Recent advances in network pharmacology applications in Chinese herbal medicine
Jian Hao*, Shi-Jun Li
1 The Fourth Central Hospital Affiliated to Nankai University, Tianjin, China.
Background
In 2007, British scientist Hopkins proposed a new discipline concept, network pharmacology, which combines pharmacology with network analysis [1].Network pharmacology is a comprehensive discipline that includes systems biology, fusion pharmacology,information networking, and computer science. Through the use of omics, high-throughput screening, molecular exchange, network analysis is used to expose the complex network signal relationship between drug genes, targets and diseases, analyze and simulate the pharmacological mechanism of drugs, and evaluate the therapeutic effects,side effects and theoretical mechanisms. Based on the network of “disease, gene, target and drugs”, network pharmacology comprehensively observes the intervention and influence of drugs on the disease network, and reveals the mystery of multi-molecular drugs synergistically acting on disease. These concepts reflect the ideas of multi-component, multi-target and system regulation, and have many similarities with the research ideas of traditional Chinese medicine (TCM), which focuses on syndrome differentiation and treatment,emphasizes the overall understanding of the etiology and pathogenesis of the pathogenesis.
Through the analysis of CNKI database, before 2010,researches of Chinese herb medicine (CHM) mainly focused on the compatibility of prescriptions, medication rules, clinical applications, etc. (Figure 1A), and after 2010, network pharmacology flourished and become a popular methodology for CHM research (Figure 1B).Therefore, this paper reviews the application fields of network pharmacology in CHM.
Process in network pharmacology research
One of the core problems of network pharmacology is to evaluate the comprehensive synergy effects of multiple targets of CHM on disease-related molecular networks. In view of the unclear targets and complex compositions of CHM, network pharmacology can be easily decomposed into 4 steps (Figure 2). (1) Identify the effective ingredients of CHM; (2) Identify the predicted or proved targets; (3) Identify the genes or proteins related to the disease and construct a disease network; (4) Determine the signal pathways and networks regulated by the target of CHM, and evaluate the impact of CHM on disease networks. The corresponding database and tools are shown in Table 1 [2-50].
The main applications of netw ork pharmacology in the researches of CHM
With increasing knowledge of the network of genes and molecular interactions, more and more researchers adopt network pharmacology in the researches of CHM. The applications of network pharmacology in CHM were systematically summarized to demonstrate the significant value in this area of researches (Figure 3).
Study the herbal mechanism
Analyze the cor e mol ecular targets and pharmacodynamic networks of CHM, explaining herbal mechanism
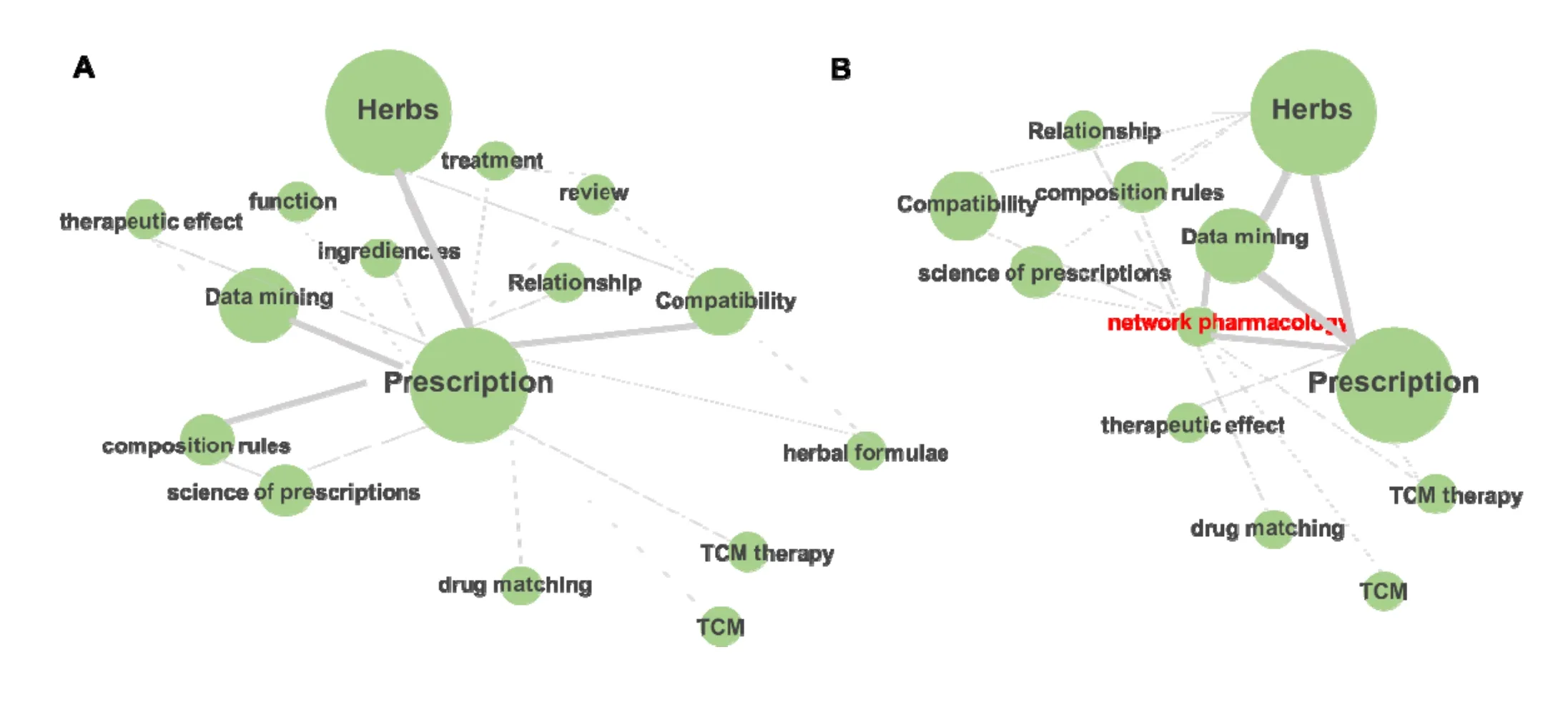
Figure 1 The main researches of Chinese herb medicine before (A) and after (B)
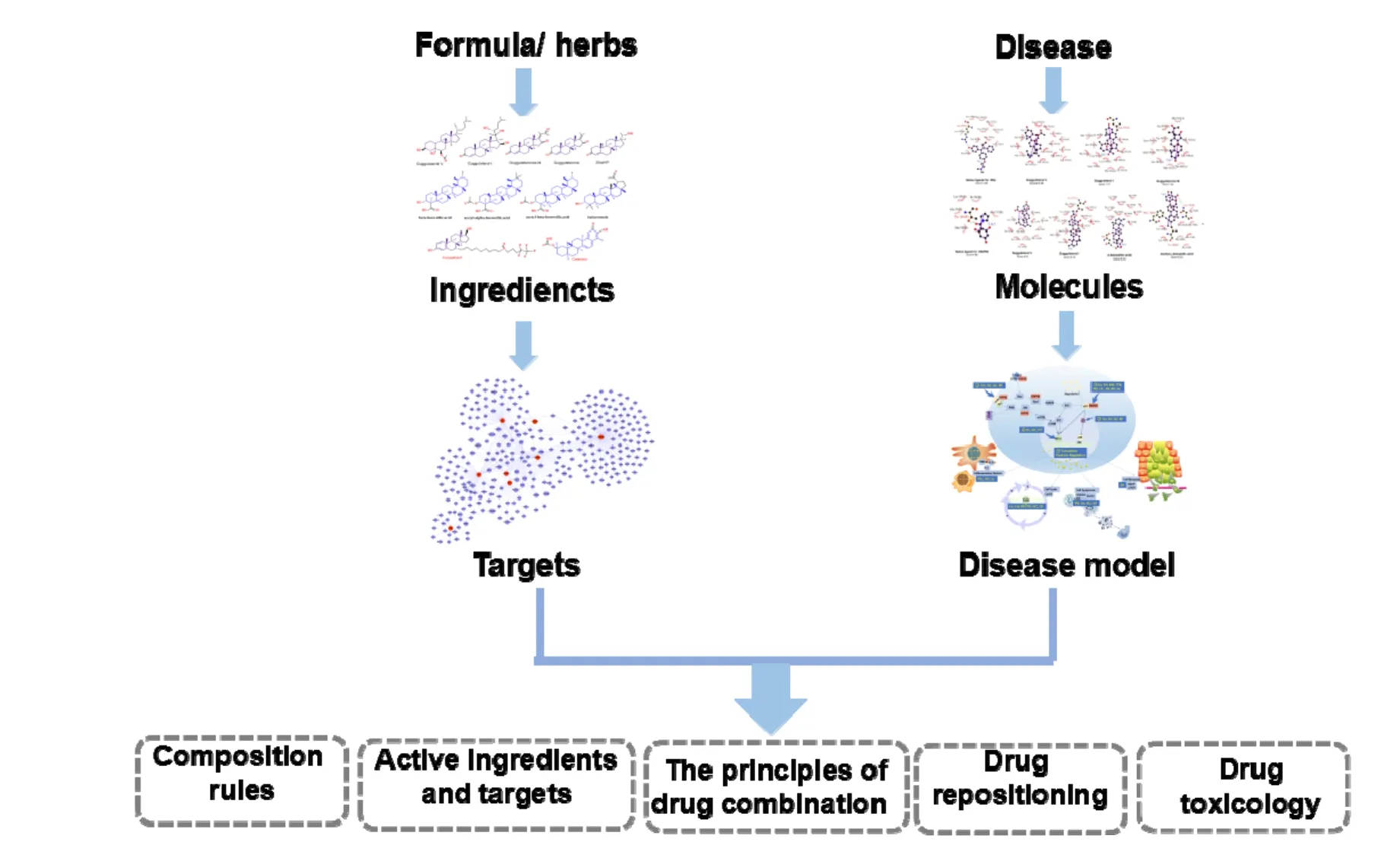
Figure 2 The main process of network pharmacology in the research of Chinese herb medicine
