Variations in microbial community during nitrogen removal by in situ oxygen-enhanced indigenous nitrogen-removal bacteria
Shi-lei Zhou ,Yue Sun ,Yi-rn Zhng ,Ting-lin Hung *,Zi-xing Li,Ki-ki Fng Chun-hu Zhng
a School of Environmental Science and Engineering,Hebei University of Science and Technology,Shijiazhuang 050018,China
b Key Laboratory of Northwest Water Resource,Environment and Ecology,Ministry of Education,Xi'an University of Architecture and Technology,Xi'an 710055,China
Abstract In this study,the enclosure system exhibited perfect nitrogen removal performance with in situ oxygen-enhanced indigenous aerobic denitrifying bacteria in an enclosure experiment.We explored changes in the microbial community during the nitrogen removal process using the MiSeq high-throughput sequencing technology.The results revealed a total of 7974 and 33653 operational taxonomic units(OTUs)for water and sediment systems,respectively,with 97%similarity.The OTUs were found to be affiliated with eight main phyla(Proteobacteria,Actinobacteria,Cyanobacteria,Bacteroidetes,Planctomycetes,Chloroflexi,Firmicutes,and Actinobacteria).The diversity of the enhanced system was found to be higher than that of the control system.Principal component analysis(PCA)revealed that significant spatial and temporal differences were exhibited in the microbial community during nitrogen removal in the enclosure experiment.Redundancy analysis(RDA)indicated that physical parameters(temperature,dissolved oxygen,and pH),nitrogen(total nitrogen and nitrate),functional genes(nirK and nirS),and dissolved organic carbon(DOC)were the most important factors affecting bacterial community function and composition.Lastly,the results suggested that the variation in the microbial community could be analyzed through the MiSeq high-throughput sequencing technology,which may provide technical support for future field tests.
Keywords:Nitrogen removal;MiSeq high-throughput sequencing technology;In situ enclosure;RDA;Drinking water reservoir
1.Introduction
Excessive nitrogen input,often in the forms of nitrate and ammonia,causes problems for water quality,resulting in water eutrophication.There is still some debate over whether nitrogen alone is the main driver of these problems,but there is no question that the increase in nitrogen loading degrades the water quality.In the past few years,more and more cities have used reservoirs as their main drinking water source,due to the pollution of groundwater and the cone of groundwater depression(Su et al.,2013).However,nutrient enrichment can cause eutrophication in reservoirs,leading to harmful algal blooms,development of hypoxic zones,and an alteration in ecosystem structure and function(Carpenter et al.,1998;Zhu et al.,2013;Shen et al.,2014).Reservoir water quality has also been affected by endogenous pollutants released into overlying water under anoxic conditions.A number of studies have previously demonstrated that anoxia can re-introduce nitrogen,phosphorus,iron,and manganese from sediments into overlying water layers(Gantzer et al.,2009;Chai et al.,2011).To effectively control the pollutants released from sediments,aerobic conditions must be maintained in the reservoir's hypolimnetic ecosystem(Beutel and Horne,1999).Over the past few decades,water-lifting aeration(WLA)technology has been developed and used effectively to increase dissolved oxygen concentration and to improve the quality of micro-polluted drinking water reservoirs(Cong et al.,2006,2009;Bryant et al.,2011;Gerling et al.,2014).As we all know,increasing nitrogen,which is the main pollutant in reservoir systems,has resulted in water eutrophication in drinking water reservoirs.Therefore,improving the self-purification of nitrogen under aerobic conditions in reservoir systems is a scientific problem that needs to be solved urgently.It is known that nitrogen removal is mainly focused on denitrification under anaerobic conditions,which makes it impractical in natural ecosystems,especially in drinking water reservoirs.However,identification of the first aerobic denitrifying bacteria by Robertson and Kuenen(1983)revealed simultaneous nitrification and denitrification,which made nitrogen removal in open water bodies easier.Subsequently,several aerobic denitrifying bacteria have been isolated from soil(Kim et al.,2008;Miyahara et al.,2010),wastewater(Neufeld et al.,2007;Jiang et al.,2012;Kim et al.,2012),and activated sludge(Su et al.,2001;Heylen et al.,2008),with most of these strains being used to treat water with high carbon and nitrogen loads.In the past few years,there have been reports on aerobic denitrification in the natural environment.These include the study by Gao et al.(2010),which showed that aerobic denitrification occurred in the surface sediment of the permeable Wadden Sea and that by Coban et al.(2015),in which the rates of aerobic denitrification in constructed wetlands were first quantified.However,little research has been conducted on the use of aerobic denitrifying bacteria to bioremediate the reservoir system.
The Zhoucun Reservoir is a large shallow eutrophic reservoir,located in northern China,with a surface area of 8.54 km2,a maximum depth of about 20 m,and a corresponding storage capacity of 84.29×106m3,which presents typical seasonal stratification characteristics.The thermal stratification is monomictic,and lasts from April to October.Fish breeding in the reservoir started in the 1980s,with the scale expanding rapidly,and the fish breeding area soon reaching 20%of the reservoir area.Net-case fish breeding has thus caused serious deterioration of the water quality in the reservoir.After the clearing up of the net-case,the water quality of the reservoir has improved,but the sediments of the reservoir have been polluted as well.Thermal stratification plays an important role in the changes of the water environment.In particular,in the stratification period(summer),the hypoxia in the bottom water caused by thermal stratification leads to the release of nutrients and reductants from the sediment.The reservoir is a typical large,shallow,eutrophic,and seasonal stratification reservoir of northern China,but there is not yet enough information available about denitrification in the surface sediments of drinking water reservoirs.In recent years,our research group has carried out a series of studies on oligotrophic aerobic denitrifying bacteria.We have not only isolated oligotrophic aerobic denitrifying bacteria from water and surface sediments,but have also explored the nitrogen removal performance in oligotrophic niches and reservoir water.Huang et al.(2012)carried out a detailed study using WLA combined with oligotrophic biofilm,and exhibited fitting nitrogen removal performances.Huang et al.(2015b)isolated and domesticated three strains of oligotrophic aerobic denitrifiers(N299,G107,and 81Y),and explored their nitrogen removal performance in a reservoir system.Zhou et al.(2016a)studied the nitrogen removal characteristics of indigenous aerobic denitrifying bacteria via in situ oxygen enhancement using WLA technology.Meanwhile,we quantified the abundance of aerobic denitrification bacteria through the functional gene(Zhou et al.,2016b)and aerobic denitrification screening medium(Zhou et al.,2016a).Moreover,when the enhancement was completed,in order to explore the nitrogen removal performance of enhanced indigenous aerobic denitrifying bacteria,we continued the in situ enclosure experiment through addition of the nitrogen source(nitrate)(Zhou et al.,2016b).However,the variations in microbial community(especially those involved in nitrogen function)in enhanced and control systems were not clear throughout the enclosure experiment period.
