Seeing the wood for the trees: towards improved quantification of glial cells in central nervous system tissue
Sinéad Healy, Jill McMahon, Una FitzGerald
Galway Neuroscience Centre, School of Natural Sciences, National University of Ireland, Galway, Ireland
Abstract The following mini-review attempts to guide researchers in the quantification of fluorescently-labelled proteins within cultured thick or chromogenically-stained proteins within thin sections of brain tissue. It follows from our examination of the utility of Fiji ImageJ thresholding and binarization algorithms. Describing how we identified the maximum intensity projection as the best of six tested for two dimensional (2D)-rendering of three-dimensional (3D) images derived from a series of z-stacked micrographs, the review summarises our comparison of 16 global and 9 local algorithms for their ability to accurately quantify the expression of astrocytic glial fibrillary acidic protein (GFAP), microglial ionized calcium binding adapter molecule 1 (IBA1) and oligodendrocyte lineage Olig2 within fixed cultured rat hippocampal brain slices. The application of these algorithms to chromogenically-stained GFAP and IBA1 within thin tissue sections, is also described. Fiji’s BioVoxxel plugin allowed categorisation of algorithms according to their sensitivity, specificity accuracy and relative quality. The Percentile algorithm was deemed best for quantifying levels of GFAP, the Li algorithm was best when quantifying IBA expression, while the Otsu algorithm was optimum for Olig2 staining, albeit with over-quantification of oligodendrocyte number when compared to a stereological approach. Also, GFAP and IBA expression in 3,3′-diaminobenzidine (DAB)/haematoxylin-stained cerebellar tissue was best quantified with Default, Isodata and Moments algorithms. The workflow presented in Figure 1 could help to improve the quality of research outcomes that are based on the quantification of protein with brain tissue.
Key Words: organotypic brain slice culture; glial cell quantification; thresholding algorithms; Fiji ImageJ; Bio-Voxxel plug-in; stereology
As evidenced by over 1100 ‘hits’ when the search string“organotypic brain slice culture” is used on Pubmed (accessed February 2018), the use of this three-dimensional (3D) tissue platform for mimicking the conditions of brain disease in vivo is increasingly popular (Jarjour et al., 2012; Humpel, 2015). While culture conditions and tissue age have been adjusted in order to provide optimum conditions for slice survival, approaches to quantification of cellular changes occurring under experimental conditions have not been developed to the same level of sophistication. Fluorescence immunocytochemistry has been the preferred technique used to identify and characterise different cell types and demonstrate expression levels of proteins within cultured brain slices. However, given that slice thickness is known to affect antibody/reagent penetration, and that there is extensive diffraction of fluorescence emitted beneath the slice surface, it is important that an agreed protocol should be in place for quantifying accurately images of key features within slices. In this way it should be possible to minimise errors of interpretation, avoid exaggerated research claims and improve consensus amongst research groups. Accepting that there is no perfect technique for quantifying protein expression within slices and that published techniques will never portray with 100% accuracy the absolute numbers of cells or levels of protein expression within test tissue samples, the following paragraphs summarise our development of a work-flow that can be used to assist glial biologists to navigate a route to best possible quantification of protein expression, given the limitations of current in vitro and image analysis protocols.
The analysis of images from thick cultured slices is the last of a multi-stage process that normally begins with fixation in paraformaldehyde after which slices are typically then exposed to primary antibodies detecting proteins of interest. Primary antibody binding is then usually visualised using tagged secondary antibodies and fluorescence confocal microscopy. The quality of outputs from tissue staining will critically depend on the quality of the tools used, the way that they are applied and the condition of the starting tissue sample. For example, strategies such as antigen retrieval via pressure cookers or detergents, may improve antibody penetration. The quantity of primary and secondary antibody applied to tissue should be optimised in order to decrease background staining. Crucially, extreme care during tissue handling during dissection and fixation, should ensure optimal starting material. Researchers can also draw on published protocols as well as guidelines recommended by antibody suppliers and in online fora. However, beyond this point, once image capture and analysis is required, best practice is far from clear.
