Altered leukocyte gene expression after traumatic spinal cord injury: clinical implications
Paige E. Herman, Ona Bloom,
1 The Feinstein Institute for Medical Research, Northwell Health, Manhasset, NY, USA
2 Departments of Physical Medicine and Rehabilitation, Molecular Medicine, Donald and Barbara Zucker School of Medicine at Hofstra Northwell, Manhasset, NY, USA
Abstract In addition to changes in motor and sensory function, individuals with spinal cord injury (SCI) experience immunological changes. These changes are clinically significant, as infections are the leading cause of death for this population. Along with increased infections, inflammation is commonly observed in persons with SCI, where it may promote many common medical consequences. These include elevated risk of cardiovascular disease,impaired wound healing, diabetes and neuropathic pain. It has also been proposed that chronic inflammation dampens neurological recovery. In order to identify therapeutic strategies to improve immune function, we need a greater understanding of the molecular changes that occur in immune cells after SCI. The purpose of this mini-review is to discuss two recent studies that used functional genomics to investigate gene expression in circulating leukocytes isolated from persons with SCI. In the future, the molecular pathways that are altered after SCI may be targeted to improve immunological function, as well as overall health and functional recovery,after SCI.
Key Words: traumatic spinal cord injury; inflammation; immune cells; functional genomics; gene expression;autoimmunity; microarray; chronic spinal cord injury
Traumatic Spinal Cord Injury (SCI)
There are an estimated 760,000–2.5 million persons living with traumatic SCI globally and more than 350,000 persons in the United States (National Spinal Cord Injury Statistical Center, 2017; Kumar et al., 2018). In the US, the average age at injury is 43 years old. The most common mechanisms of injury are motor vehicle accident (38%), followed by falls (32%),violence (14%), sports (8%) and other (8%) (National Spinal Cord Injury Statistical Center, 2017). The most common levels of injury are cervical (54%), thoracic (35%), and lumbar (10%).Unfortunately, there is no Food and Drug Administration(FDA) approved pharmacological therapy for SCI and living with SCI therefore imposes significant physical, psychosocial and economic burdens for patients and their families.
The gold standard clinical exam to classify motor and sensory function after SCI is the International Standards for Neurological Classification of SCI (ISNCSCI) exam (Kirshblum and Waring, 2014; Ahuja et al., 2017). The ISNCSCI exam tests motor and sensory function throughout the body and based on exam findings, spinal cord injuries are classified with the American Spinal Injury Association (ASIA) Impairment Scale(AIS) grades A–E (Table 1): A indicates a clinically complete injury with no motor or sensory function preserved in sacral segments S4–5, B–D indicate incomplete injuries, with some motor or sensory function preserved below the neurological level of injury, and E indicates a normal exam (Table 1) (Kirshblum and Waring, 2014). Different tools are used to reflect abilities to perform activities of daily living after SCI. One widely used tool is the Spinal Cord Independence Measure,which is a standardized, validated rating scale that measures the functional status or improvement in performing daily activities for people with SCI and has domains related to self-care, respiration, bowel and bladder care, and mobility (Catz et al., 1997;Itzkovich et al., 2007). Another more recently developed tool that was designed to be used as a patient reported outcome is the SCI-Functional Index (SCI-FI), which describes aspects and modes of mobility, self-care, and fine motor function (Jette et al., 2012; Fyffe et al., 2016).
Despite advances in clinical care, the National SCI Statistical Center (NSCISC) estimates that life expectancy for persons with SCI remains lower than for able-bodied persons and depends on age at injury, injury severity, injury level, ventilator use, general comorbidities and other factors (National Spinal Cord Injury Statistical Center, 2017). Life expectancy after SCI also changes with time from initial injury (National Spinal Cord Injury Statistical Center, 2017). For example, data from the NSCISC estimates that life expectancy for a person with a cervical level injury sustained at age 25 who survived the initial 24 hours after their injury (without ventilator use) was 31–36 years, while life expectancy for an able-bodied person at age 25 was 54.8 years (Ahuja et al., 2017; National Spinal Cord Injury Statistical Center, 2017). Data from the NSCISC estimates that life expectancy for a person with a cervical level injury sustained at age 70 who survived the initial 24 hours after their injury (without ventilator use) was 6.5–8 years, while life expectancy for an able-bodied person at age 70 was 15.6 years(National Spinal Cord Injury Statistical Center, 2017).