Therefore,it is necessary to explore the changes in microbial community during the nitrogen removal process based on the MiSeq high-throughput sequencing technology.A previous study found perfect nitrogen removal performance(Zhou et al.,2016b).The objectives of this study were(1)to investigate the diversity and composition of the microbial community by using the MiSeq high-throughput sequencing technology;(2)to determine spatial and temporal changes of the microbial community(especially,denitrifying bacteria)during the enclosure experiment period;and(3)to investigate the relationship between the microbial community structure and environmental driving factors throughout the experimental period through redundancy analysis(RDA).The results of this study can enrich our understanding of the relationship between the bacterial community(especially,denitrifying bacteria)structure and environmental factors,and provide references for research into,protection of,and pollution control of drinking water reservoir environments.
2.Materials and methods
2.1.Experimental system
In order to explore the nitrogen removal characteristics of indigenous aerobic denitrifying bacteria via in situ oxygen enhancement and the changes of the microbial community during the nitrogen removal process,we implemented the enclosure experiment system in the Zhoucun Reservoir(34°56′43′N,117°40′54′E)of Zaozhuang City,in Shandong Province.The details of the enhanced and control systems are as follows:
Enhanced system:The enhanced system was used to simulate the operational environment of WLA,and was placed in the Zhoucun Reservoir with a steel pipe(with an inner diameter of 1.0 m).The enclosure system was filled with approximately 11 m3of raw water in a polyethylene body(with a height of 14.0 m)to simulate natural reservoir conditions.Compressed air was released into the bottom of the enclosure system in the form of small bubbles,which maintained low levels of dissolved oxygen(DO)concentration(4-5 mg/L)through direct mixing and oxygenation(Zhou et al.,2016b).The low DO conditions of the enhanced system lasted one month;after that the indigenous aerobic denitrifying bacteria had been enhanced completely.Next,nitrate was added to the enclosure system to maintain the nitrogen concentration at 2-2.5 mg/L.After two days,the enclosure system reached a stable state.The nitrogen removal performance and the changes in the microbial community of water and surface sediment systems in the enclosure experiment were investigated.
Control system:All the reservoir water except for that in the enclosure system was considered the control system(Zhou et al.,2016b).
2.2.Sample handling and collection
In this study,water samples(2.0 L for water depths of 0.5 m,7.5 m,and 13 m in the enhanced and control systems)and surface sediment samples(50 mL at different periods in the enhanced and control systems)were collected throughout the enclosure experiment.Surface sediment samples were collected at a layer of 0-2 cm using a sterilized Petersen stainless steel grab sampler(Huang et al.,2015a).All samples were kept in an icebox and shipped to the water research laboratory within 6 h.
2.3.DNA extraction
In order to obtain the total DNA,2 L of water sample,filtered with a 0.22-μm cellulose-acetate filter,was collected,and approximately 50 mL of surface sediment(with a depth of 0-2 cm)was collected as well.Microbial DNA was extracted from sediment samples using the E.Z.N.A.®Soil DNA Kit(Omega Bio-tek,Norcross,GA,USA)according to the manufacturer's protocols.DNA was purified using the AxyPrep DNA Gel Extraction Kit(Axygen Biosciences,Union City,CA,USA)according to the manufacturer's instructions.The extracted DNA was stored at-80°C for polymerase chain reaction(PCR)amplification analysis.
2.4.Quantification of nirS and nirK genes abundance
Quantitative PCR was used to estimate the numbers ofnirSandnirKcopies in water and sediment systems collected during the enclosure experiment.Primers used fornirK(Hallin and Lindgren,1999) quantification were F1aCu,5′-ATYGGCGGVCAYGGCGA-3′,and R3Cu,5′-GCCTCGATCAGRTTRTGGTT-3'.Primers used fornirS(Throb¨ack et al.,2004)quantification were cd3aF,5′-GTSAACGTSAAGGARACSGG-3′, and R3cd, 5′-GASTTCGGRTGSGT CTTGA-3'.Real-time PCR(qPCR)was performed on an Applied Biosystems(ABI)7500 real-time system (Life Technologies,USA)using a qPCR kit,following the method described by Huang et al.(2015a).
2.5.Microbial community analysis
To explore the microbial community composition of water and surface sediment systems in the enhanced and control systems,MiSeq high-throughput sequencing was performed at Shanghai Majorbio Bio-pharm Technology Co.,Ltd.DNA extracted from surface sediment samples(as described above)was amplified by PCR using primers 27F(5′-AGAGTTTGATCCTGGCTCAG-3′) and 338R (5′-TGCTGCCTC CCGTAGGAGT-3′)targeting the V2 regions of bacterial 16S rRNA genes(Ravel et al.,2011).The purified amplicons were sequenced on an Illumina MiSeq platform.The amplicons were sequences shorter than 200 base pairs and low-quality sequences(with a quality score lower than 25)were removed(Quince et al.,2011).The taxonomic classification of effective sequences was determined using the Ribosomal Database Project(RDP).
2.6.Instrumental analysis
The environment parameters of the sampling site were measured in situ at 0.5-m increments using a multi-parameter water quality analyzer(Hydrolab DS5,HACH Company,USA).Specifically,parameters including temperature,dissolved oxygen(DO),pH,oxidation-reduction potential(ORP),electrical conductivity(EC),and Chlorophyll-a(Chl-a)were measured.The water parameters of enclosure experiments were measured using a spectrophotometer(DR6000,HACH Company,USA)(CNEPA,2002).Total nitrogen(TN)and total phosphorus(TP)of surface sediment were determined by persulfate(Zhou et al.,2016a).The moisture content(MC)was determined by weighing,following the method described by Huang et al.(2015a).The aerobic denitrifier concentration(a),in colony-forming units per mL(cfu/mL),was calculated with a plate count(Zhou et al.,2016b).
2.7.Statistical analysis
The rarefaction curves(RC),abundance-based coverage estimators(ACE),and community diversity indices(the Chao richness estimator,the Shannon index,and the Simpson index)were calculated by MOTHUR(Loman et al.,2012).The difference in composition of species was compared with this method(Fouts et al.,2012).Principal component analysis(PCA)and RDA were performed using the Canoco(5.0)project.In our study,the length of the maximum gradient was less than 3.0,and thus RDA was chosen to construct the reˇlationship between species and environmental factors(Smilauer and Lepˇs,2014).One-way analysis of variance(ANOVA)was used to determine the significance of difference(P<0.05)using the software statistical product and service solutions(SPSS)20.
3.Results and discussion
3.1.Variations in environmental factors and functional genes
As shown in Fig.1 and Table 1,Zhou et al.(2016a)studied the nitrogen removal characteristics of indigenous aerobic denitrifying bacteria via in situ oxygen enhancement using WLA technology.The values of temperature and DO concentrations revealed similar variations for enhanced and control systems throughout the enclosure experiment period.