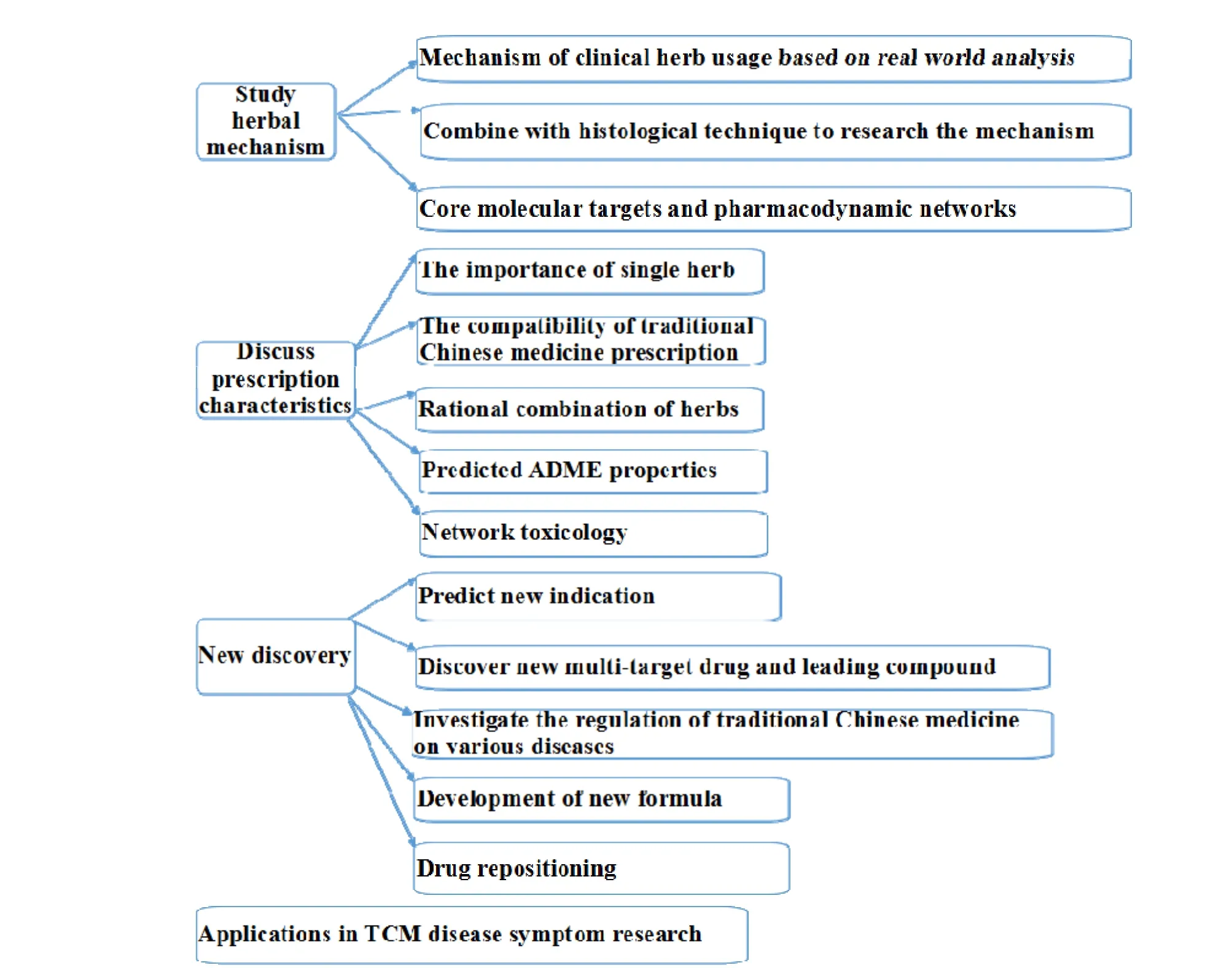
Figure 3 The main applications of network pharmacology in the researches of Chinese herb medicine

Table 1 The main databases and tools for traditional Chinese medicine research
The traditional discovery of the herbal active ingredients generally uses animal or cell models and conducts multiple separation and extraction and activity tests to screen chemical components and determine the active site;Or based on the metabolic law, infer prodrugs or secondary metabolites to determine the active ingredient of the herbs. Although high-throughput drug screening technology can be screened quickly and efficiently on a large scale, it still requires a lot of work to determine the appropriate active ingredient, which is low in efficiency,large in workload, and costly. Network pharmacology can predict key targets, active ingredients at a holistic level,based on multiple interactions between drugs, in vivo molecules characters, and biological progresses, through targeted separation, virtual screening, and molecular docking, etc.
This technology integrates drug interaction networks into biological networks to provide a more intuitive understanding of the interaction between drugs and organisms, helping to research drugs at an overall level. It is of great benefits for drug discovery and optimized design. For example, Lei-Hong Wu et al. [51] screened the molecular structure and target data provided by the Food and Drug Administration (FDA), and predicted 22 chemical constituents of aconites, reflecting the"multi-component, multi-target" characteristics of CHM.Li et al. [52] predicted the targets of known chemical constituents of Gegen Qinlian Decoction (Shanghan Zabing Lun, Zhong-Jing Zhang, 200 A.D.-205 A.D.) and clustered the targets according to the FDA-approved diabetes drugs. They inferred 19 major active componentsin Gegen Qinlian Decoction, and verified in cell experiments. Wang et al. [53] established a quantitative content-weighted ingredient-target network based on the qualitative identification of active ingredient groups. Taking Xue Sai Tong injection as the research object, the composition-weighted index of each component's target in the network and the composition-weighted index were used to comprehensively evaluate the pharmacodynamic contribution of each component. The results showed that notoginsenoside R1, ginsenoside Rg1, Rb1, Rd and Re were the main active components in Xue Sai Tong injection, and the results were also verified in the rat myocardial infarction model.
Combine with histological technique to research the mechanism of herbs
With the rapid development of omics technology, the combination of proteomics [54], metabolomics [55], gene chip [56] and network pharmacology has gradually increased. At first, the bio-effect spectrum of experimental animals before and after modeling and herb intervention was obtained by using omics technology.Then, the key targets and pathways were screened out and matched with network model to explain the active ingredients and mechanism of herb more accurately.Xiang et al used molecular docking, pathway enrichment analysis, network analysis and other methods, combined with metabolomics, serum medicinal chemistry,histopathology, immunohistochemistry results to clarify the active ingredients of Dahuang (Rhubarb) for the treatment of renal fibrosis and the mechanism of action of molecules [57].
