ldenti fication of salinity-related genes in ENO2 mutant (eno2–) of Arabidopsis thaliana
ZHANG Yong-hua, CHEN Chao, SHl Zi-han, CHENG Hui-mei, BlNG Jie, MA Xiao-feng, ZHENG Chao-xing, Ll Hong-jie, ZHANG Gen-fa
1 Beijing Key Laboratory of Gene Resource and Molecular Development/College of Life Sciences, Beijing Normal University,Beijing 100875, P.R.China
2 Beijing Normal University, Zhuhai 519087, P.R.China
3 National Key Facility for Crop Gene Resources and Genetic Improvement/Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, P.R.China
RESEARCH ARTICLE
ldenti fication of salinity-related genes in ENO2 mutant (eno2–) of Arabidopsis thaliana
ZHANG Yong-hua1, CHEN Chao2, SHl Zi-han1, CHENG Hui-mei1, BlNG Jie1, MA Xiao-feng1, ZHENG Chao-xing1, Ll Hong-jie3, ZHANG Gen-fa1
1 Beijing Key Laboratory of Gene Resource and Molecular Development/College of Life Sciences, Beijing Normal University,Beijing 100875, P.R.China
2 Beijing Normal University, Zhuhai 519087, P.R.China
3 National Key Facility for Crop Gene Resources and Genetic Improvement/Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, P.R.China
Abiotic stress poses a great threat to plant growth and can lead to huge losses in yield. Gene enolase2 (ENO2) is important in resistance to abiotic stress in various organisms. ENO2 T-DNA insertion mutant (eno2–) plants of Arabidopsis thaliana showed complete susceptibility to sodium chloride treatment when were analyzed either as whole plants or by measuring root growth during NaCl treatment. Quantitative real-time RT-PCR (RT-qPCR) was performed to investigate the expression pro file of ENO2 in response to NaCl stress in Arabidopsis. The transcript level of ENO2 was rapidly elevated in 300 mmol L–1NaCl treatment. ENO2 also responded to 300 mmol L–1NaCl treatment at the protein level. To illuminate the mechanism underlying ENO2 resistance to salt at the transcriptional level, we studied the wild-type and eno2–Arabidopsis lines that were treated with 300 mmol L–1NaCl for 18 h using 454 GS FLX, which resulted in an expressed sequence tag (EST)dataset. A total of 961 up-regulated and 746 down-regulated differentially expressed genes (DEGs) were identi fied in the pairwise comparison WT-18 h:eno2–-18 h. The DEGs were identi fied and functionally annotated using the databases of Gene Ontology (GO) and the Kyoto encyclopedia of genes and genomes (KEGG). The identi fied unigenes were subjected to GO analysis to determine biological, molecular, and cellular functions. The biological process was enriched in a total of 20 GO terms, the cellular component was enriched in 13 GO terms, and the molecular function was enriched in 11 GO terms. Using KEGG mapping, DEGs with pathway annotations contributed to 115 pathways. The top 3 pathways based on a statistical analysis were biosynthesis of the secondary metabolites (KO01110), plant-pathogen interactions (KO04626),and plant hormone signal transduction (KO04075). Based on these results, ENO2 contributes to increased resistance to abiotic stress. In particular, ENO2 is involved in some of the metabolic stress response pathways in Arabidopsis. Our work also demonstrates that this EST dataset will be a powerful resource for further studies of ENO2, such as functional analyses,investigations of biological roles, and molecular breeding. Additionally, 3-phosphoglycerate kinase (PGK), 3-phosphoglycerate kinase 1 (PGK1), triosephosphate isomerase (TPI), and pyruvate kinase (PK) in glycolysis interactions with ENO2 were veri fied using the yeast two-hybrid experiment, and ENO2 may regulate the expression of PGK, PGK1, TPI, and PK. Taken together, the results from this study re flects that ENO2 gene has an important role in the response to the high salt stress.
ENO2, NaCl tolerance, abiotic stress, 454 GS FLX sequencing, GO, KEGG
1. lntroduction
In nature, plants are exposed to complex environmental conditions with different abiotic stresses such as drought,high temperature, chilling injury, metal toxicity, salinity,hypoxia, and water logging, which in fluence plant growth and development (Bray et al. 2000; Bent and Mackey 2007; Ahmad and Prasad 2012; Shi et al. 2013; Parimalan et al. 2014). Abiotic stresses were estimated to cause decline of up to 50–70% in major crop productivities (Mittler 2006). Therefore, abiotic stresses are serious problem in agriculture worldwide. In the past decades, studies on the molecular basis of the plant response to these abiotic stresses have been a major focus of research. A number of genes/pathways and regulatory networks were responsive to the tolerance to abiotic stresses in plants (Mickelbart et al. 2015). Numerous studies have indicated that NAC transcription factors (TFs), AP2/EREBP, MYB, WRKY or bZIP families, and some TF genes are effective in the improvement of stress resistance in model plant species and crops of economic importance (Shao et al. 2015; Wang et al.2016). The effects of hormones in abiotic stress responses have been examined (Pieterse et al. 2012; Liu et al. 2013).