Traumatic SCI Compromises More Than Motor and Sensory FunctionInfections (genitourinary or respiratory) are the leading cause of death for persons with SCI and are also the most common cause of re-hospitalization after SCI (DeVivo et al., 1993;Cardenas et al., 2004; DeJong et al., 2013; National Spinal Cord Injury Statistical Center, 2017). Recent data has also identified infections as an independent risk factor for poor neurological recovery (Failli et al., 2012). Along with increased infection rates, inflammation is commonly observed in the SCI population and may play a role in the pathogenesis of many common medical consequences of SCI, including accelerated atherogenesis and osteoporosis, impaired wound healing,type II diabetes, and neuropathic pain (Bauman and Spungen,2000). In humans, elevated numbers of inflammatory cells in the spinal cord were identified years after initial injury (Fleming et al., 2006). Persistent elevated levels of intraspinal and systemic inflammatory mediators may also oppose neurological recovery (Schwab et al., 2014). Impaired immune cell function ex vivo has also been reported from persons living with SCI(Nash, 1994; Campagnolo et al., 2000). Autoreactive antibodies have also been described in persons with SCI (Schwab et al.,2014).
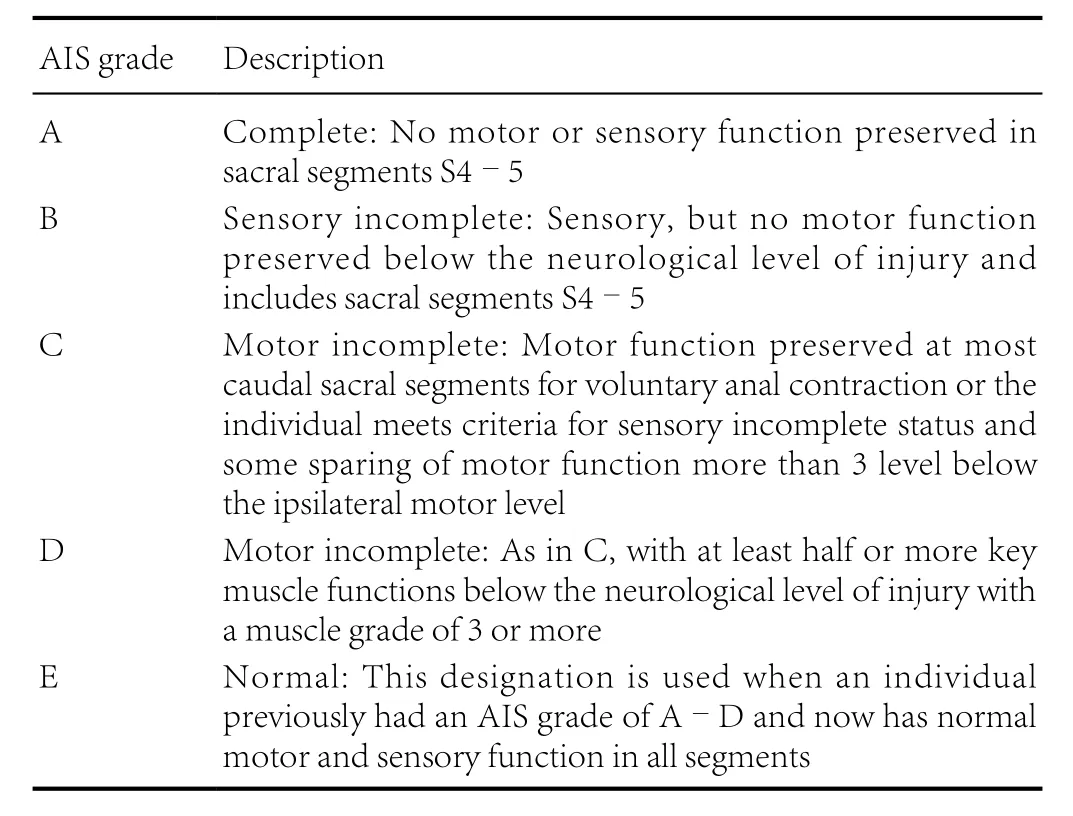
Table 1 American Spinal Injury Association Impairment Scale (AIS)
Impaired Neuroimmune Interactions after SCI May Contribute to Increased Rates of Infection and Chronic Inflammation
The autonomic nervous system (ANS) innervates most visceral organs (heart, lung, liver, and intestine) and also regulates immune system function (spleen, lymph nodes, bone marrow) (Figure 1). In able-bodied people, the ANS regulates homeostasis through synergistic and coordinated activation of the sympathetic nervous system (SNS) and parasympathetic nervous system (PNS) (Pavlov and Tracey, 2017). In persons with SCI, impaired ANS regulation of organ systems below the injury level are associated with serious medical consequences, ranging from autonomic dysreflexia to adverse changes in metabolism (Bauman and Spungen, 2000). A growing body of evidence shows that the ANS also regulates immune system function. Both the PNS and SNS innervate immune organs,such as the spleen and lymph nodes, and there are many points of regulated interaction between