Research the charact eristics and mechanism of clinical herb usage based on real world analysis
Real-world research is a clinical study of a large sample,open, non-randomized, non-intervention, and placebofree drug for the purpose of collecting clinical real-time medication data from the practicality of clinical trials.The research scope is wider, the data processing is more complicated, and it is more representative and truer to reflect the safety and effectiveness of the drug in the clinic. By analyzing the rules of clinical prescriptions, we found out the survival-related drugs and prescriptions,and further analyzed the main components and mechanisms of their effects through network pharmacology. This kind of real-world-based clinical prescription drug exploration can better discover the application of clinical prescriptions, and at the same time,according to the prediction of the drug mechanism by network pharmacology, it can better understand the characteristics of the prescription herbs. Meanwhile, the analysis of the mechanism based on real-world clinical drug prescriptions is closer to the actual situation of herb usage, and its guiding significance is very important. An example of its appliaction on the research field of cancer is as following. At first, we analyze the effect of CHM on advanced cancer treatment in the real-world situation. If we prove that CHM is effective for patients with advanced cancer, we further apply bivariate correlation analysis to find out which herbs are related to patient survival using correlation analysis. Next, we analyze the mechanisms by which these drugs treat advanced tumors through network pharmacology. At present, we have analyzed the main drugs, components, targets and mechanisms related to survival in breast cancer [58], liver cancer [59], gastric cancer [60], and colorectal cancer [61]using network pharmacology.
Discuss the prescription characteristics
Investigate the importance of single herb in formula
The prescription is based on the compatibility principle of Jun Chen Zuo Shi in TCM theory, but the drug effects may change in different diseases. By studying the role of Xi Huang formula (Wai Ke Zheng Zhi Quan Sheng Ji,Qing-Ren Wang, 1740) in the treatment of breast cancer,we found that Ruxiang (Olibanum) and Moyao (Myrrh)are the main drug in Xi Huang formula, which can directly inhibits breast cancer, but the direct inhibition of Shexiang (Musk) and Niuhuang (Calculus bovis) is not clear. This finding challenges the original compatibility principle of Xi Huang formula, in which Shexiang (Musk)and Niuhuang (Calculus bovis) were considered as the main drugs. Further, through network pharmacological analysis, it was found that ER (estrogen) receptor and HSP90 (Heat shock protein 90) are the main targets for inhibiting breast cancer in Ruxiang (Olibanum) and Moyao (Myrrh). It has been confirmed by molecular biology experiments that Ruxiang (Olibanum) and Moyao(Myrrh) can block the binding of estrogen receptor to HSP90, promote the degradation of ER receptor, and block the transport of ER receptor into the nucleus. [62].