Enolase (ENO) is highly expressed in certain organisms during glycolysis, a vital metabolic pathway (Holland et al.1978). It functions in the transformation of 2-phosphoglycerate to phosphoenolpyruvate (Reed et al. 1996). Some ENO genes encoding this enzyme have been cloned from various plant species, including Mesembryanthemum crystallinum L., Arabidopsis (van der Straeten et al. 1991), Castor bean Latin (Blakeley et al. 1994), Oryza sativa L. (Umeda et al.1994), Lycopersicon esculentum Mill (van der Straeten et al.1991), Zea mays L. (Lal et al. 1991), and Nicotiana tabacum L. (Voll et al. 2009). In Arabidopsis, ENO2 is the highly expressed enolase (Andriotis et al. 2010). As a bifunctional locus, it encodes AtENO2 and AtMBP-1 (A. thaliana cMycbinding protein) as a positive regulator in response to ABA treatment (Kang et al. 2013). To date, AtMBP-1 has been identi fied as an ENO2 transcription repressor that mediates glycolysis (Eremina et al. 2015). ENO2 is also required for abiotic stress responses (Sachs et al. 1980; Lee et al.2002; Ndimba et al. 2005; Yan et al. 2005; Jiang et al. 2007;Barkla et al. 2009). Therefore, research investigating the ENO2 gene is important owing to its involvement in vital glycolysis and stress tolerance pathways as functional gene or transcription factor in plants.
We generated the ENO2 T-DNA insertion mutant (eno2–).Compared to the wild type (WT), this mutant displayed some phenotypic changes, such as reduced shoot and root growth,impaired male gametophyte organogenesis, and shortened siliques, as well as a reduced number of seeds. Our results were different from that of Eremina et al. (2015) who found that the development of female and male organs and the male gametophyte function required ENO2, and the fertility is reduced as a consequence of defects in ENO2. Bray (1997)suggested that the genotype, species, and phenotypic traits all affected the responses to abiotic stresses. Barkla et al. (2009) showed that a single point mutation (G325S)allele exhibited a reduction in ENO protein speci fically in tonoplast that was induced by salinity. In this study, we found that eno2–plants were completely susceptible to NaCl stress. Thus, we studied the expression pro files of AtENO2 during NaCl stress treatment using RT-qPCR and Western blotting. The results demonstrated that the transcript levels of AtENO2 were rapidly increased in response to treatment with 300 mmol L–1NaCl. ENO2 also responds to 300 mmol L–1NaCl stress at the protein level. To further unravel the mechanism of AtENO2 in responses to abiotic stress in plants, we adopted the 454 GS FLX sequencing method because it facilitates the identi fication and quanti fication of numerous transcripts (Rothberg et al. 2008). Global analysis of high-throughput sequencing in species exposed to different stress conditions will facilitate the identi fication of speci fic pathways and genes responsible for tolerance to a particular stress (Rangan et al. 2014). To generate a global perspective of the salinity-induced alterations at the transcriptomic level and the salinity-responsive metabolic pathways in the WT and eno2–mutant Arabidopsis, 454 GS FLX sequencing was performed with the leaves collected from the salinity-stressed WT and eno2–plants using the Ion Proton platform. We identi fied many differentially expressed genes (DEGs), which were functionally annotated using the GO and KEGG databases. The analyzed DEGs were associated with plant-pathogen interactions, plant hormone signal transduction, and the glycolysis pathway in KEGG.Beyond that, we determined whether ENO2 interacts with PGK, PGK1, TPI, PK, and GAPC1, which are also the key enzymes of glycolytic pathway. Based on these findings, we explored the effect of ENO2 on its interaction protein genes by RT-qPCR. This study improves our understanding of ENO2 and supports its importance in the survival of plants exposed to abiotic stress.
2. Materials and methods
2.1. Plant materials and stress treatment
The eno2–mutant plants were obtained progressively from T0through T3by the addition of 50 μg mL–1of kanamycin (Kan)to the Murashige and Skoog (MS) medium. After surfacesterilizing with 0.1% (w/v) HgCl2solution, seeds were allowed vernalization in the dark for at least 3 d at 4°C. Then, they were sown on MS plates (pH 5.8) containing 3% sucrose as the source of carbon and 0.7% (w/v) agar for solidi fication.The plants were grown at 22°C with a photoperiod of 16-h light/8-h dark and a relative humidity of 70%.