The parameters for the entire water layer(0.5 m,5 m,7.5 m,10 m,and 13 m)were analyzed.The temperature showed a decrease from 11.85 ± 0.02°C and 11.92 ± 0.02°C(Nov.24)to 4.10 ± 0.04°C and 4.09 ± 0.11°C(Dec.30),respectively.The DO concentration in the enhanced system ranged from 4.00±0.44 mg/L(Nov.24)to 10.51±0.26 mg/L(Dec.30),owing to the mixed state of the reservoir,whereas that in the control system increased from 8.06±0.14 mg/L(Nov.24)to 10.46±0.65 mg/L(Dec.30)(Fig.1(a)and(b)).The TN concentration of the enhanced system decreased from 2.27±0.05 mg/L(Nov.24)to 1.40±0.05 mg/L(Dec.30)with a 38.24%removal rate,whereas in the control system it increased from 1.29±0.05 mg/L(Nov.24)to 1.42±0.03 mg/L(Dec.30)(Fig.2(a)and(b)).The nitrate concentration decreased from 1.70±0.01 mg/L(Nov.24)to 0.80±0.06 mg/L(Dec.30)in the enhanced system,whereas in the control system it increased from 0.74±0.01 mg/L(Nov.21)to 0.90±0.00 mg/L(Dec.30)(Fig.3(a)and(b)).There was no nitrite accumulation in the enclosure system(Zhou et al.,2016b).Both aerobic denitrifying bacteria(Fig.4(a)and(b))and denitrification functional genes(nirSandnirK)of the enhanced system(Table 1)increased and were higher than those in the controlsystem during the enclosure experiment.Meanwhile,compared with the control system(from 1468 to 2721 μg/L),the sediment of the enhanced system also showed significant nitrogen removal(from 2751 to 2095μg/L)(Zhou etal.,2016b).The total organic carbon(TOC)of the enhanced system also fit with regard to removal performances.
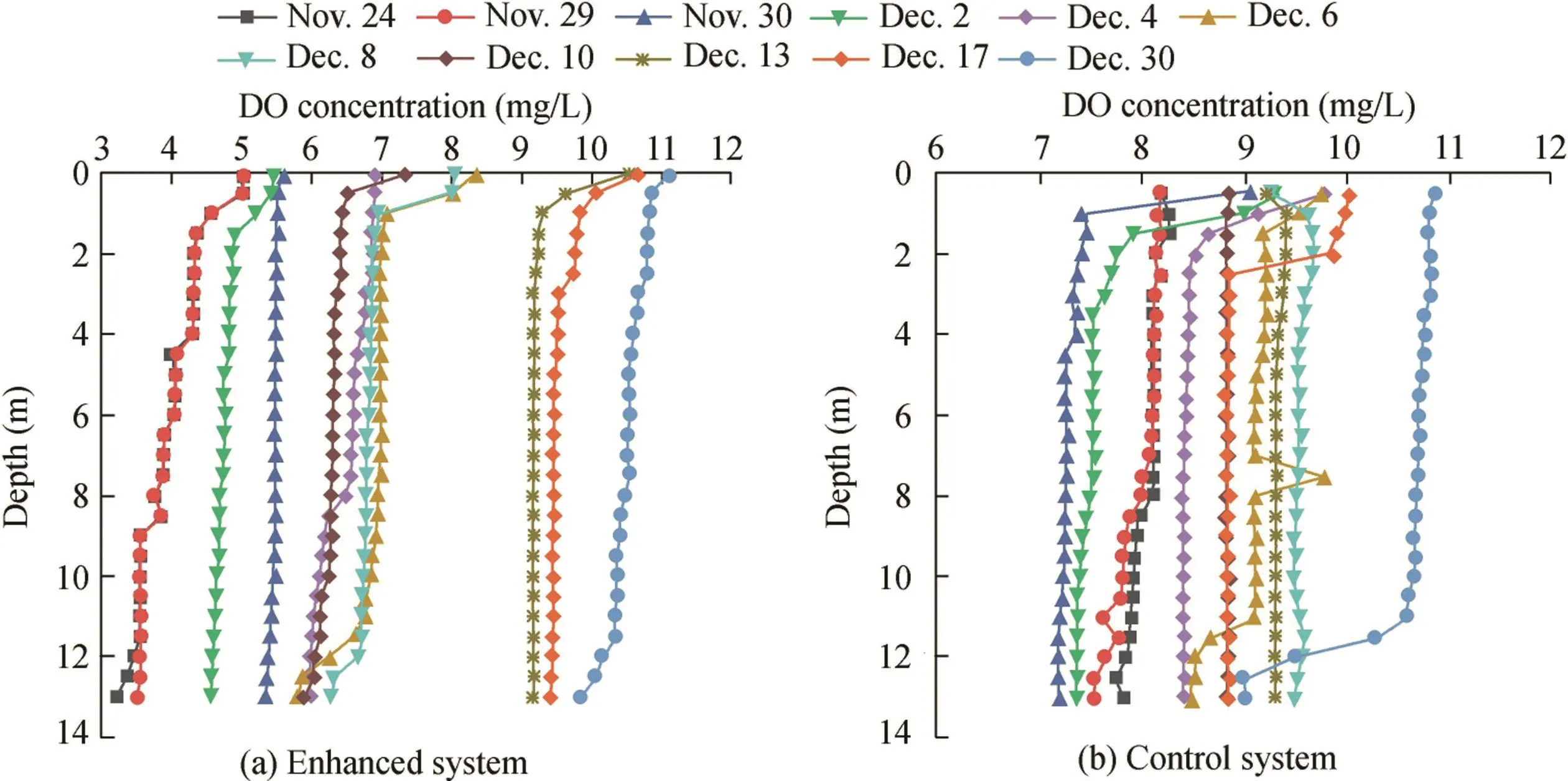
Fig.1.Variations of DO in control and enhanced systems in Zhoucun Reservoir.
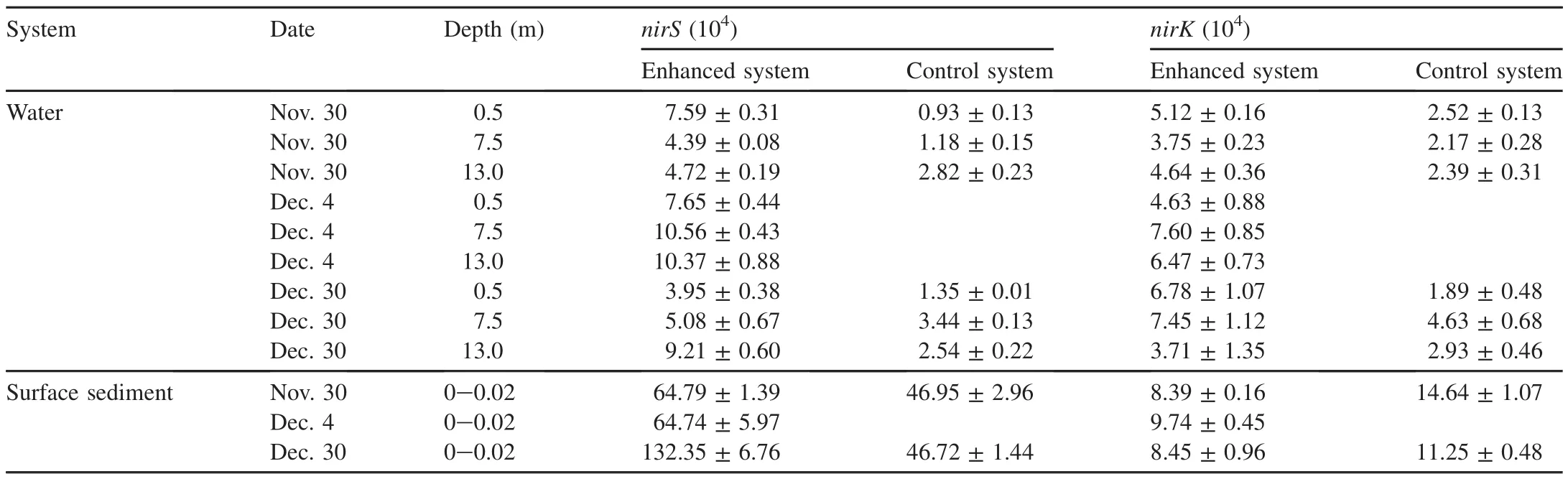
Table 1 Changes in nirS and nirK functional genes in water and surface sediment systems.