Study the compatibility of CHM prescriptions
The compatibility of CHM prescriptions mainly focuses on the aspects of “medicinality”, “Jun Chen Zuo Shi”,“impassioned harmony” and “micro-inhibition but multi-effects”. There are synergistic, additive,antagonistic and attenuating effects after combining different herbs, which lead to multi-component,sequential amplification and multi-target advantages.Previous researches often explained the functions of formula according to the correlation between the active ingredients and the effective targets. The more the active ingredients and the targets, the more important position in the prescription, and vice versa. Tao et al [63] calculated 58 active constituents and their associated 32 protein targets in Yu Jin Fang. Through compound-target-disease network analysis, 7 of the 9 most important active ingredients were found from Yu Jin (Curcuma Aromatica),thus reflecting the importance of Yu Jin (Curcuma Aromatica); the potential target of Zhi Zi (Cape jasmine)was second only to Yu Jin (Curcuma Aromatica),indicating synergy with Yu Jin (Curcuma Aromatica); the number of targets calculated by musk and borneol was less, which suggested that the two herbs might not directly act on the disease, but reduced the toxic side effects of Yu Jin (Curcuma Aromatica) and Zhi Zi (Cape jasmine), or promoted the distribution of the main active ingredient in the target organ. Through network analysis,it was found that the central, near-central, topological coefficient, shortest path and other parameters of Dan Shen (Salvia Miltiorrhiza) and San Qi (Pseudo-Ginseng)chemical components were similar, which showed the possible mechanisms that the two herbs could synergistically enhance pharmacodynamic effects of each other.
Guide rational combination of herbs
The ancient book recorded that some herbs could not be used in combination because of incompatibility. The eighteen incompatible medicaments, the nineteen medicaments of mutual restraint and the contraindication during pregnancy, which the practitioners have been abiding by, have a long history. The eighteen incompatible medicaments, the nineteen medicaments of mutual restraint are taboo principles in the CHM compatibility. It is mainly believed that the opposite herbal compatibility may cause side effects even toxicity.Based on these theories, Gua Lou (Trichosanthes) and Wu Tou (Aconitum) cannot be used together. However,according to the modern pharmacology, some herbs in these records are not absolute incompatibility. The combination of some herbs in particular compability condition including different dosage and ratio has synergistic effects, especially the application of active ingredients in the modern researches. Through network pharmacology analysis and molecular biology experiments, we found that higenamine, the main active ingredient of Wu Tou (Aconitum), could synergistically promote the anti-breast cancer effects of cucurbitacin B,the active ingredient of Gua Lou (Fructus Trichosanthis).This combination broke the incompatibility between Wu Tou (Aconitum) and Gua Lou (Fructus Trichosanthis)[64].
Predicted absorption, distribution, metabolism,excretion (ADME) properties
Metabolic process of the herbs is very complicated. It used to be a practical and way to find the effective ingredients based on the in vivo process. With the development of the network pharmacology, it is a simple and quick method to perform virtual screening by calculation method and identify the effective component group. Ekins et al. [65] summarized some of the models currently used in computer-constructed predictive compound ADME properties in the 220th Annual Meeting of the American Chemical Society, including Stephen Johnson's application of regression tree and neural network methods to predict drug-protein binding,William Egan Model for passive intestinal absorption of drugs predicting, a semi-empirical quantitative method to predict the permeability of blood-brain barriers used by Bernd Beck, etc., but a common deficiency of these methods was based on two-dimensional structure. VolSurf software convert the relevant information extracted from the 3D structure into several easily understood and interpreted descriptors (parameters). Variable prediction model is used then to construct a 3D structure of the compound to explain its biological behavior [66, 67]. By calculating the ADME virtual high-throughput screening of herbs, it provides predictions for the study of metabolites of CHM, making the research more targeted and purposeful. At present, through the integration of multiple types of data, integrated oral bioavailability screening, ADME characteristics analysis,pharmacodynamic pattern recognition, target prediction,network analysis and other tools [68], a variety of database and analysis platform are established, which is of great significance for the discovery and evaluation of biologically active natural products.
Network toxicology
The cause of the toxicity of the formula is complicated.At present, the research on prescription toxicology is mainly to understand the adverse reactions of prescriptions through various toxicity tests and to guide rational drug use. There are currently thousands of formulae, more than 10,000 kinds of herbs. However, we know very little about the medicinal properties and its toxicity or compatibility.
Network pharmacology, from a holistic perspective,comprehensively prospects to understand the molecular mechanisms of disease and the mechanism of action of drugs. Some domestic scholars have proposed the drug CIPHER method based on large-scale prediction of disease-causing genes and drug targets. The predicted drug target spectrum contains drug targets and off-target effects. The clustering characteristics of the target spectrum can be used to discover the side effects of drugs[69]. In addition, a comprehensive and dynamic drug toxicity evaluation method was established based on the metabolic network analysis method to study the nephrotoxicity of CHM containing aristolochic acid [70].
New discovery
Predict new indications for herbs or formula
The ingredients in herbs can influence multiple disease-related targets through interrelated signaling pathways. By constructing a drug-target-signal pathway-disease network model, new possible indications for herbs or formula can be speculated [71, 72]. For example, by analyzing the main components and targets of Yu Jin Fang, the authors found that it not only acted on cardiovascular disease-related signaling pathways, but also on PI3K/AKT signaling pathways and ERK signaling pathways, which are tumor and cerebrovascular diseases. Therefore,Yu Jin Fang was found to have a good effect on tumor and cerebrovascular diseases in addition to cardiovascular and vascular diseases [71]. In addition to anti-influenza, Re-du-ning injection could also be used for clinical treatment of tuberculosis, diabetes, cancer,cardiovascular disease and immune system diseases [73];The anti-Alzheimer's disease herbal ingredients not only inhibited the classic targets, also affected some targets of the inflammation, cancer and diabetes [74].
Discover n ew multi-target drug and lead ing compound
As a summary of clinical experience, CHM has clear pharmacodynamic effects, and shows good development prospects in discovering new multi-targeted drugs. Gu et al. [75] analyzed the principal components of 197, 201 natural compounds and found that these compounds had high chemical structure overlap with FDA-approved drugs, indicating the great potential of natural compounds to develop into lead compounds. At the same time, the collected natural compounds were docked with the 332 FDA-approved protein targets and obtained 10 most potent compounds. Cardiovascular disease is a hot trend in network pharmacology research. Gu et al [76]constructed a cardiovascular disease herbal database(CVDHD, https://pkuxxj-pku-edu-cn.vpn.seu.edu. Cn/),containing 35230 compounds and their molecular properties, molecular docking results with 2395 protein targets, and correlation analysis with related diseases,pathways, and biological indicators.