To measure root growth, seedlings were grown on MS plates supplemented with appropriate concentrations of NaCl. After 10 d, root length of seedlings was measured.For evaluation of salinity resistance, 30-d-old plants were further grown in a mixture of soil and vermiculite (3:1) for treatment with 300 mmol L–1NaCl.
To study the expression pro file of ENO2 (GenBank accession no. NM_129209.3) under the salt-stress, wild-type Arabidopsis seedlings at the 4–6 leaf stage were transferred to the plastic pots containing a mixture of vermiculite and perlite (3:1, v/v) and watered with the Hoagland nutrient solution supplemented with 100, 150, 200, or 300 mmol L–1NaCl for 0, 2, 4, 6, 8, 10, 12, 24, 36, or 48 h. To study the expression levels of ENO1, ENO2 and ENO3 under the salt stress, 30-d-old WT and eno2–plants were treated with 300 mmol L–1NaCl for 0, 18, or 24 h. To survey the expression pro file of ENO2 or ENO2 in the presence of cold, heat,drought, and salt stresses, 30-d-old WT plants were treated with 4°C, 28°C, 20% PEG 600, or 300 mmol L–1NaCl.
2.2. RNA extraction
Total RNA was puri fied from the leaves grown under the control or different salt stress conditions using the One-Step RNA Reagent (TransGen, Beijing, China). The RNA from each sample (2.5 μg) was used for reverse transcription with a cDNA Synthesis Kit (TransGen, Beijing, China) and then used in quantitative real-time RT-PCR (RT-qPCR)analysis. More than 10 μg of RNA per sample were puri fied using an RNeasy Plant Mini Kit (Qiagen, Valencia, CA,Spain). Total RNA was quanti fied using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) and a NanoDrop spectrophotometer (Thermo Fisher Scienti fic,Wilmington, DE, USA). These RNA are designed for 454 GS FLX sequencing.
2.3. Quantitative real-time RT-PCR analysis
Quantitative real-time RT-PCR was performed on an ABI 7500 System (Applied Biosystems, Foster City, CA, USA)with the SYBR green detection using a TransStart®Green qPCR SuperMix Kit (TransGen, Beijing, China). The ENO2 gene was analyzed in triplicate and at least three biological replicates. The average threshold cycle (Ct) was then calculated for each sample. The 2–ΔΔCtmethod was used to calculate the relative expression levels. The expression of β-actin was measured in parallel as an endogenous control.For examining the expression pro file of ENO2 under salt stress conditions, the control sample served as the calibrator with a nominal value of 1. The primers used in RT-qPCR for ENO1, ENO2, ENO3, CNGCs, AUX1, AHP, ATFBA7,and β-actin are shown in Appendix A.
2.4. Protein extraction
Proteins from the WT and eno2–mutant leaves were extracted in 50 mmol L–1Tris-HCl, 150 mmol L–1NaCl,10% glycerol, 0.1% Nonidet P-40, 5 mmol L–1dithiothreitol(DTT), and 0.1% Triton X-100 with vigorous vortexing. After incubation on ice for 30 min, the supernatant was produced by centrifugation at 13 000×g and 4°C for 15 min. The protein concentration was determined using the bicinchoninic acid(BCA) assay (Dingguo Changsheng, Beijing, China).
2.5. Western blotting analysis
The protein samples (20 μg each) were separated on 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) prior to transferring to nitrocellulose membranes (Pall, New York, NY, USA). Rabbit polyclonal antibody was prepared by BGI-Tech Solutions Co., Ltd. (BGI-Tech, Shenzhen, China). The membrane was blocked by Tris-buffered saline (TBS) containing 5%milk for 1 h at room temperature. The primary antibody(rabbit poly-clonal ENO2 and mouse β-actin antibodies) was prepared in the TBS buffer (1:3 000). After washing in the TBST buffer (TBS containing 0.05% Tween-20) to remove unbound antibodies, the blot was incubated with the goat anti-rabbit IgG conjugated to horseradish peroxidase and sheep anti-mouse secondary antibodies in TBST buffer at a dilution of 1:3 000, and the results were visualized using enhanced chemiluminescent reagents (Pierce, Appleton,WI, USA).