the nervous and immune systems throughout the body (Pavlov and Tracey, 2017). Based on these physiological interactions, a causal relationship has been proposed between SCI level-dependent changes in the ANS and changes in immune system function that include chronic inflammation, changes in adaptive immunity and immunosuppression (Schwab et al., 2014). SNS fibers exit the spinal cord and innervate organs of the immune system at T5,and persons with spinal cord injuries rostral to T5 have the most immunological symptoms (Campagnolo et al., 2000;Failli et al., 2012; Schwab et al., 2014).
What are the Molecular Causes of Immunological Symptoms after SCI?
Despite the medical importance of immune function after SCI, to date there have been only two original research studies that systematically investigated gene expression in immune cells isolated from persons with chronic SCI (Saltzman et al.,2013; Herman et al., 2018). The first study by Saltzman and colleagues compared whole blood gene expression in men with chronic motor complete (AIS A) SCI (at least one year from initial injury) to able-bodied (AB) men (N = 13, 7 respectively) (Saltzman et al., 2013). RNA was extracted from whole blood and gene expression levels were quantified by microarray using the HG-U133A Plus 2.0 GeneChip (Affymetrix). The study identified 1453 genes that were differentially expressed in men with SCI. Using Ingenuity Pathway Analysis to provide a functional description of differentially expressed genes, Saltzman and colleagues focused their discussion on a gene network related to lymphoid tissue structure, which is regulated by the master transcription factor nuclear factor kappa B (NF-κB). They showed enhanced expression of the autoimmunity-promoting cytokines B-cell maturation antigen(BCMA), B cell activating factor of the tumor necrosis factor(TNF) family (BAFF) and a proliferation-inducing ligand(APRIL) in this network by microarray and qPCR, consistent with sparse but continued literature demonstrating serological signs of autoimmunity after SCI, such as anti-central nervous system (CNS) antibodies (Saltzman et al., 2013; Schwab et al.,2014). Other key genes of the pathway included several members of the TNF receptor superfamily.
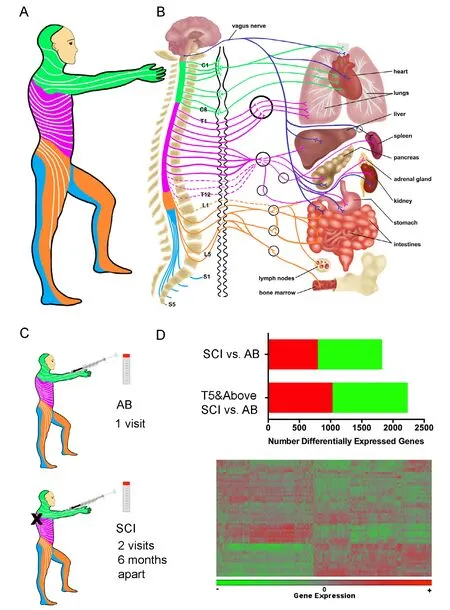
Figure 1 Neuroimmune interactions relevant to spinal cord injury (SCI).