Network pharmacology has also been applied to the discovery of new leading compounds from natural products. Sun et al. used network pharmacology and comparative proteomics to synthesize anti-cancer monomer component U12 with ursodeoxycholic acid as the leading compounds; The animal experiments have confirmed that anticancer activity of U12 is superior to ursodeoxycholic acid, and side effects of U12 are less than fluorouracil [77].
Development of new formula
Network pharmacology explores the relevance of drugs-diseases from a holistic and systematic perspective,discovers drug targets on biological networks. It is essentially a strategy for the development of new drugs.There are two main methods for discovering new drugs by using network pharmacology. One is the “new drug use” method, and the other is to develop single-molecule multi-target drugs by mining key nodes and functional modules in the network, or multi-molecules [78, 79].CHM contains a huge potential for new drug development. Traditional prescriptions are an inexhaustible source of multi-target drugs. At present,drug research and development based on CHM will focus on the active ingredients. However, the efficacy of CHM is often not the result of a single herb, but is achieved by the synergy of multiple herbs. According to the basic characteristics of the role of CHM, it is needed to establish a network of CHM pharmacodynamic components, a target network of pharmacodynamic components, a network of pharmacodynamic components,etc., and explore the adjustment, integration and optimization of multi-component, multi-target and multi-link [80].
Build multi-level network model to investigate the regulation of CHM on various disease.
The emergence of network pharmacology has opened up a new way to explore the potential pharmacodynamics of CHM and their prescriptions, their targets and their mechanisms from the level of systems biology and network biology. By constructing a“ingredients-target-signal pathway-disease” multi-level network model, network pharmacology research can dynamically understand the active components and targets of drugs, and explore the possible indications,which is somewhat similar with the idea of "same treatment to different diseases" of TCM. Shao Li [81]proposed a distance-based mutual information model, to uncover the combination rule embedded in herbal formulae and found that Liu Wei Di Huang Wan (Xiao Er Yao Zheng Zhi Jue, Yi Qian, 1735) acted on a common network target underlying different diseases, and captured the “one formula, different diseases” relationship from a co-module viewpoint based on multilayer networks of herb-biomolecule-disease.
Drug repositioning
Drug repositioning refers to the discovery of new indications or new uses of a marketed drug [82].Traditional single target drug development has exposed limitations. Compared with the development of new drugs, repositioning the clinically reliable drugs can not only effectively reduce the cost of research and development, shorten the cycle, but also effectively control the safety and pharmacokinetics. The number of herbs and prescriptions written in ancient and modern literatures have been clinically proven safe and effective.Moreover, the same prescription has therapeutic effects on different diseases. Therefore, CHM is the source of new drug development and drug repositioning of multi-target drugs [83, 84].
The multi-level research strategy of network pharmacology coincides with the cognition of TCM's overall healing balance, which provides new hope for traditional prescriptions to explore new drug compatibility, with a view "implementing the old drug new use". For example, Wang Yi [85] searched nine components of compound Dan Shen Fang, obtained target information, screened OMIM database for extracting cardiovascular related disease gene data, and constructed active ingredient-gene-cardiovascular disease network model, and found 9 activities. The components all had effects on multiple targets, involving various diseases such as hypoglycemia and diabetes. It indicated that each active ingredient had a regulatory effect on different gene groups, and the shared genes were linked to different gene groups, indicating the synergistic effects between different targets. Compound Dan Shen had a new research prospect of clinical indications. Xiao-hua Zhang established a pharmacophore model of L-type calcium channel antagonist, which was obtained through database evaluation by analyzing 12 compounds from NCBI that inhibited the L-type calcium channel of New Zealand rabbit heart. This model was used to screen the listed drugs and TCMS with potential L-type calcium channel antagonism in Drugbank and TCM for re-positioning and evaluation of antihypertensive effects. This strategy was helpful to promote drug repositioning in TCM [86].
Applications in TCM disease symptom research
The most distinctive diagnosis and treatment model of TCM is the overall combination of “diseasesymptoms-formula”. TCM syndrome research can use the classical method of network pharmacology to screen the typical clinical syndrome information. Li Shao et al. [87]conducted research on the biological basis of syndromes from the perspective of biomolecular networks, and formed a systemic "disease, syndrome and prescription"system, conducted the "phenotype network -biomolecular network - drug network" research framework, and further put forward the concept of "signs biomolecular network". Taking the basic syndrome system of "cold and hot" of TCM as an example, the research team established a cold and heat syndrome biomolecular network based on the neuroendocrineimmune system, and found that the cold syndrome biomolecular network was based on the functional modules of hormones, the hot syndromebiomolecular network was mainly composed of functional modules of cytokines, and the neurotransmitter functional modules were distributed in both cold and hot networks.; It was found that the bio-molecular network of cold-heat syndrome had the scale-free nature, that was, the function realization of the network mainly depended on some key nodes, which were expected to become network markers of cold syndrome and heat syndrome; It was also found that the network could better characterize the different biological effects of the cold formula Qing Luo formula and the warm formula Wen Luo formula. In addition, the researchers also found two gene expression patterns of energy metabolism and immune regulation network imbalance in cold syndrome and hot syndrome patients[88]. The researchers verifyed the key nodes of the network, found the potential biomarkers of patients with cold syndrome and heat syndrome [89]; they also carryed out the research on the microbes of tongue coating of TCM "cold and hot" syndrome for the first time. The high-throughput sequencing and bioinformatics analysis of the tongue-skin microbial group constructed a differential microbial network of "cold and hot"syndrome, suggesting that the microbial community of tongue coating is a new type of biomarker for distinguishing patients with "cold, hot" syndrome [90].The above researches applied the syndrome network to syndrome objectification and individualized diagnosis and treatment, which provides a new idea for the basic research of syndrome biology.