2.6. Library preparation and sequencing
Total RNA from the WT and eno2–plants treated with 300 mmol L–1NaCl for 18 h was converted to cDNA. After removing the rRNA, the samples were sequenced using the 454 GS FLX platform. Brie fly, mRNA was puri fied from 8 µg of total RNA, a mixture of RNA from WT or eno2–leaves exposed to high salinity of 300 mmol L–1for 18 h, using oligo(dT) magnetic beads. The cDNA fragments produced by divalent cations under elevated temperature were subjected to an end repair process and adapter ligation.These products were puri fied and enriched to create the final cDNA library. Additionally, Ion Proton sequencing was performed by BGI Tech Solutions Co., Ltd. (Shenzhen,China).
2.7. Differential gene expression analysis
In the 454 GS FLX study, we used the reads per kb per million reads (RPKM) method to analyze the gene expression pro files. Using a 0.1% or less false discovery rate (FDR),genes that exhibited a change of two-fold or greater were regarded as differentially expressed genes (DEGs). A DEG was considered up-regulated if its expression level in the eno2–sample was signi ficantly higher than those in the WT sample, while a down-regulated DEG was identi fied if the expression level in the WT was greater than that in the eno2–sample.
2.8. Analyses of Gene Ontology (GO) and pathway
The assembled sequences were compared against the public non-redundant protein sequences (Nr) database.According to the obtained Nr annotation, all DEG annotation information was collected in the Blast2GO software (Conesa et al. 2005). GO terms are assigned to query sequences,resulting in a group of genes that were cataloged in the transcriptome for separate ontology vocabularies, biological processes, molecular functions, and cellular components(Altschul 1993). Furthermore, pathway-based analysis improves our understanding of the biological functions of genes, and all DEGs were mapped to terms in the Kyoto encyclopedia of genes and genomes (KEGG) database.
2.9. Yeast two-hybrid assays
To assess protein interactions, yeast two-hybrid assay was carried out following the instructions of BD Matchmaker Library Construction & Screening Kits (Clontech, PaloAlto,CA, USA). The full-length cDNAs of PGK, PGK1, TPI, PK,GAPC1, and ENO2 were ampli fied separately. The primers used are shown in Appendix A. The cDNAs of PGK, PGK1,TPI, PK, and GAPC1 were cloned into pGADT7 to be used as the prey vector pGADT7-PGK, pGADT7-PGK1, pGADT7-TPI, pGADT7-PK, and pGADT7-GAPC1. The cDNA of ENO2 was cloned into pGBKT7 to be used as the bait vector pGBKT7-ENO2. The resulting pGADT7-PGK, pGADT7-PGK1, pGADT7-TPI, pGADT7-PK, pGADT7-GAPC1, and each bait vector were co-transformed into yeast strain AH109. The resulting bait vector and empty prey vector were also co-transformed into yeast strain AH109. The transformed colonies were identi fied on yeast SD/-Trp-Leu(DDO) and SD/-Trp-Leu-His-Ade (QDO) medium.
2.10. Statistical analysis
All the experiments were repeated at least three times, and all the data are presented as means±standard deviations(SD). One-way analysis of variance (ANOVA) was performed for all the data and the signi ficance of difference in values (P<0.05 or P<0.01) was determined by the Fisher’s least signi ficant difference (LSD) test using SPSS 20.0 for Windows.
3. Results
3.1. The WT exhibited superior salinity tolerance compared to eno2–
Salt stress had an obvious effect on the root growth at the 10th day after germination in the WT and eno2–plants.The root growth was signi ficantly different between the WT and eno2–, even though they were grown under normal conditions. Compared to the WT, the root length of eno2–plant had reduced by half under regular growth condition (Fig. 1-A), with the eno2–plants exhibiting a shorter root length. Root elongation of the WT and eno2–was signi ficantly reduced even by 50 mmol L–1NaCl, and the eno2–root length was only 1/4 of WT root length, which indicates increased sensitivity to NaCl stress (Fig. 1-A).Root elongation decreased in the WT and eno2–on MS medium supplemented with 100 mmol L–1NaCl. In particularly, the eno2–root growth was greatly inhibited,suggesting the eno2–was much more sensitive to salt than the WT (Fig. 1-A). Thus, the root growth of eno2–plants was greatly inhibited under the same conditions. The eno2–mutant was sensitive to salt stress based on root growth.This result indicates that ENO2 is involved in the response of Arabidopsis to salt stress. Thus, the comparison of the tolerance to NaCl between the eno2–mutant and the WT was conducted by treatment of 30-d-old plants grown under normal conditions with 300 mmol L–1NaCl for 21 d.