In order to provide a deeper understanding of the molecular mechanisms underlying immunological symptoms after SCI,we set out to perform a larger (by participant number) and deeper (by analytical scope) study of gene expression in leukocytes of persons with chronic SCI (Herman et al., 2018). We recruited adults with chronic SCI, (at least one year from initial injury) (Figure 1). In this study, 58% of participants had cervical, 36% had thoracic and 6% had lumbar spinal cord level injuries (N = 25 male, 6 female). For comparison, we recruited 26 able-bodied persons (N = 19 male, 7 female). Participants in this study mostly had AIS A injuries, followed by D, C and B respectively. Since we were studying immune function, persons with clinically significant infections were excluded from participating in the study. We collected data from participants about their medical histories and current medication regimens.Participants were asked to contribute blood samples at two study visits held 6 months apart. Most participants (N = 24/31)completed both study visits, and able-bodied adult participants without SCI (N = 26) were used as the comparison group.
RNA was extracted from whole blood and gene expression levels were quantified by microarray using the using the HT-12v4 Expression BeadChip (Herman et al., 2018). We first determined that within the SCI group, gene expression did not differ significantly between the study visits that occurred 6 months apart. We then compared the transcriptional profiles of individuals living with chronic SCI to able-bodied individuals. Hierarchical clustering of differentially expressed genes showed global differences in leukocytes from persons with SCI as compared to able-bodied participants. We discovered 1815 genes that were differentially expressed between the groups(false discovery rate (FDR) = 0.05) (Figure 1). Participants with SCI levels rostral to T5, where SNS disruption would occur, had more differentially expressed genes (N = 2226)compared to able-bodied participants. For the complete list of differentially expressed genes, see Supplementary Table 1 in Herman et al. (2018).
Characterization of Differentially Expressed Genes in Persons with High Spinal Cord Level Injuries
Immunologists have recently developed a series of empirical modules comprised of genes co-expressed in blood cells across different disease states (Chaussabel et al., 2008). Modules were created by experimental observation of gene expression in blood of patients with many different inflammatory or autoimmune diseases genes, so that genes that were expressed in a consistently coordinated manner across diseases, for example,up- or down-regulated together, are clustered together into a module (Chaussabel et al., 2008). This modular analysis assumes that genes that change coordinately in expression across different diseases likely have related or coordinated biological functions and also presents an opportunity to compare changes in the gene modules across disease states (Chaussabel et al.,2008). Modules are named according to the known function of genes within it and unnamed modules contain genes whose function is not yet determined. We assigned the genes differentially expressed in persons with SCI levels rostral to T5 to the previously established modules (Chaussabel et al., 2008).We found an under-expression of modules related to many immune cell types including Natural Killer (NK) cells, B cells and T cells. Conversely, we identified elevation of a monocyte module, as well as elevation of six different inflammation modules. For a list of all differentially expressed genes by module,see Supplementary Table 1 in Herman et al. (2018).
NK cells are innate immune system lymphocytes responsible for killing virally infected cells or tumor cells, and they also have modulating effects on other immune cell types.Dramatic downregulation of NK cell genes and elevation of pro-inflammatory genes in persons with SCI levels rostral to T5 were reflected by many independent bioinformatics approaches. For example, killer inhibitory receptors (KIRs) are most commonly found on NK cells and are also found on other immune cell subsets. KIRs can activate or inhibit NK cells from killing their target cells. In this study, we found that 5 of the 36 most significantly downregulated genes in persons with SCI levels rostral to T5 were KIR3DL1, KIR3DL3, KIR2DL4,KIR2DL1 and KIR2DS5. For a complete list of significantly up- and down-regulated genes, see Supplementary Table 1 in Herman et al. (2018). We used the bioinformatics resource“Enrichr”, which allows a gene list to be analyzed simultaneously by multiple bioinformatics tools to reveal the functions of differentially expressed genes (Chen et al., 2013). The Kyoto Encyclopedia of Genes and Genomes (KEGG) bioinformatics platform also showed that the “NK cell mediated cytotoxicity” pathway was significantly enriched in genes differentially expressed in participants with SCI at levels rostral to T5. For a list of pathways significantly enriched in differentially genes, see Supplementary Table 1 in Herman et al. (2018).