Liu Zhongdi et al. [91] used two kinds of basic diagnostic syndromes of rheumatoid arthritis (RA),containing cold syndrome and heat syndrome, to compare the genes and metabolites of patients and healthy people to obtain different substances through metabolomics and gene chip technology. They analyzed the biomolecular network corresponding to cold syndrome and heat syndrome, and explained the biological basis of TCM cold and heat syndrome. Niu Xuyan et al. [92] analyzed the drug target network and biomolecular network of RA heat syndrome type, and concluded that RA heat syndrome had a common signal pathway, which might be the "drug-certification" treatment of RA heat syndrome.Network pharmacology had also achieved in the study of syndromes of other diseases, such as RA deficiency syndrome [93], liver cancer deficiency syndrome [94],coronary heart disease, phlegm and blood stasis syndrome[95], blood stasis syndrome [96] and strokes of the wind and phlegm blocking evidence [97] and other aspects.Biomolecular network and drug target network are mainly based on a large number of databases and algorithms as the cornerstone of predictive research, providing a new channel for TCM syndrome differentiation and drug research.
Perspective and shortage
Traditional prescriptions are an inexhaustible source of multi-target drugs. According to the basic characteristics of CHM, establishing a network of Chinese medicine pharmacodynamic components and targets will provide new ideas for drug discovery. Network pharmacology explores the relevance of drugs-diseases from a holistic and systematic perspective, discovers drug targets on biological networks, and clarifies the mechanism. It is essentially a strategy for the research of CHM. However,there are still some disadvantages needs to be perfected.First, the biggest problem is the lack of a complete database containing CHM, Chinese medicine ingredients and biological target. In the most existing databases, the information is not complete, especially lacking the information of animal-related drugs.
Second, the resent researches lack the dynamic investigation of the relationship between drugs and diseases. Disease is a process of progress and a dynamic process. The prediction of the corresponding disease target through network pharmacology does not truly reflect the characteristics of the disease progression process. Last but not least, the research method is not uniform. Such as inconsistent database selection results in incomplete drug composition. The molecular model construction of the disease lacks objective criteria. Even,the values of drug likeindex and oral bioavailability are inconsistent.
Conclusion
The characteristics of multi-component and multi-target of CHM are in line with the research strategy of network pharmacology. With the continuous development of network biology and informatics, network pharmacology provides new methods for the researches of CHM, which has large scientific and clinical value. It provides key technical support for drug research and development,clinical diagnosis and personalized treatment, and also provides a new way for the modernization and internationalization of CHM.
1. Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol 2008, 4:682.
2. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014, 6: 13.
3. TCM Database@Taiwan. The world's largest traditional Chinese medicine database for drug screening in silico. PLoS ONE 2014, 6: e15939.
4. Gaulton A, Bellis LJ, Bento AP, et al. ChEMBL: a large-scale bioactivity database for drug discovery.Nucleic Acids Res 2011, 40: D1100-D1107.
5. Banerjee P, Erehman J, Gohlke BO, et al. Super Natural II-a database of natural products. Nucleic Acids Res 2014, 43: D935-D939.
6. Xue R, Fang Z, Zhang M, et al. TCMID: traditional Chinese medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Res 2012, 41: D1089-D1095.
7. Chen X, Ji ZL, Chen YZ. TTD: therapeutic target database. Nucleic Acids Res 2002, 30: 412-415.
8. Hao Y, Li Y, Hong K, et al. HIT: linking herbal active ingredients to targets. Nucleic Acids Res 2011,39: D1055-D1059.
9. Szklarczyk D, Santos A, von Mering C, et al.STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res 2016, 44: D380-D384.
10. Gao Z, Li H, Zhang H, et al. PDTD: a web-accessible protein database for drug target identification. BMC bioinformatics 2008, 9: 104.
11. Cheng D, Knox C, Young N, et al. PolySearch: a web-based text mining system for extracting relationships between human diseases, genes,mutations, drugs and metabolites. Nucleic Acids Res 2008, 36: W399-W405.
12. Wishart DS, Knox C, Guo AC, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 2006, 34:D668-D672.
13. Liu Z, Guo F, Wang Y, et al. Batman-TCM: a bioinformatics analysis tool for molecular mechanism of traditional Chinese medicine. Sci Rep 2016, 6: 21146.
14. Goodman J. Computer software review: Reaxys. J Chem Inf Model 2009, 49: 2897-2898.
15. Fang YC, Huang HC, Chen HH, et al.TCMGeneDIT: a database for associated traditional Chinese medicine, gene and disease information using text mining. BMC Complement Altern Med.2008, 8: 58.
16. Zhang R, Yu S, Bai H, et al. TCM-Mesh: The database and analytical system for network pharmacology analysis for TCM preparations. Sci Rep 2017, 7: 2821.
17. Gong J, Cai C, Liu X, et al. ChemMapper: a versatile web server for exploring pharmacology and chemical structure association based on molecular 3D similarity method. Bioinformatics 2013, 29:1827-1829.
18. Liu X, Ouyang S, Yu B, et al. PharmMapper server:a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res 2010, 38: W609-W614.
19. Benson DA, Karsch-Mizrachi I, Lipman DJ, et al.GenBank. Nucleic Acids Res 2008, 36: D25.
20. Becker KG, Barnes KC, Bright TJ, et al. The genetic association database. Nature genetics 2004, 36: 431.21. Rebhan M, Chalifa-Caspi V, Prilusky J, et al.GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics 1998, 14:656-664.
22. Hamosh A, Scott AF, Amberger JS, et al. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 2005, 33: D514-D517.
23. Piñero J, Queralt-Rosinach N, Bravo À, et al.DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes.Database 2015, 2015: bav028.
24. Wang W, Yang J, Muntz R. STING: A statistical information grid approach to spatial data mining.International Conference on Very Large Data Bases 1997, 97: 186-195.
25. Stark C, Breitkreutz BJ, Reguly T, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res 2006, 34: D535-D539.
26. Baolin L, Bo H. HPRD: a high performance RDF database. International Conference on Network and Parallel Computing. Springer, Berlin, Heidelberg,2007: 364-374.
27. Beuming T, Skrabanek L, Niv MY, et al. PDZBase:a protein–protein interaction database for PDZ-domains. Bioinformatics 2004, 21: 827-828.
28. Chatr-Aryamontri A, Ceol A, Palazzi LM, et al.MINT: the Molecular INTeraction database. Nucleic Acids Res, 2006, 35: D572-D574.
29. McDowall MD, Scott MS, Barton GJ. PIPs: human protein–protein interaction prediction database.Nucleic Acids Res 2008, 37: D651-D656.