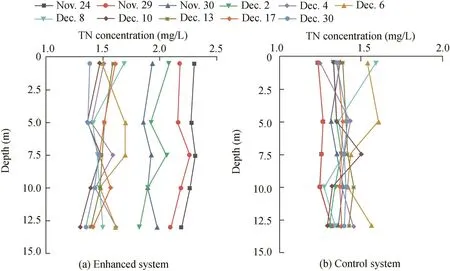
Fig.2.Variations of TN in control and enhanced systems in Zhoucun Reservoir.
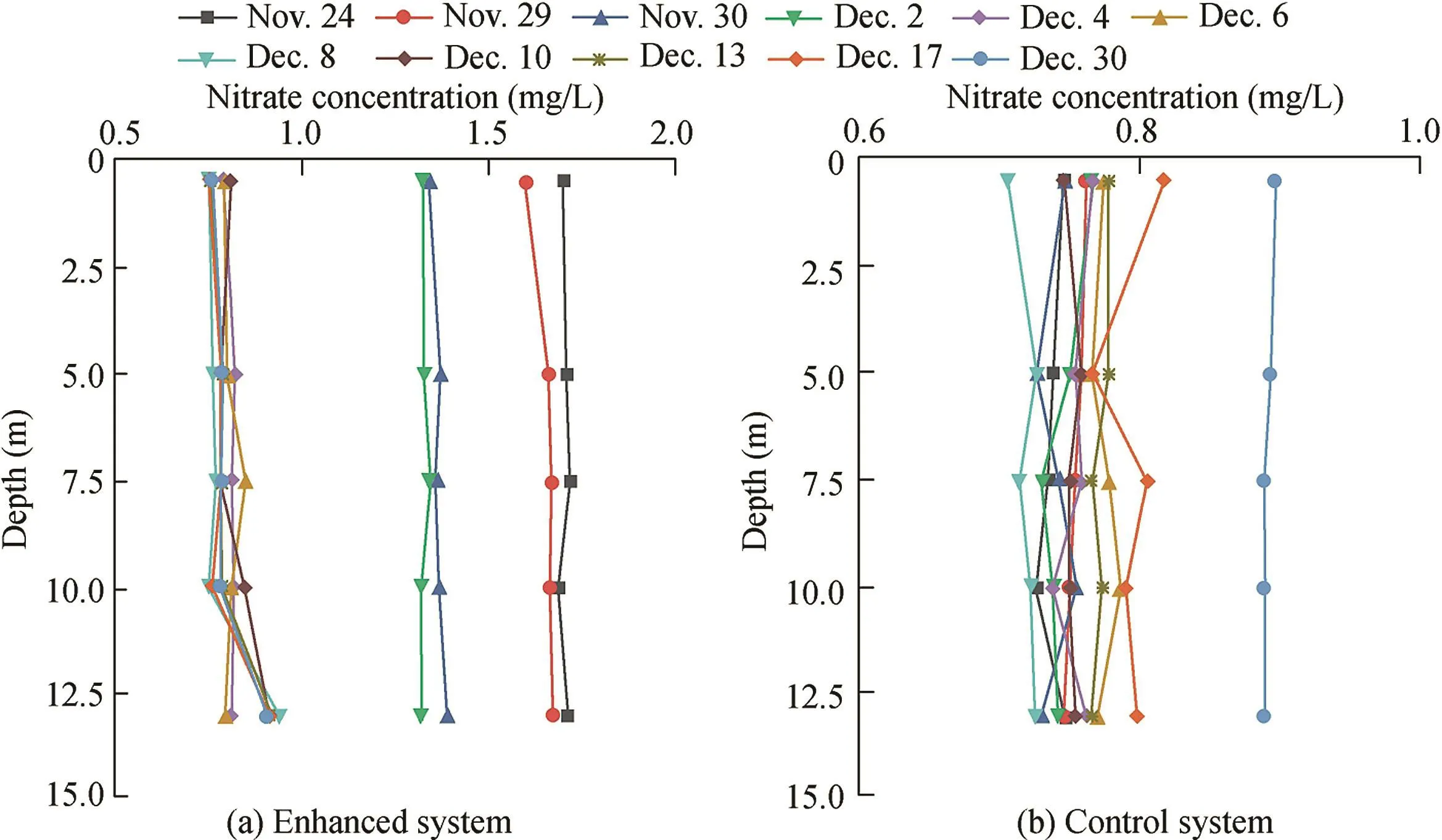
Fig.3.Variations of nitrate in control and enhanced systems in Zhoucun Reservoir.
3.2.MiSeq high-throughput sequencing overview of bacteria
The microbial diversity and community structure of water and surface sediment systems were investigated using MiSeq high-throughput sequencing.After removing low-quality sequences,a total of 184024 sequences with an average length of 315 base pairs and 65945 sequences with an average length of 327 base pairs for water and surface sediment systems,respectively,were obtained during the enclosure experiment.After this,quality trimming for 20 samples(15 samples for the water system and five samples for the surface sediment system)was carried out in triplicate.Each sample generated 334 to 723 and 1262 to 2504 operational taxonomic units(OTUs)for water and surface sediment systems,respectively.The coverage ranged from 0.9782 to 0.9875 and from 0.9186 to 0.9518 for water and sediment systems,respectively,at 97%similarity(Table 2).The largest number of OTUs was found in the enhanced system for water and surface sediment systems throughout the experimental period.The abundance-based coverage estimator(ACE)and Chao diversity estimators for water and surface sediment systems presented significant seasonal variation throughout the experiment period,and the diversity of water and surface sediment systems in the enhanced system were evidently higher than those in the control system.However,the Shannon richness and Simpson richness of water and surface sediment systems exhibited a similar trend throughout the experiment period.

Fig.4.Variations of aerobic denitrifier in control and enhanced systems in Zhoucun Reservoir.