Conversely, several pro-inflammatory genes were among the top 50 upregulated differentially expressed genes in study participants with SCI rostral to T5, including JAK2, a protein tyrosine Janus kinase family member. JAK2, as well as other elevated genes, such as the beta-adrenergic receptor, are targets of FDA-approved drugs. Toll like receptors (TLR) are innate pattern recognition receptors critical for maintaining immunity against pathogens and TLR activation promotes production of inflammatory mediators (Foster and Medzhitov, 2009). TLR modulating agents are also in clinical trials for indications such as autoimmune disease and inflammation (Gao et al., 2017).We also discovered that TLR signaling pathways were highly enriched among genes upregulated in participants with SCI rostral to T5. Interestingly, there are TLR modulating agents in clinical trial for sepsis, which is associated with infections and is a major cause of mortality for persons with SCI (Gao et al.,2017; National Spinal Cord Injury Statistical Center, 2017).Adrenomedullin, a vasodilator that is increased in sepsis, was also upregulated in participants with SCI rostral to T5 (see Supplementary Table 1 in Herman et al. (2018)). A recent phase I clinical trial of anti-adrenomedullin antibodies was performed, with the hopes of developing them as a therapeutic agent in sepsis (Geven et al., 2018a, b). Many immune cells,including NK cells, are highly responsive to adrenaline and another gene of particular interest that was upregulated in participants with SCI rostral to T5 is the beta-2 adrenergic receptor. A recent study demonstrated that elevated noradrenergic signaling leads to immunosuppression in an animal model of acute SCI (Pruss et al., 2017).
We identified transcription factors that are known to regulate genes differentially expressed in individuals with SCI at levels ≥ T5 using the TRANSFAC database (Matys et al., 2006;Chen et al., 2013). The top 10 transcription factor identified by TRANSFAC were: E2F1, FOXC1, PPARg, PITX2, NRF1,STAT3, GATA2, LEF1, GATA3, and GFl1. Many of these transcription factors are elevated in individuals with metabolic syndrome. For example, E2F1 and STAT3 binding motifs were also upregulated in circulating blood cells isolated from insulin resistant Latino adults with metabolic syndrome (Tangen et al.,2013). This was consistent with data generated independently by the PANTHER bioinformatics platform, which identified the insulin pathway as enriched among upregulated differentially expressed genes in individuals with SCI at levels T5 and above (see Figure 4 in Herman et al. (2018)). Members of the peroxisome proliferator-activated receptors (PPAR) family of nuclear hormone receptors are transcription factors widely considered in the treatment of type II diabetes and in particular, PPAR gamma is expressed mostly in adipose (Bugge and Holst, 2017). PPARs also play a role in secondary inflammation after SCI and clinically used anti-inflammatory agonists of PPAR had beneficial effects in rodent models of SCI (McTigue et al., 2007; Genovese et al., 2009; Zhang et al., 2010, 2011; Li et al., 2013, 2016, 2017; Bugge and Holst, 2017). An inhaled version of DNAzyme that targets GATA3, which is critical for T helper type 2 (TH2) T cell development, was used in a phase I study of asthma (Homburg et al., 2015; Krug et al., 2015). Elevated expression of LEF1, which is a key regulator of the WNT pathway, was identified to associate with worse outcomes in B cell acute lymphoblastic leukemia (ALL) patients, while LEF1 inactivation was associated with T cell ALL (Gutierrez et al.,2010; Kuhnl et al., 2011).