30. Xenarios I, Rice DW, Salwinski L, et al. DIP: the database of interacting proteins. Nucleic Acids Res 2000, 28: 289-291.
31. Chen JY, Mamidipalli SR, Huan T. HAPPI: an online database of comprehensive human annotated and predicted protein interactions. BMC genomics 2009, 10: S16.
32. Sapkota A, Liu X, Zhao XM, et al. DIPOS: database of interacting proteins in Oryza sativa. Molecular BioSystems 2011, 7: 2615-2621.
33. Nishimura D. BioCarta. Biotech Software & Internet Report: The Computer Software Journal for Scient,2001, 2: 117-120.
34. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000, 28:27-30.
35. Zhao S, Li S. Network-based relating pharmacological and genomic spaces for drug target identification. PloS one 2010, 5: e11764.
36. Reardon S. Project ranks billions of drug interactions:drugable. com predicts mechanisms through computation. Nature 2013, 503: 449-451.
37. Chen YZ, Zhi DG. Ligand–protein inverse docking and its potential use in the computer search of protein targets of a small molecule. Proteins 2001,43: 217-226.
38. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading.J Comput Chem 2010, 31: 455-461.
39. Drews J. Drug discovery: a historical perspective.Science 2000, 287: 1960-1964.
40. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003, 13: 2498-2504.
41. Batagelj V, Mrvar A. Pajek-program for large network analysis. Connections 1998, 21: 47-57.
42. Jacknin L, Ericson MA. Virtual navigator, and inertial angular measurement system: U.S. Patent 5,854,843. 1998-12-29.
43. Thomas S, Bonchev D. A survey of current software for network analysis in molecular biology. Human genomics 2010, 4: 353.
44. Rohani VA, Hock OS. On social network web sites:definition, features, architectures and analysis tools.J Comp Engin 2009, 1: 3-11.
45. Hagberg A, Schult D, Swart P, et al. Networkx.High productivity software for complex networks.Webová strá nka https://networkx. lanl. gov/wiki,2013.
46. Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome biology 2003, 4: R22.
47. Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. International AAAI Conference on Weblogs and Social Media 2009, 8: 361-362.
48. Adar E. GUESS: a language and interface for graph exploration. Proceedings of the SIGCHI conference on Human Factors in computing systems. 2006:791-800.
49. Borgatti SP, Everett MG, Freeman LC. Ucinet for Windows: Software for social network analysis.2002.
50. Mueller LAJ, Kugler KG, Dander A, et al. QuACN:an R package for analyzing complex biological networks quantitatively. Bioinformatics 2010, 27:140-141.
51. Wu LH, Gao XM, Wang LL, et al. Prediction of multi-component target of aconite and network pharmacology research. Chin J Tradit Chin Med 2011, 36: 2907-2910.
52. Li H, Zhao L, Zhang B, et al. A network pharmacology approach to determine active compounds and action mechanisms of ge-gen-qin-lian decoction for treatment of type 2 diabetes. Evid Based Complement Alternat Med 2014, 2014: 495840.
53. Wang L, Li Z, Shao Q, et al. Dissecting active ingredients of Chinese medicine by content-weighted ingredient-target network. Mol Biosyst 2014, 10: 1905-1911.
54. Li X, Wu L, Liu W, et al. A network pharmacology study of Chinese medicine QiShenYiQi to reveal its underlying multi-compound, multi-target,multi-pathway mode of action. PLoS One 2014, 9:e95004/1.
55. Xiang Z, Sun H, Cai X, et al. The study on the material basis and the mechanism for anti-renal interstitial fibrosis efficacy of rhubarb through integration of metabonomics and network pharmacology. Mol Biosyst 2015, 11: 1067-1078.
56. Sun H, Zhang A, Yan G, et al. Proteomics study on the hepatoprotective effects of traditional Chinese medicine formulae Yin-Chen-Hao-Tang by a combination of two-dimensional polyacrylamide gel electrophoresis and matrix-assisted laser desorption/ionization-time of flight mass spectrometry. J Pharm Biomed Anal 2013, 75: 173179.
57. Chen S, Wu S, Li W, et al. Investigation of the therapeutic effectiveness of active components in Sini decoction by a comprehensive GC/LC-MS based metabolomics and network pharmacology approaches. Mol Biosyst 2014, 10: 3310-3321.
58. Mao Y, Hao J, Jin ZQ, et al. Network pharmacology-based prediction of active ingredients and potential targets of Chinese herbs on metastatic breast cancer patients. Oncotarget 2017: 15351.
59. Gao L, Wang XD, Niu YY, et al. Molecular targets of Chinese herbs: a clinical study of hepatoma based on network pharmacology. Sci Rep 2016, 6: 24944.
60. Gao L, Hao J, Niu YY, et al. Network pharmacology dissection ofmultiscale mechanisms of herbal medicines in stage IV gastric adenocarcinoma treatment. Medicine 2016, 95.
61. Zhu HX, Hao J, Chen D, et al. Molecular targets of Chinese herbs: a clinical study of metastatic colorectal cancer based on network pharmacology.Sci Rep 2018, 8: 7238.
62. Hao J, Jin Z, Zhu H, et al. Antiestrogenic activity of the Xi-Huang formula for breast cancer by targeting the estrogen receptor α. Cell Physiol Biochem 2018,47: 2199-2215.
63. Tao W, Xu X, Wang X, et al. Network pharmacology-based prediction of the active ingredients and potential targets of Chinese herbal Radix Curcumae formula for application to cardiovascular disease. J Ethnopharmacol 2013, 145:1-10.
64. Jin ZQ, Hao J, Yang X, et al. Higenamine enhances the antitumor effects of cucurbitacin B in breast cancer by inhibiting the interaction of AKT and CDK2. Oncol Rep 2018, 40: 2127-2136.
65. Ekins S, Rose J. In silico ADME / Tox : the state of the art. J Mol Graph Mod 2002, 20: 305.
66. Cruciani G, Pastor M, Guba W. VolSurf: a new tool for the pharmacokinetic optimization of lead compounds. Eur J Pharm Sci 2000, 11: 29.
67. Zhuang XM, Xiao JH, Zhang ZQ, et al. VolSurf software and its application in virtual high-throughput screening oournal Pharm Toxic 2005, 19: 156.