Rarefaction curves,Shannon-Wiener curves,and Rankabundance curves for water and surface sediment systems at 97% similarity revealed that MiSeq high-throughput sequencing could supply enough information to explore microbial community variations.All OTUs with 97%similarity were assigned to eight phyla each for water and surface sediment systems.
3.3.Spatial and temporal changes of microbial community in water and surface sediment systems
As shown in Fig.5 and Fig.6,(X_Y_Zrefers to the system_period_location;X=T and C are the enhanced and control systems,respectively;Y=1,2,and 3 are Nov.30,Dec.4,and Dec.30 during the enclosure experiment,respectively;andZ=0.5,7.5,13,and Nare the water depths of0.5 m,7.5 m,13 m,and surface sediment,respectively).In the water system(Fig.5(a)),the main phyla included Proteobacteria(dominant 1,43.99%±11.17%,andP=0.00 for the enhanced system;28.96%±1.67%,andP=0.00 for the control system),Actinobacteria(dominant 2,22.16%±3.83%,andP=0.00 for the enhanced system;24.72%±5.32%,andP=0.00 for the control system),Cyanobacteria(dominant3,19.78%±11.73%for the enhanced system,and 27.34%±3.95%for the control system),Bacteroidetes(dominant 4,4.88%±1.35%for the enhanced system,and 6.71%±2.71%for the control system),and Planctomycetes(dominant 5,3.22%±1.64%for the enhanced system,and 5.77%±1.70%for the control system).
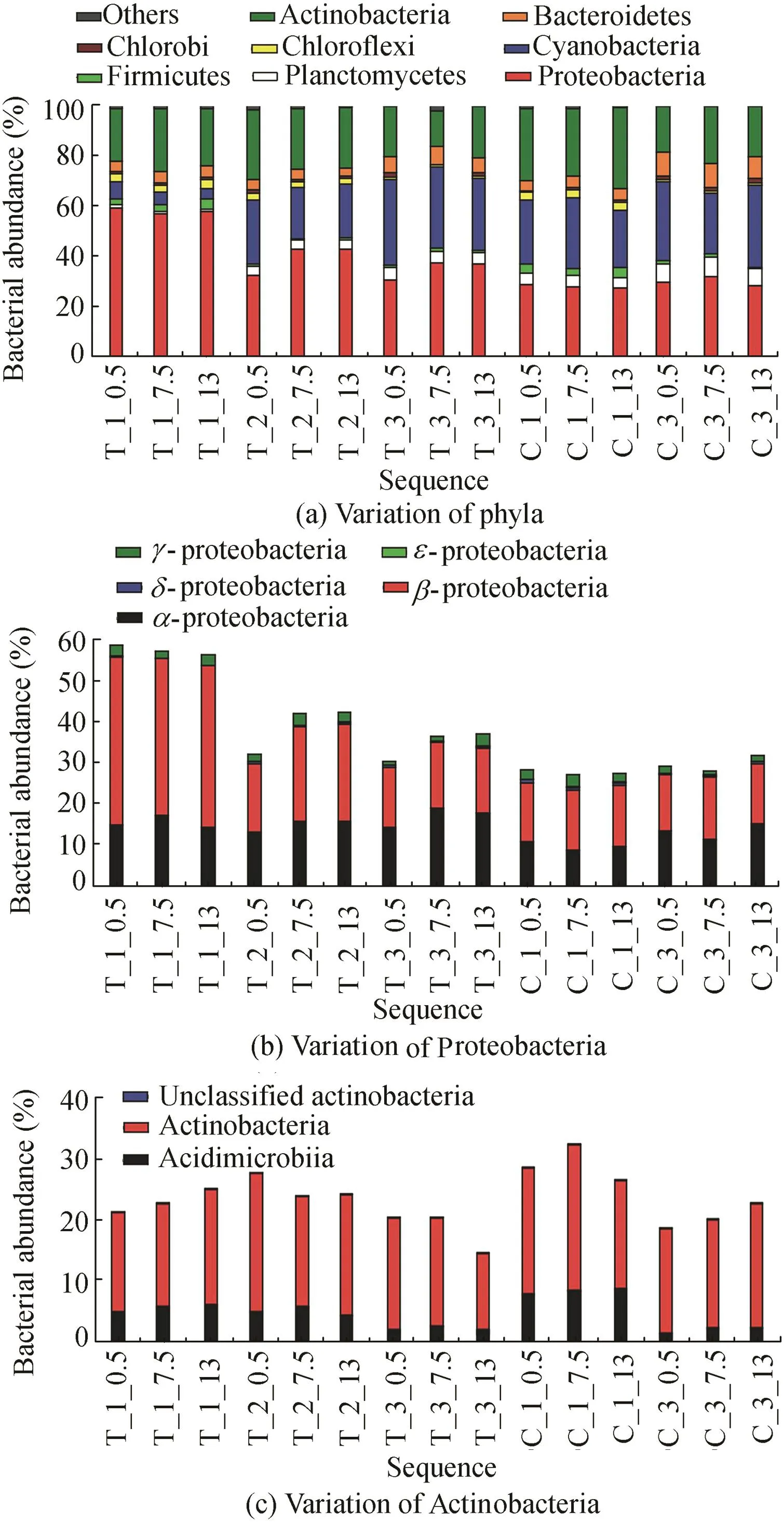
Fig.5.Taxonomic distributions of sequences in water body in enhanced and control systems.
Meanwhile,for the phylum Proteobacteria,β-proteobacteria(dominant 1,25.63%±11.13%for the enhanced system,and 14.76%±0.48%for the control system)was the largest class,with α-proteobacteria at 15.60% ± 1.92%for the enhanced system and 11.27%±2.39%for the control system(Fig.5(b));for the phylum Actinobacteria level(Fig.5(c)),the main classes were Acidimicrobiia(from 5.36%to 1.87%for the enhanced system,and from 8.09%to 1.69%for the control system)and Actinobacteria(18.16%±2.81%for the enhanced system,and 19.80%±2.59%for the control system).Based on the analysis of microbial communities,in the enhanced system,unclassifiedComamonadaceaedecreased from 17.79%±4.32%(at the beginning)to 2.07%±0.67%(at the end);hgcI clade remained at 15.44%±3.23%(for the whole experiment),CL500-29 marine group decreased from 7.45%±0.53%(at the beginning)to 1.44%±0.40%(at the end),andLactococcusranged from 2.03%±0.35%(at the beginning)to 0.71%±0.21%(at the end).
In the surface sediment system(Fig.6(a)),the main phyla included Proteobacteria(dominant 1,44.22%±2.17%for the enhanced system,which was consistent with the change of the water system,and 31.02%±8.38%for the control system),Bacteroidetes(dominant 2,11.37%±0.34%for the enhanced system,and 17.30%±2.18%for the control system),Chloroflexi(dominant 3,6.61%±1.52%for the enhanced system,and 5.25%±0.03%for the control system),Firmicutes(dominant 4,4.71%±0.20%for the enhanced system,and 4.61%±0.29%for the control system),and Actinobacteria(dominant 5,2.62%±0.27%for the enhanced system,and 3.88%±1.78%for the control system).