Fig. 1 Estimation of the salt stress tolerance of the wild-type(WT) and ENO2 T-DNA insertion mutant (eno2–) plants. A,root growth of the WT and eno2– plants in response to 0, 50,or 100 mmol L–1 NaCl for 10 d. B, salt tolerance of the WT and eno2– plants. 7-d-old seedlings were transferred to soil for an additional 23 d of normal growth (upper panel) and subjected to progressive salinity by the application of 300 mmol L–1 NaCl for 13 d (middle panel) and for 21 d (lower panel).
During the period of salt stress, most WT plants and almost all the eno2–plants displayed severe withering(Fig. 1-B). At least 40 strains of the WT and eno2–plants were treated separately with NaCl. We randomly chose a plate from the reproducible results (Fig. 1-B). This suggests that the loss-of-function of ENO2 compromised salt tolerance. Thus, ENO2 promotes salt tolerance in plants.
3.2. Salinity-responsive transcription and protein changes in the WT plants
To determine whether ENO2 expression is induced by salt stress, RT-qPCR analysis was carried out using total mRNA that were puri fied from the 30-d-old plants grown either under 100, 150, 200, or 300 mmol L–1NaCl treatment or normal condition. The mRNA levels in ENO2 increased in the plants exposed to 300 mmol L–1NaCl by approximately 11-fold at 18 h. The expression of ENO2 at 6 h exposure at 150 mmol L–1NaCl took the second place. ENO2 mRNA showed lower induction from 0.3- to 10-fold by the treatment of other salt conditions. However, The ENO2 mRNA levels were reduced to a low level approximately equaling the control at 24 h (Fig. 2-A). Arabidopsis contains three ENOs: ENO1, ENO2, and ENO3/ENOc. Consequently, we also studied ENO1 and ENO3 for the comparison to ENO2 between the WT and eno2–plants. The ENO2 transcript level was the highest, followed by ENO1 and ENO3 (Fig. 2-B).Among the three ENO genes, ENO2 was clearly the most important in response to salt stress. To con firm whether the expression of ENO2 was induced by other abiotic stresses in addition to salt, we performed RT-qPCR analysis to detect the expression pattern of ENO2. The transcript level of ENO2 increased dramatically upon exposure to salt. Salt was a more effective trigger for the transcription of ENO2 in comparison to cold, heat, and drought (Fig. 2-B). This indicates that ENO2 functions in the rapid response to salt stress in Arabidopsis. With an extended response time,it is possible that other defense genes or pathways play vital roles in resistance to salt damage. Moreover, we evaluated changes in ENO2 protein expression in the WT plants suffering from cold, heat, drought, or salt stress by Western blotting. Clear differences were observed in the abundance of ENO2 protein in response to these abiotic stresses. Analysis of the control and the cold, heat, drought,or salt-stressed samples revealed that ENO2 protein was signi ficantly altered in response to short-term salt stress.Likewise, cold, heat, or drought stress also enhanced the levels of ENO2, albeit to a lesser degree than salt stress. However, ENO2 accumulation was not obviously affected by drought stress at 24 h (Fig. 2-D). These results demonstrated that ENO2 is involved in the response to salt stress in Arabidopsis.
3.3. Sequence assembly and mapping statistics
It should be noted that, after pre-processing, reads shorter than 30 bp were excluded; after trimming the adapter sequence, each of sequences that were less than the length of the set threshold was also excluded (Fig. 3). The raw data were then converted to clean data, which were used for subsequent analyses.
Table 1 shows an overview of the genome sequencing and mapping results. RNA sequencing on a 454 GS-FLX provided 2.71 and 2.77 billion base pairs from the WT and eno2–plants, respectively. In each dataset, 18.88 and 19.25 million sequences matched perfectly with known genes.A total of 17 260 006 (63.76%) and 17 754 183 (64.08%)unique matches in each dataset were also obtained. The annotated reference assembly was downloaded for the reference genes (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/TAIR10_blastsets/TAIR10_cdna_20101214_updated).
For the WT, out of 27 071 011 total reads, 27 004 513(99.75%) were mapped to unique positions in the genome,and 26 530 577 (98.00%) reads showed unique matches to the genome. For eno2–, of 27 707 416 total reads, 27 636 225(99.74%) reads were mapped to unique positions in the genome, and 27 242 599 (98.32%) reads showed unique matches to the genome. The annotated reference was downloaded from TAIR10.