The challenge of correctly and accurately analysing complex images may be further exacerbated by problematic features such as non-stationary, correlated and non-Gaussian noise (a type of generalised statistical noise that has a non-normal probability density function), ambient illumination (dark room illumination may vary between total and medium darkness depending light from computer monitors, etc.), busyness of grey levels within the object and the background, inadequate or inappropriate contrast (contrast adjusts the shape of the intensity histogram of an image and adjustments may over- or under-saturate the intensity information and truncate information) and inter-image variability arising from specimen and staining variability (Sezgin and Sankur, 2004).
In an excellent review of biological imaging software tools, Eliceiri et al. (2012) described the use of ImageJ, Fiji, BioImageXD,Icy, CellProfiler, Vaa3D programmes to analyse tissue images. In our recent study, we report on the use of Brocher’s BioVoxxel plugin, available in Fiji, to evaluate systematically 25 threshold-based segmentation algorithms for suitability in analysing fluorescent and chromogenic images of glial cells within cultured hippocampal slices and in thin brain tissue sections (Healy et al.,2018). Exploiting slices from a model of iron accumulation in rat hippocampus (Healy et al., 2016), six different projection algorithms were first compared to determine which one best rendered 2D images from a series of z-stacked micrographs of fluorescently-stained cells. The maximum intensity projection algorithm was found to be the best at preserving sharpness and quality of detail,when compared to the standard deviation, mean, sum, minimum and median algorithms.
The accuracy of image segmentation and feature extraction,the process whereby each pixel is analysed to determine if it is part of the background or part of the object-of-interest, depends critically on the application of the most appropriate binarisation algorithm (Johnson and Walker, 2015). Researchers might feel that they are capable of making an objective assessment, by eye,of the quality of image binarisation and to test this idea, we compared three manually-chosen thresholds to the Default algorithm(http://imagej.net/Auto, Threshold#Default, March 2017), when applied to two different images each of astrocytes, microglia and oligodendrocytes (Healy et al., 2018). For all glial cells, the auto Default algorithm was found to produce the best quality output binarisation. However, in the case of microglia, the output was considered sub-optimal, leading us to complete a more thorough exploration of approaches to image segmentation. Instead of opting for the ‘lowest common denominator’, researchers can exploit one of Fiji’s 16 global or 9 local algorithms, in order to yield the most accurate imaging data. Depending on underlying methodology, the 16 global algorithms, which compute a single threshold using complete image information, are classed as histogram shape (Intermodes, Mean, Minimum), Cluster (Default,Isodata, MinError, Otsu), Entropy (Shanbhag, Li, Yen, Huang,RenyiEntropy, MaxEntroy), Attribute (Moments, Triangle),or miscellaneous (Percentile). The nine local algorithms, which compute various thresholds from different partitions of the image, are Bernsen, Contrast, Mean, Median, Midgrey, Niblack,Otsu, Sauvola and Phansalker. The reader is referred to Healy et al. (2018) for a detailed listing of references for each algorithm.
Motivated by the desire to produce the most accurate data possible, a researcher may try to carry out a subjective assessment of the suitability of the above algorithms for analysing experimental images. Indeed, our first attempt at doing this, for GFAP-stained astrocytes did allow us to immediately dismiss many algorithms e.g., the MinError, Triangle, Yen, Max Entropy, Minimum, RenyiEntropy and Intermodes algorithms. However, distinguishing between others, all of which appeared to do the job equally well, was a challenge. In what we believe is the first reported instance of its use in the analysis of images of glial cells, we exploited Fiji’s BioVoxxel plugin (Brocher, 2014), a semi-quantitative, colour-coded method, to evaluate the relative merit of the 25 thresholding algorithms. The object-of-interest is coloured yellow-to-red, while background is blue-tocyan (Figure 1). A single reference intensity value per image,chosen by the user, is used to classify each pixel as an object or non-object (i.e., background), giving 4 categories for each pixel:object-of-interest/cellular staining, (true positive; TP), background (true negative; TN), over-estimated (false positive; FP)and under-estimated (false negative; FN). A word of caution is necessary at this point, as the reference value chosen by the user has the potential to introduce bias. The potential for bias can be mitigated by using a battery of test images for evaluation,or by selecting and comparing multiple reference points from the same image. Users must be able to justify a carefully chosen reference point on which subsequent evaluations can safely be based. Ideally, researchers would be able to access positive control tissue containing a cell population that has been genetically altered such that it emits a relevant strong fluorescent signal in situ. The corollary to this would be a negative-control sample,for example tissue that has been stained using secondary antibody only. Alternatively, areas of tissue section that are known to be negative for the protein of interest (e.g., inter-cellular spaces). For further details of managing reference values, the reader is referred to Healy et al. (2018).