Meanwhile,for phylum Proteobacteria(Fig.6(b)),β-proteobacteria was the main class(21.40%±2.53%for the enhanced system,and 5.81%-11.70%for the control system),which was consistent with the water system.The δ-proteobacteria reached 13.51%and 13.41%for the enhanced and control systems,respectively.In the enhanced system,vadin HA17 norank(dominant 1)remained at 6.15%±0.61%,GIF9 norankdecreased from 5.29%to 2.29%,andFerribacteriumincreased from 4.74%to 9.35%.In the control system,the main bacteria both showed a decrease(vadin HA17 norank,from 14.81%to 9.63%,andCandidate division OP8 norank,from 8.07%to 2.66%).The main classes were Acidimicrobiia(Fig.6(c))(from 0.24%to 41%for the enhanced system,and from 0.21%to 0.69%for the control system),and Actinobacteria(from 0.94%to 0.81%for the enhanced system,and from 0.84%to 1.73%for the control system).
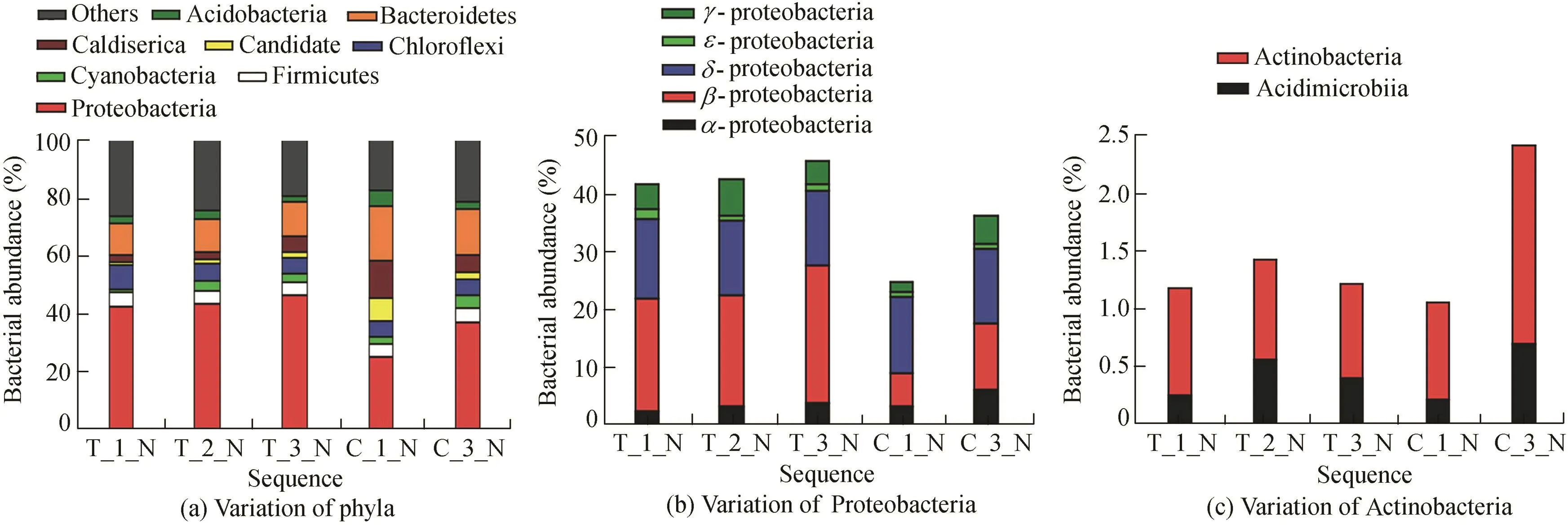
Fig.6.Taxonomic distributions of sequences in surface sediment in enhanced and control systems.
3.4.Changes of denitrification microbial diversity
The aerobic denitrification microbial compositions for the water and surface sediment systems showed significant differences in the enhanced and control systems during the enclosure experiment(as shown in Table 3).There were several bacteria in the enclosure experiment that exhibited evident spatial-temporal variations.For example,Yan et al.(2015)showed in detail that a large amount of Proteobacteria(such asHydrogenophagaandAcidovorax)were involved in nitrogen cycling;Heylen et al.(2008)isolatedAcidovorax caeni(denitrifying bacteria)from activated sludge in Belgium;Heylen et al.(2006)isolatedDechloromonas(denitrifying bacteria)from activated sludge in a municipal wastewater treatment plant;Rhodocyclaceae(denitrifying bacteria,ThaueraandMethylophaga)was isolated from wastewater(Neufeld et al.,2007;Jiang et al.,2012;Kim et al.,2012);Zoogloeasp.299(aerobic denitrifying bacteria)was isolated from reservoirs with perfect nitrogen performances(Huang et al.,2015a);and Patureau et al.(2000)isolatedThiobacillussp.from environmental samples.
3.4.1.Water system
The denitrifying bacteria of the enhanced system(Acidovorax,Bacillus,Dechloromonas,Hydrogenophaga,Pseudomonas,Rhodocyclaceae,Sphingomonas,Thiobacillus,andZoogloea)mostly decreased during the enclosure experiment.Sphingomonaswas seen as an exception,displaying an increase from 0.11%to 0.43%(Tables 3 and 4)during the enclosure experiment.The denitrifying bacteria showed a decrease(Bacillus:from 0.49%to 0.17%;Pseudomonas:from 0.47%to 0.21%)in the control system(Tables 3 and 4).
3.4.2.Surface sediment system
In the enhanced system(Tables 3 and 4),Acidovorax(from 0.05%to 0.12%),Dechloromonas(from 1.50%to 2.76%),Hydrogenophaga(from 0.01%to 0.28%),Rhodocyclaceae(from 0.66%to 0.87%),andZoogloea(from 0 to 0.03%)presented an increase throughout the enclosure experiment period,whereas the abundance ofThiobacillusshowed minimal change(4.20%±0.39%).In the control system(Tables 3 and 4),variations in denitrifying bacteria presented the same trend.Dechloromonasincreased from 0.17%to 0.91%,Bacillusincreased from 0.13%to 0.26%,andThiobacillusincreased from 0.32%to 1.51%.
The changes in denitrifying bacteria have an important influence on the nitrogen cycle. However, aerobic denitrification functional bacteria are rarely cultured and studied in the natural ecosystem,especially in water and sediment systems of a drinking water reservoir.Therefore,studies(such as those based on high-throughput GeoChip functional gene microarray analysis,denitrification activity,denitrifying enzymes,and nitrogen metabolic genes)involved in the mechanism of nitrogen removal are necessary to further investigate the community structure and function of denitrifying bacteria.