68. Xu HY, Tang SH, Chen JX, et al. Research ideas and strategies of traditional Chinese medicine“group-effect relationship” based on metabolomics.World Sci Technol Mod Tradit Chin Med Mater Med 2011, 13( 1) : 30
69. Li S, Zhang ZQ, Wu LJ, et al. Understanding ZHENG in traditional Chinese medicine in the context of neuro- endocrine- immune network. IET Syst Biol 2007, 1: 51-60.
70. Zhou MM, Liu P, Jia W, et al. Evaluation of therapeutic effects of TCM based on metabolic variation. World Sci Technol Mod Tradit Chin Med Mater Med 2006, 8: 113-119.
71. Sun Y, Zhu R, Ye H, et al. Towards a bioinformatics analysis of anti-Alzheimer′s herbal medicines from a target network perspective. Brief Bioinform 2013, 14: 327-343.
72. Gu J, Gui Y, Chen L, et al. Use of natural products as chemical library for drug discovery and network pharmacology. PLoS One 2013, 8: e62839.
73. Zhang XZ, Xiao W, Xu XJ, et al. Study on the molecular mechanism of the Reduning Injection on the Influenza virus using the network pharmacology method. Acta Phys Chim Sin 2013, 29: 1415-1420.
74. Agis-Torres A, Sollhuber M, Fernandez M, et al.Multi-target-directed ligands and other therapeutic strategies in the search of a real solution for Alzheimer’s disease. Curr Neuropharmacol 2014, 12:2-36.
75. Gu J, Gui Y, Chen L, et al. Use of natural products as chemical library for drug discovery and network pharmacology. PLoS One 2013, 8: e62839.
76. Gu J, Gui Y, Chen L, et al. CVDHD: a cardiovascular disease herbal database for drug discovery and network pharmacology. J Cheminf 2013, 5: 51.
77. Xu Y, Luo Q, Lin T, et al. U12, a UDCA derivative,acts as an anti-hepatoma drug lead and inhibits the m TOR/S6K1 and cyclin/CDK complex pathways.PLoS One 2014, 9: e113479.
78. Liu ZH, Chen SL. ER regulates an evolutionarily conserved apoptosis pathway. Biochem Biophys Res Commun 2010, 400: 34-38.
79. Wang Y, Gao XM, Zhang BL, et al. Building methodology for discovering and developing Chinese medicine based on network biology.China J Chin Mater Med 2011, 36: 228-231.
80. Li S, Zhang B, Zhang NB. Network target for screening synergistic drug combinations with application to traditional Chinese medicine. BMC Syst Biol 2011, 5: S10.
81. Li S, Zhang B, Jiang D, et al. Herb network construction and co-module analysis for uncovering the combination rule of traditional Chinese herbal formulae. BMC Bioinformatics 2010, 11: S6.
82. Li JW, Vederas JC. Drug discovery and natural products: end of anera or an endless frontier?Science 2009, 325: 161-165.
83. Li X, Wu LH, Fan YH, et al. Network pharmacology study of the main active ingredients of compound Danshen. Chin J Tradit Chin Mede 2011, 36: 2912-2915.
84. Liang YX, He YS, Jiang LZ, et al. L-type calcium channel channel antihypertensive drugs were found in traditional Chinese medicine using relocation strategy. Chin J Tradit Chin Med 2015, 40:2650-2654.
85. Li X, Wu L, Fan X, et al. Network pharmacology study on major active compounds of Fufang Danshen formula. Zhongguo Zhong yao Zazhi 2011,36: 2911-2915.
86. Zhang X, Gu J, Cao L, et al. Network pharmacology study on the mechanism of traditional Chinese medicine for upper respiratory tract infection. Mol Biosyst 2014, 10: 2517-2525.
87. Li S, Zhang ZQ, Wu LJ, et al. Understanding ZHENG in traditional Chinesemedicine in the context ofneuro-endocrine-immune network. IET Syst Biol 2007, 1: 51-60.
88. Ma T, Tan C, Zhang H, et al. Bridging the gap between traditional Chinese medicine and systems biology: the connection of cold syndrome and NEI network. Mol Biosyst 2010, 6: 613-619.
89. Li S. Network systems underlying traditional Chinese medicine syndrome and herb formula. Curr Bioinform 2009, 4: 188-196.
90. Zhang B, Wang X, Li S. An integrative platform of TCM network pharmacology and its application on a herbal formula, Qing-Luo-Yin. Evid Based Complement Alternat Med 2013, 2013: 456747.
91. Liu ZD, Jiang W, Tan Y, et al. Network pharmacology research strategy corresponding to rheumatoid arthritis cold and heat syndrome. Chin J Tradit Chin Med 2015, 30: 3191-3195.
92. Niu XY, Li J, Lu C, et al. Network pharmacology research on the mechanism of "drug-certification"for rheumatoid arthritis syndrome. Chin J Experim Formul 2012, 18: 299.
93. Wang M, Chen G, Lu C, et al. Rheumatoid arthritis with deficiency patternin traditionalChinese medicine shows correlationwith cold and hotpatterns in gene expression pro files. Evid Based Complement Alternat Med 2013, 20: 248650.
94. Chen QL, Lu YY, Zhang GB, et al. Characteristic analysis from excessiveto deficient syndromes in hepatocarcinoma underlying miRNA array data.Evid Based Complement Alternat Med 2013, 20:324636.
95. Li Z, Zhang W, Wang Y, et al. A New Model of TCM Syndrome Research in Network Syndrome.Tianjin J Tradit Chin Med 2013, 6:787-790.
96. Li X, Wu LH, Fan YH, et al. Pharmacological study on the main active ingredients of compound Danshen prescription. Chin J Tradit Chin Med 2011,36: 2911.
97. Dai W, Liu X, Zhang Z, et al. A two-level model for the analysis of syn drome of acute ischemic stroke:from diagnostic model to molecular mechanism.Evid Based Complement Alternat Med 2013, 2013:293010.
 Traditional Medicine Research2018年6期
Traditional Medicine Research2018年6期
- Traditional Medicine Research的其它文章
- The role of Si-Jun-Zi decoction in the management of gastric precancerous lesions and its mechanism
- A network pharmacology approach to investigate the mechanisms of Si-Jun-Zi decoction in the treatment of gastric precancerous lesions
- Analysis of the action mechanism of Fang Ji Huang Qi decoction in treating rheumatoid arthritis by network pharmacology
- Network pharmacology-based approach to investigate the mechanisms of Yiyi Fuzi Baijiang Powder in the treatment of malignant tumors
- Camel milk could be helpful in the treatment of asthma
