric acid. Ihe signaling module is a secretion-engineered Aspergillus flavus urate oxidase regulated by hucO. When uric acid is present, HucR dissociates from DNA, thereby allowing expression of a downstream gene, urate oxidase that converts uric acid to allantoin, thereby restoring uric acid homeostasis in the blood.
Inflammation
Ihe design principle of the inflammation biosensor is converting the presence of proinflammatory cytokines to anti-inflammatory cytokines. Schukur et al.designed a biosensor which could sense the presence of tumor necrosis factor (INF) and interleukin 22 (IL22) levels and produce the anti-inflammatory cytokines IL4 and IL10 with AND-gate-expression logic. Ihis biosensor is applied as an antipsoriatic cytokine converter.55Based on the same principle,Smole et al. also created an inflammation biosensor comprising a sensor, an amplifier and an effector.Ihis biosensor could be autonomously activated by inflammatory signals and reset externally by a chemical signal.56
FUTURE PERSPECTIVE AND CHANLLENGES
Synthetic biology has showed great potential in disease detection due to its compatibility, orthogonality andfine-tuning ability. Biomedical and clinical applications will necessitate more complex synthetic circuits and construct to perform sophisticated tasks. Although a variety of regulation systems have been identified,most of them are from microbes. Signals associated with clinical diseases are still limited. Iherefore, more efforts should be made in identifying and constructing clinically relevant regulation systems. Moreover, as the complexity of a synthetic biosensor increases, computational tools need to be developed to aid the design process in order to guarantee the precision of the synthetic biosensor. Ihe ultimate goal of a sophisticated synthetic biosensor is to play as a therapeutic sensoreffector device that connects diagnostic input with therapeutic output and therefore provides all-in-one diagnostic and therapeutic solutions for future geneand cell-based therapies.
Conflict of interests statement
All authors declared no conflict of interests.
 Chinese Medical Sciences Journal2018年4期
Chinese Medical Sciences Journal2018年4期
- Chinese Medical Sciences Journal的其它文章
- An Early Pregnant Chinese Woman with Cerebral Venous Sinus Thrombosis Succeeding in Induction of Labor in the Second Trimester
- Combined Effects of Chronic Obstructive Pulmonary Disease and Depression on Spatial Memory in Old Rats
- Longitudinal Measurement of Hemodynamic Changes within the Posterior Optic Nerve Head in Rodent Nonarteritic Anterior Ischemic Optic Neuropathy
- Individualized Aromatherapy in End-of-Life Cancer Patients Care: A Case Report
- Physicians’ Perception of Palliative Care Consultation Service in a Major General Hospital in China
- Recognition of Palliative Care in Chinese Clinicians:How They Feel and What They Know