We also confirmed several genes first identified by Saltzman and colleagues as differentially expressed in whole blood of persons with SCI in participants with SCI rostral to T5 (Saltzman et al., 2013). These included CD36, which is a platelet thrombospondin receptor, the autoimmune promoting receptor B cell maturation antigen (BCMA/TNFRSF17), inositol-trihophate-3-kinase B (ITPKB) that regulates inositol metabolism,MAP Kinase Activating Death Domain (MADD) that interacts with the TNF-alpha receptor 1 and interacting partners of the NF-κB pathway. Saltzman and colleagues also implicated the PPAR gamma pathway, as we discussed above. We did not find evidence of elevated the cytokines APRIL (TNFSF13) or BAFF(TNFSF13B). These differences are likely due to relatively small participant group sizes with different clinical and demographic characteristics.
Novel Mechanistic Insights into the Causes Underlying Symptoms of Immune Dysfunction in Persons Living with SCI
Here we discovered broad changes in innate and adaptive immune cell genes. Modular and pathway analyses identified downregulation of NK cells genes, which we were particularly interested in, because NK cells are critical for fighting pathogens, previous studies showed NK cell defects ex vivo from persons with tetraplegia, and infections are the leading cause of mortality for persons with SCI (DeVivo et al., 1993; Nash,1994; Campagnolo et al., 2000). Modular analysis also identified upregulation of inflammatory genes. Multiple pathway analyses identified elevated TLR pathway genes, which promote inflammation. Other elevated inflammatory genes we discovered in persons with SCI included targets of FDA-approved drugs, including JAK2 and the beta-adrenergic receptor, which has been implicated in neuroimmune signaling in able-bodied and SCI populations (Schwab et al., 2014; Pavlov and Tracey, 2017). Transcription factors regulating genes differentially expressed in persons with SCI rostral to T5 included PPARg, which is a clinical target in diabetes and has been implicated in inflammation in animal models of SCI. Finally,there were some common observations between the 2 studies discussed, including upregulation of the autoimmunity promoting cytokine BCMA and in a broader sense, the upregulation of pro-inflammatory signaling pathways.
Going forward, we intend to identify the specific immune cell subsets in which inflammatory genes are elevated. We will also perform mechanistic studies to evaluate functional changes in NK cells (and other leukocyte subsets) from persons with SCI to determine which of the differentially expressed gene pathways is critical for maintaining health, like eliminating viral infections or promoting inflammation. These data may also be relevant to the increasing number of clinical studies or trials that aim to promote functional recovery for persons living with chronic SCI, where inflammation may play an antagonistic role. Taken together, these data suggest potential mechanisms for therapeutic strategies to improve immune function, promote survival, health and perhaps even functional recovery in persons living with SCI.
Acknowledgments: The authors wish to thank the study participants. The authors also thank their colleagues who co-authored the study discussed here.
Author contributions: PEH performed data analysis, interpretation, created figures and co-authored the manuscript. OB designed the project,performed data analysis, interpretation, created figures and co-authored the manuscript.
Conflicts of interest: The authors declare that they do not have any conflicts of interest.
Financial support: This research was supported by grants from the Craig H.Neilsen Foundation, the NY State Empire Clinical Research Program, and the NY State Spinal Cord Injury Research Board (grants to OB).
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open access statement: This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-Non-Commercial-ShareAlike 4.0 License, which allows others to remix,tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewers: Tetsuro Tamaki, Tokai University School of Medicine, Japan; Anup Dutt Sharma, AxoSim Technologies LLC, USA.
Additional file: Open peer review reports 1, 2.
- 中国神经再生研究(英文版)的其它文章
- Seeing the wood for the trees: towards improved quantification of glial cells in central nervous system tissue
- 3′-Daidzein sulfonate sodium protects against memory impairment and hippocampal damage caused by chronic cerebral hypoperfusion
- The novel chalcone analog L2H17 protects retinal ganglion cells from oxidative stress-induced apoptosis
- Treatment with NADPH oxidase inhibitor apocynin alleviates diabetic neuropathic pain in rats
- Saikosaponin a increases interleukin-10 expression and inhibits scar formation after sciatic nerve injury
- Various changes in cryopreserved acellular nerve allografts at −80° C