To evaluate the performance of each algorithm, the number of pixels assigned to each category is counted, and the following metrics are calculated: sensitivity, specificity, accuracy and relative quality. Incorporating a multiplication (100×) of the product of each of the following four calculations, sensitivity is computed by dividing TP by (TP + FN), specificity, by dividing TN by (TN + FP), accuracy, by dividing (TP + TN) by (TP +FP + FN), and relative quality, by dividing TP by (TP + FP +FN) (Figure 1).
On applying Brocher’s plugin to selected reference images of glial fibrillary acidic protein (GFAP)-stained astrocytes within cultured hippocampal slices, and using one-way analysis of variance (ANOVA) followed by post-hoc comparisons using Dunnett’s test for statistical analyses, the Percentile algorithm yielded the highest scores for sensitivity, specificity, accuracy and relative quality, although the Huang, Li and Mean algorithms performed almost equally well. These four algorithms performed significantly better than the other global and all of the local algorithms. To illustrate the potential importance of identifying the best algorithms to use in the context of measuring astrocyte reactivity, we applied the Percentile, Mean, Otsu,Sauvola and Bernsen algorithms to images of GFAP-stained control and ferrocene-treated cultured hippocampal slices.Data generated using the Otsu algorithm suggested a significant(although small) increase in astrocyte reactivity, while the other four algorithms indicated no change (Healy et al., 2018).
Replicating the approach used to quantify changes in GFAP staining, the performance of 25 algorithms in analysing images of IBA1-stained microglia was also assessed using the BioVoxxel plugin and the same statistical tools. This time the Li algorithm returned the highest score across all four metrics and the Huang,Mean, Triangle and Phansalker algorithms performed almost equally well and would be deemed acceptable in this context. In contrast, the remaining 20 algorithms performed significantly less well.
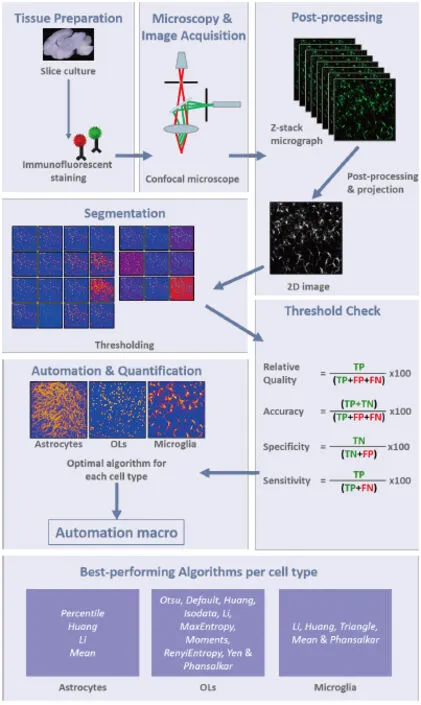
Figure 1 Image analysis workflow.Z-stack images of immunofluorescently stained ex vivo brain slice cultures are acquired using a laser scanning confocal microscope. After post-processing (background subtraction and despeckling) the stacks are converted to maximum intensity projections and analysis of automatic threshold algorithms is then carried out using the Biovoxxel plug-in.Optimal projection and thresholding methods for each glial cell type are summarised in in the bottom pane and all steps are automated in a macro. TP: True positive; TN: true negative; FP: false positive; FNL false negative; OLs: oligodendrocytes.
When images generated using antibodies to nucleus-localised Olig2 were similarly analysed, eight global (Otsu, Default,Huang, Isodata, Li, MaxEntropy, Moments, RenyiEntropy,Yen) and one local (Phansalker) thresholding algorthims performed almost equally well, but the Otsu algorithm performed the best across all four metrics.