Fig. 2 Expression of enolase (ENO) in response to different abiotic stresses in the wild-type (WT) and ENO2 T-DNA insertion mutant(eno2–) plants. A, expression of ENO2 under salt stress as determined by quantitative real-time RT-PCR (RT-qPCR) analysis. B,the expression of ENO1, ENO2, and ENO3 under the salt stress as determined by RT-qPCR analysis. C, the expression of ENO2 in the presence of cold, heat, drought, and salt stresses, as revealed by RT-qPCR analysis. A–C, data are the means±standard deviation (SD) of three replicates. D, the expression of ENO2 in response to cold, heat, drought and salt stresses, as revealed by Western blot analysis. * and **, signi ficances at P<0.05 and P<0.01, respectively.
All unigenes were further classi fied with speci fic annotations using standardized Gene Ontology (GO)vocabulary. The available biological information was analyzed at P≤0.01 by pairwise comparisons, i.e., WT-18 h:eno2–-18 h.
3.4. Functional annotation (GO analysis)
Biological process was enriched in a total of 20 GO terms,cellular component in 13 GO terms, and molecular function in 11 GO terms (Fig. 4).
GO: Biological processBiological process enrichment was observed for 3 668 genes in 20 GO terms, ranging from 4 to 742 genes. Most identi fied unigenes were predicted in the metabolic process (742 genes), cellular process (697 genes), response to stimulus (568 genes), and singleorganism process (372 genes). In addition, there were identi fied unigenes predicted in the response to stress (67 genes), response to osmotic stress (40 genes), response to metal ion (27 genes), response to abiotic stimulus (21 genes), cellular response to stress (12 genes), response to biotic stimulus (12 genes), response to salt stress (2 genes),and cellular response to stimulus (2 genes). This finding suggests that the defense system and metabolic pathways are activated.
GO: Cellular componentCellular component enrichment was observed for 3 254 genes ranging from 2 to 905 genes in 13 GO terms. Most identi fied unigenes were associated with cell (905 genes), cell part (905 genes), organelle (575 genes), and membrane (354 genes).
GO: Molecular functionMolecular function enrichment was observed for 1 689 genes with a range of 1 to 720 genes in 11 GO terms. Most of the unigenes were involved in binding (720 genes), followed by catalytic activity (657 genes).
3.5. Differential gene expression between eno2– and the WT

Fig. 3 Sequence length distribution of the wild-type (WT) and ENO2 T-DNA insertion mutant (eno2–) libraries. A, size distribution of 454 sequencing reads after trimming of adapter and short reads (<30 bases) for the Arabidopsis WT. B, size distribution of 454 sequencing reads after trimming of adapter and short reads (<30 bases) for Arabidopsis eno2–.
Differential gene expression during the progression of stress reveals stress responsiveness and its putative role in stress tolerance. The DEGs are genes with a log2(fold change) ratio≥1 and false discovery rate (FDR)≤0.001 based on the DESeq software (Fig. 5). A total of 961 sequences were categorized as up-regulated genes and 746 sequences were categorized as down-regulated genes in eno2–compared to the WT. The detected fold changes(log2ratio) in gene expression ranged from –10.6 to 12.8.The most up-regulated known gene was AT1G67265.1,which encodes ROTUNDIFOLIA like 21, followed by AT1G29510.1, encoding a SAUR-like auxin-responsive protein family. The most down-regulated known genes|log2ratio (eno2–-18 h/WT-18 h)≥10| were AT5G11140.1,which encodes Arabidopsis phospholipase-like protein(PEARLI 4); AT3G21520.1, encoding a DUF679 domain membrane protein 1 (AtDMP1); AT1G42050.1 and AT1G42040.1, encoding transposable element genes;AT2G26400.1, encoding acireductone dioxygenase 3(ARD3); AT1G33760.1, encoding an integrase-type DNA-binding superfamily protein; AT2G23830.1, encoding a papD-like superfamily protein; and AT5G02780.1, encoding glutathione transferase lambda 1 (GSTL1). In addition,many well-characterized salt response genes were found among these DEGs. For example, AT3G09260.1 encoding the β-glucosidase 23, AT2G24850.1 encoding the tyrosine aminotransferase 3, and AT1G19210.1 encoding the ethylene-responsive transcription factor ERF017 were up-regulated. AT3G48360.1 encoding the BTB and TAZ domain protein 2 and AT1G71030.1 encoding the v-myb avian myeloblastosis viral oncogene homolog (MYB)-related transcription factor were down-regulated.