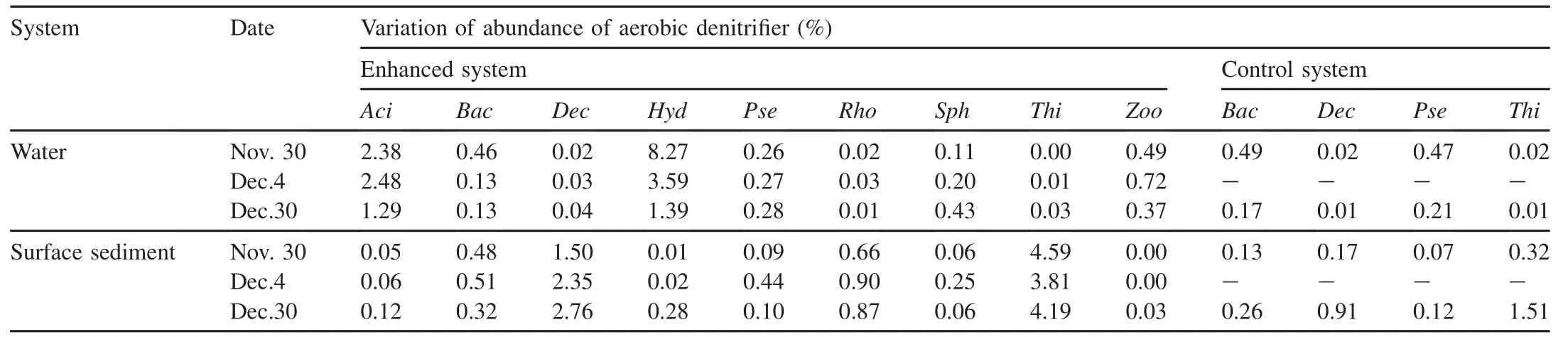
Table 3 Variations of abundance of aerobic denitrifiers in enhanced and control systems.
Table 4 Information of aerobic denitrification bacteria(high-throughput sequence).
3.5.Microbial community structure comparison
In order to explore the bacterial community relationship between water and surface sediment samples during the enclosure experiment period in the Zhoucun Reservoir,community structure comparisons were carried out with PCA for different samples during different periods based on Bray-Curtis dissimilarity(Ligi et al.,2014).The PCA results for water and surface sediment systems are shown in Fig.7(a)and(b),respectively.Results revealed that the first two principle components(PCA axis 1 and PCA axis 2)explained 63.13%and 64.16%of the variability for water and surface sediment systems,respectively.The accumulated contribution ratios of PCA axis 1 and PCA axis 2 for the water system achieved 41.62%and 21.51%,respectively(Fig.7(a)),whereas for the surface sediment system,the accumulated contribution ratios of PCA axis 1 and PCA axis 2 achieved 43.74%and 20.42%,respectively(Fig.7(b)).Microflora of water and sediment systems was well separated in different samples throughout the experiment period.
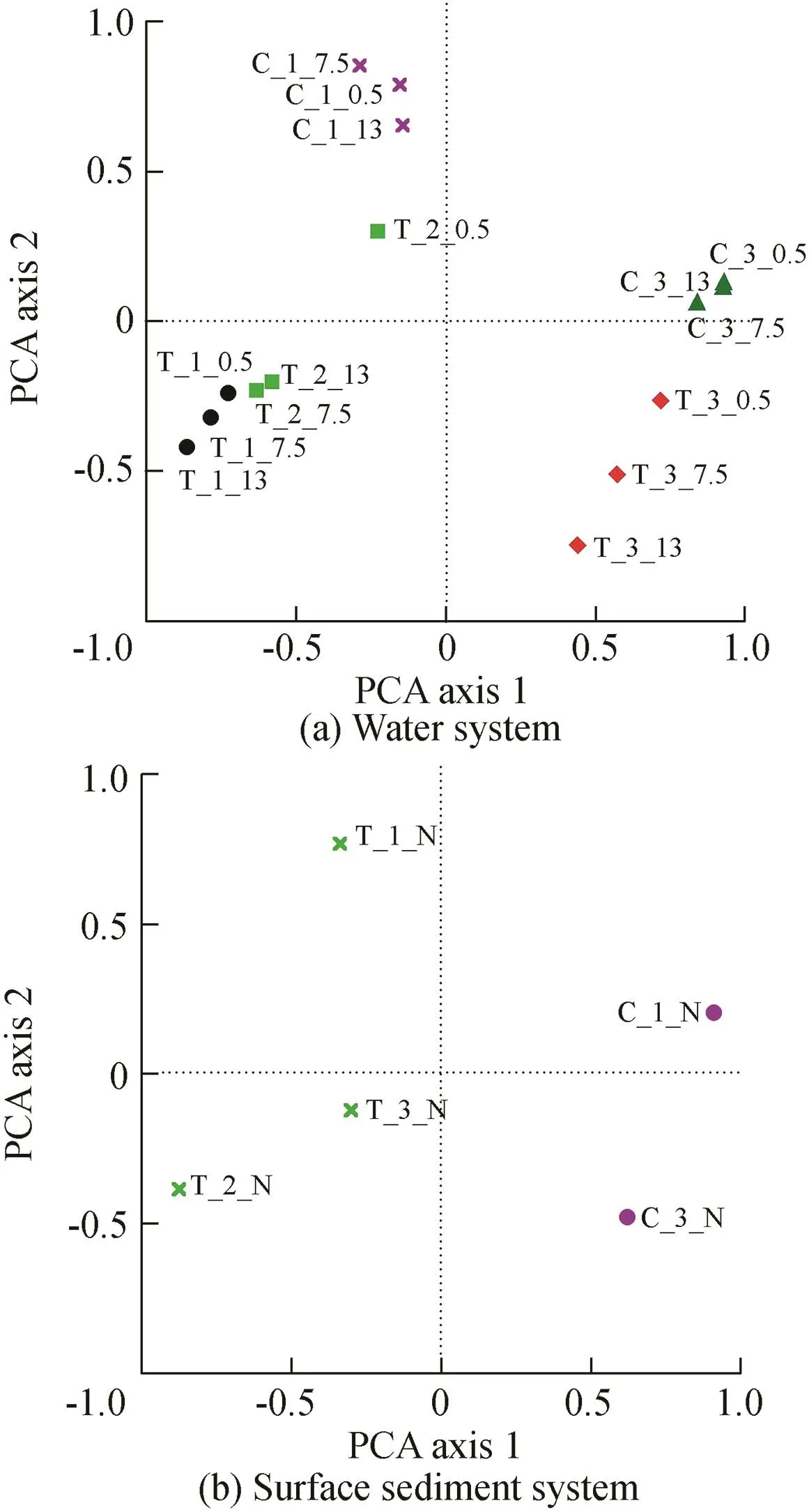
Fig.7.PCA of water and surface sediment systems in Zhoucun Reservoir.