Ionized calcium binding adapter molecule 1 (IBA1) or GFAP staining yield complex images of glia, since features present within the cell body and extended processes are identified. This makes the counting of individual cells a challenge. Such limitations are absent when nuclear stains such as the Olig2 stain for oligodendrocyte, are used. After identifying the Otsu algorithm as the optimum auto-thresholding algorithm for fluorescent Olig2-stained cells in hippocampal slices, we were interested in determining how this algorithm compared to a design-based,unbiased, stereological approach (Schmitz and Hof, 2005).Using a cohort of images generated following treatment of cultured slices with vehicle or ferrocene, the relationship between the two counting methods was assessed by calculating a Pearson correlation coefficient. Results indicated a moderate positive correlation between the two methods (r = 0.55, n = 11, P =0.01). Moreover, the relative loss of oligodendrocytes reported following application of the Otsu algorithm, was similar when stereology was used. However, in keeping with previous reports(Schmitz and Hof, 2005; Howard and Reed, 2010), the stereological approach generated an oligodendrocyte count that was significantly lower (approximately 30% lower) than that produced using auto-thresholding.
Chromogenically-stained tissue represents an altogether different challenge when it comes to automatic quantification of positive staining. There is considerably less contrast between object and background, when compared to fluorescently-stained features. In an extension of the above-described study of stained thick brain tissue slices, and in order to explore the potential utility of automated thresholding for use on chromogenically-stained thin tissue sections, we completed a preliminary image analysis of IBA1- and GFAP-stained microglia and astrocytes in 3,3′-diaminobenzidine (DAB)/Haematoxylin-stained adult rat cerebellar tissue. The Default, Isodata and Moments algorithms performed best for microglia and astrocytes (Healy et al., 2018), none of which were amongst the best group of algorithms identified when images of fluorescently-stained microglia or astrocytes were analyzed.
In summary, the lack of detail regarding quantification and analysis of cells within stained organotypic tissue slices, prompted us to develop a new proposed workflow (Figure 1). We believe that this workflow could be useful to researchers aiming to efficiently quantify positive staining of glial cells, as it highlights different algorithms that were found by us to be optimal for each glial cell type. These algorithms could be used as a starting point by researchers developing protocols within different labs. Beginning with image acquisition, protocol development moves on to generation of an auto-thresholded maximum-intensity projection before image binarisation using thresholding algorithms that have been validated using the colour-coded BioVoxxel plugin. Once optimal binarisation and thresholding algorithms have been established for the particular protein-of-interest, the workflow finishes with relevant quantification methodology,using software such as ImageJ (platform-independent freeware developed by the National Institutes of Health, USA). While automated thresholding and evaluated algorithms should prevent skewing of results due to subjective bias, researchers should be aware that there is not a ‘one-size-fits-all’ binarisation algorithm for glial cells. Indeed, algorithms that work for fluorescently-stained cells may not be suitable for tissue stained using chromogenic methods. Furthermore, automated cell counting may inflate cell number moderately, when compared to stereological methods. Nonetheless, we have presented what we believe is an improved approach to the quantification of stained tissue that could assist in raising the quality of research outcomes that are shared with the scientific community as a whole.
Acknowledgments: We also acknowledge the facilities and scientific and technical assistance of the Centre for Microscopy & Imaging at the National University of Ireland Galway (www.imaging.nuigalway.ie), a facility that is funded by NUIG and the Irish Government’s Programme for Research in Third Level Institutions, Cycles 4 and 5, National Development Plan 2007–2013.
Author contributions: SH and UF wrote the manuscript. JM produced the figure.
Conflicts of interest: None declared.
Financial support: The work was supported by a grant from Thomas Crawford Hayes Research Fund; the NUI Galway College of Science scholarship to SH; a grant from NUI Galway Foundation Office to JM.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open access statement: This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-Non-Commercial-ShareAlike 4.0 License, which allows others to remix, tweak,and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewers: Ilias Kazanis, University of Cambridge, UK; Randall D. McKinnon, Robert Wood Johnson Medical School, USA.
Additional file: Open peer review reports 1, 2.
- 中国神经再生研究(英文版)的其它文章
- 3′-Daidzein sulfonate sodium protects against memory impairment and hippocampal damage caused by chronic cerebral hypoperfusion
- The novel chalcone analog L2H17 protects retinal ganglion cells from oxidative stress-induced apoptosis
- Treatment with NADPH oxidase inhibitor apocynin alleviates diabetic neuropathic pain in rats
- Saikosaponin a increases interleukin-10 expression and inhibits scar formation after sciatic nerve injury
- Various changes in cryopreserved acellular nerve allografts at −80° C
- Patterns of cortical reorganization in facial synkinesis: a task functional magnetic resonance imaging study