3.6. eno2–-speci fic gene expression pro file
In an attempt to determine the eno2–-associated expression of DEGs, the expression of 7 genes was assessed. Among them, DEGs, such as AT1G09930.1 (oligopeptide transporter 2), AT1G21850.1 (SKU5 similar 8 protein), AT1G68765.1(protein IDA), AT2G39530.1 (UPF0497 membrane protein),AT4G11170.1 (similar to several A. thaliana disease resistance proteins), AT5G11140.1 (phospholipase-like(PEARLI 4) family protein), and AT5G24110.1 (WRKY DNA-binding protein 30), were detected. Results of GO analysis indicated that 6 of these genes were enriched in the categories of response to abiotic stimulus, metabolic process, light stimulus, response to hormone stimulus,defense response, response to osmotic stress, response to stress, and transcription DNA-dependent. The KEGG analysis revealed that AT1G21850.1, AT4G11170.1, and AT5G24110.1, which function as an L-ascorbate oxidase[EC:1.10.3.3], the disease resistance protein RPS2, and the WRKY transcription factor 29, were separately enriched in PATH: map00423, PATH: map13457, and PATH: map13424, respectively.

3.7. KEGG analysis
To identify the biological pathways that were altered by ENO2 in response to salt stress, up- and down-regulated DEGs were assigned to KEGG pathways with enrichment statistics (Fig. 6). In the KEGG mapping, the DEGs with pathway annotations contributed to 115 pathways. The top 3 pathways (sorted by the q-value) were biosynthesis of the secondary metabolites (PATH: map01110), plantpathogen interaction (PATH: map04626), and plant hormone signal transduction(PATH: map04075). The pathways with the largest number of genes were sorted as follows: biosynthesis of the secondary metabolites with 159 (16.56%) DEGs,plant-pathogen interaction with 131 (13.65%) DEGs, and plant hormone signal transduction with 90 (9.38%) DEGs. However, there was no map of biosynthesis of the secondary metabolites in the KEGG database for DEGs identi fied.Consequently, we focused on map04626 and map04075 because both of them are important stress adaptation pathways.
Numerous salinity-responsive DEGs in the contrasting WT-18 h and eno2–-18 h were mapped onto the plant-pathogen interaction pathways using the KEGG database.The mapping revealed that the genes involved in the plant-pathogen interaction processes were signi ficantly affected during salinity stress, followed by kinase,transcription factors, defensive protein and genes involved in signal transduction(Fig. 7). The following 15 up-regulated protein families were identi fied, including cyclic nucleotide gated channel (CNGCs), calcium-dependent protein kinase(CDPK, EC:2.7.11.1), calmodulin/calcium-binding protein (CMLCaM/CML), mitogenactivated protein kinase kinase 1 (MEKK1, EC:2.7.11.25), WRKY transcription factor 25/29/33 (WRKY25/29/33), pathogenesis-related protein 1 (PR1), RPM1-interacting protein 4 (RIN4), disease resistance protein RPM1 (RPM1), disease resistance protein RPS2 (RPS2), heat shock protein 90 kDa beta (HSP90), jasmonate ZIM domain-containing protein (JAZ), and transcription factor MYC2 (MYC2). Only 2 protein families were down-regulated: respiratory burst oxidase (Rboh, EC:1.6.3.-1.11.1.-) and somatic embryogenesis receptor kinase (BKK1, EC:2.7.10.1 2.7.11.1).Interestingly, 5 protein families were up- and down-regulated simultaneously: chitin elicitor receptor kinase 1 (CERK1), LRR receptor-like serine/threonine-protein kinase FLS2 (FLS2, EC:2.7.11.1), brassinosteroid insensitive 1-associated receptor kinase 1 (BAK1, EC:2.7.10.1 2.7.11.1), WRKY transcription factor 22 (WRKY22),and serine/threonine-protein kinase PBS1 (PBS1, EC:2.7.11.1).