Particularly in the water system,samples from the same system showed tighter clustering,whereas others were relatively distributed.The samples of the enhanced system were mostly located on the negative side of PCA axis 2(from quadrant 3 to quadrant 4),whereas the samples of the control system were located on the positive side of PCA axis 2(from quadrant 2 to quadrant 1)(Fig.7(a)).Moreover,the variations of all the water samples throughout the enclosure experiment could be further explored using the PCA axis 1 and PCA axis 2 values of the water samples in enhanced and control systems.In the sediment system,the samples of the enhanced system were all located on the negative side of PCA axis 1(from quadrant 2 to quadrant 3),whereas the samples of the control system were located on the positive side of PCA axis 1(from quadrant 1 to quadrant 4)(Fig.7(b)).The low PCA axis 2 and high PCA axis 1 values suggest that samples of different systems throughout the experiment period in the Zhoucun Reservoir may be mainly dominated by PCA axis 1.Meanwhile,the samples of sediment systems exhibited evident distribution in the enclosure experiment period based on the values of PCA axis 1 and PCA axis 2.
The results of Venn analysis(Fouts et al.,2012)in the enhanced and control systems suggested different and rapidly changing bacterial communities throughout the experiment period.The results showed that the numbers of shared genera for the enhanced and control systems showed a decrease in the water system(from 361 OTUs to 277 OTUs in the enhanced system,and from 323 OTUs to 202 OTUs in the control system).Meanwhile,in the sediment system,the number of shared genera for the enhanced system ranged from 1505 OTUs(at the beginning)to 1451 OTUs(in the middle)to 1087 OTUs(at the end),whereas for the control system they increased from 1452 OTUs(at the beginning)to 1962 OTUs(at the end).Hierarchical clustering based on OTU information was also generated.The samples of water and surface sediment for the enhanced and control systems from the same period were tightly grouped,and could be well separated in different periods.
3.6.Environmental effect on microbial community
RDA was used to study the relationship between the microbial community and environmental variables based on genus level(Fig.8),which can provide the missing link between diversity and activity.In detail,the separation locations of water samples and surface sediment samples of the enhanced and control systems had a better resolution at the genus.Therefore,the different bacterial communities for the enhanced and control systems were well discriminated at the genus level(Fig.8(a)and(b)).
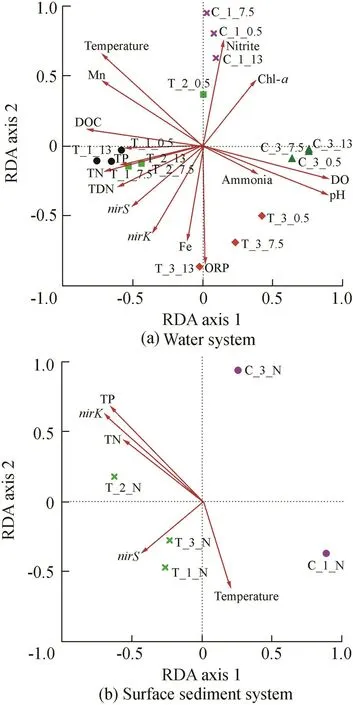
Fig.8.RDA of water and surface sediment systems in Zhoucun Reservoir.
For the water system(Fig.8(a)),the first two RDA dimensions according to the 16 parameters explained 62.69%of the microbial community variation.The accumulated contribution ratios of RDA axis 1 and RDA axis 2 could explain 41.85%and 20.84%,respectively.The lower RDA axis 2 and higher RDA axis 1 scores suggest that samples of different systems throughout the experiment period may be mainly dominated by RDA axis 1.As shown in Table 5,physical and chemical parameters including temperature,pH,DO,nitrate,TN,total dissolved nitrogen(TDN),dissolved organic carbon(DOC),Mn,TP,andnirSsignificantly influenced water bacterial community composition(Fig.8(a)).For the surface sediment system(Fig.8(b)),the first two RDA dimensions explained 72.55%of the bacterial community variation(RDA axis 1:52.63%;RDA axis 2:19.92%;Fig.8(b)),and RDA analysis revealed that temperature,sediment total nitrogen(STN),sediment total phosphorus(STP),andnirKwere the critical environmental factors influencing the spatial and temporal variations of the bacterial community.
In order to explore the effects of environmental factors on nitrogen-function(N-function)microbial community,RDA was carried out during the enclosure experiment period.The sample-environmental variations of water and surface sediment systems(Fig.9(a)and(b))were explored,respectively.The first two RDA dimensions could explain 84.31%and 93.73%for the water and surface sediment systems of the total fitting cumulative variable,respectively,indicating that these results are credible.Meanwhile,the samples of the same system showed tighter clustering,whereas the others were relatively distributed.The lower RDA axis 2 and higher RDA axis 1 scores suggest that samples of different systems throughout the experiment period may be mainly dominated by RDA axis 1.As shown in Fig.9(a)and(b)(sample-environmental variation),the enhanced system and control system were located on the positive and negative sides of RDA axis 1(water)and the negative and positive sides of RDA axis 1(sediment),respectively.It is clear that the environment variations(TN,DOC,nirK,andnirS)were the most important factors affecting the N-function bacterial community and composition for water and sediment systems,respectively.

Table 5 Critical environmental factors influencing spatial and temporal variations for water and surface sediment systems.
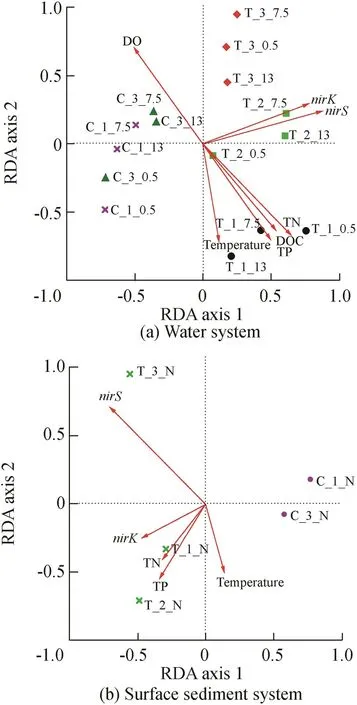
Fig.9.RDA of water and surface sediment systems based on N-function genus distribution.
4.Conclusions
The results showed perfect nitrogen removal and pollutant inhibition in the enhanced system.The microbial diversity and community structure of the water and surface sediment systems were investigated using MiSeq high-throughput sequencing,respectively.The microbial community of the water and surface sediment systems presented spatial and temporal changes in the enhanced and control systems,respectively.RDA was carried out throughout the enclosure experiment period.Based on the results,temperature,DO,TN,nitrate,nirK,nirS,and DOC were shown to play key roles in affecting bacterial community function and composition.The changes in the microbial community during the nitrogen removal process were investigated successfully with the MiSeq high-throughput sequencing technology.This can provide a theoretical basis and technical support for bioremediation of micro-polluted reservoir water.
 Water Science and Engineering2018年4期
Water Science and Engineering2018年4期
- Water Science and Engineering的其它文章
- Monitoring models for base flow effect and daily variation of dam seepage elements considering time lag effect
- Numerical models and theoretical analysis of supercritical bend flow
- A regional suspended load yield estimation model for ungauged watersheds
- Free-surface long wave propagation over linear and parabolic transition shelves
- A comparative study of pseudo-static slope stability analysis using different design codes
- A simple permanent deformation model of rockfill materials