Mapping of the DEGs onto plant hormone signal transduction pathways using the KEGG database revealed a clear effect on the following hormones during salinity stress: auxin, cytokinin, gibberellin, abscisic acid, ethylene, brassinosteroid,jasmonic acid, and salicylic acid (Fig. 8). During auxin biosynthesis, most protein or protein families were changed. Three proteins, auxin in flux carrier (AUX1 LAX family), transport inhibitor response 1 (TIR1), and auxin responsive GH3 gene family (GH3) were up-regulated. Auxin response factor (ARF) was down-regulated.Auxin-responsive protein IAA (IAA) and SAUR family protein (SAUR) were up- and down-regulated simultaneously. In cytokinin biosynthesis, histidine-containing phosphotransfer protein (AHP), the two-component response regulator ARR-B family (ARR-B), and the two-component response regulator ARR-A family (ARR-A)were down-regulated. Histidine kinase 2/3/4 (cytokinin receptor, AHK2_3_4,EC:2.7.13.3) was up- and down-regulated. In the gibberellin biosynthesis, only the gibberellin receptor GID1 (GID1) was up-regulated. In abscisic acid biosynthesis,the abscisic acid receptor PYR/PYL family (PYL) was down-regulated. Protein phosphatase 2C (PP2C, EC:3.1.3.16), serine/threonineprotein kinase SRK2 (SNRK2, EC:2.7.11.1), and ABA responsive element binding factor (ABF) were up- and down-regulated. In ethylene biosynthesis, serine/threonineprotein kinase CTR1 (CTR1, EC:2.7.11.1) was downregulated, but ethylene-responsive transcription factor 1(ERF1) was up-regulated. In brassinosteroid biosynthesis,brassinosteroid insensitive 1-associated receptor kinase 1(BAK1, EC:2.7.10.1 2.7.11.1) and protein brassinosteroid insensitive 1 (BRI1, EC:2.7.10.1 2.7.11.1) were up- and down-regulated. Brassinosteroid resistant 1/2 (BZR1_2)was down-regulated, and xyloglucan:xyloglucosyl transferase TCH4 (TCH4, EC:2.4.1.207) was up-regulated.In jasmonic acid biosynthesis, jasmonate ZIM domaincontaining protein (JAZ) and the transcription factor MYC2(MYC2) were up-regulated. In salicylic acid biosynthesis,the transcription factor TGA (TGA) was up- and downregulated, and pathogenesis-related protein 1 (PR1) was up-regulated.

Fig. 4 Distribution of identi fied unigenes from wild-type (WT)-18 h:ENO2 T-DNA insertion mutant (eno2–)-18 h in the Gene Ontology (GO) term biological process, cellular component, and molecular function identi fied using the GO database (http://www.geneontology.org/).

Fig. 5 Analysis of differentially expressed genes (DEGs) in the wild-type (WT)-18 h and ENO2 T-DNA insertion mutant(eno2–)-18 h mutant libraries. The red and green dots denote signi ficantly different expression levels in each dataset (2-fold change), whereas the blue dot indicates no signi ficant difference in gene expression. RPKM, reads per kilobase per million mapped reads.
Because ENO2 catalyzes a key step in glycolysis, we examined glycolysis/gluconeogenesis (PATH: map00010)in 4 (0.42%) DEGs: AT2G36530.1 (ENO2), AT1G47860.1(LINE), AT4G26520.1 (ATFBA7), and AT4G26270.1 (PFK3)(Fig. 9). The most down-regulated gene was AT2G36530.1(ENO2 gene). This result showed that ENO2 was basically not expressed. Moreover, TFs (transcription factors) that are responsible for the expression of multiple downstream target genes (LINE and ATFBA7) in front of stress conditions were down-regulated. This result may be associated with the reduced salinity tolerance. Additionally, remedial key genes are critical during salt stress. The up-regulation of PFK3 could in fluence the phosphorylation events controlling the stress signaling process. Such genetic modi fications will lead to the activation of numerous down-stream response genes and provide salinity tolerance in plants.

Fig. 6 Scatter plot of Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment statistics. The rich factor is the ratio of differentially expressed genes (DEGs) parsed in a given pathway to the total number of genes parsed in the same pathway. A greater rich factor indicates greater intensity. The q-value is the corrected P-value, ranging from 0 to 1; a lower q-value indicates greater intensity. This figure shows the degree of enrichment for the top 20 pathways.
3.8. Validation of the identi fied DEGs through RT-qPCR
To con firm the accuracy of the 454 GS FLX sequencing results, the CNGCs (Fig. 7), AUX1 (Fig. 8), AHP (Fig. 8),and ATFBA7 (Fig. 9) randomly selected from above the three pathways were validated by the RT-qPCR. The differential expression levels of these genes observed by RT-qPCR were consistent with the results of 454 GS FLX sequencing (Fig. 10).
3.9. ENO2 regulated the expression of PGK, PGK1,TPI, and PK was involved in the glycolytic pathway
To determine whether ENO2 interacts with PGK, PGK1,TPI, PK, and GAPC1, we performed yeast two-hybrid experiment. The resulting prey and bait vectors were co-transformed into the yeast strain AH109. The yeast transformants were isolated and assessed for growth on SD/-Trp-Leu (DDO) and SD/-Trp-Leu-His-Ade (QDO)medium. The results demonstrated that ENO2 interacted with PGK, PGK1, TPI, and PK (Fig. 11-A). The expression of PGK and PGK1 genes, as indicated by the RT-qPCR, had an apparent reduction in the eno2–plants, but the expression of TPI and PK genes was not signi ficantly different between the WT and eno2–plants under regular growth condition.The expression of PGK, PGK1, TPI, and PK genes were affected obviously in the eno2–plants after salt treatment(Fig. 11-B–E).
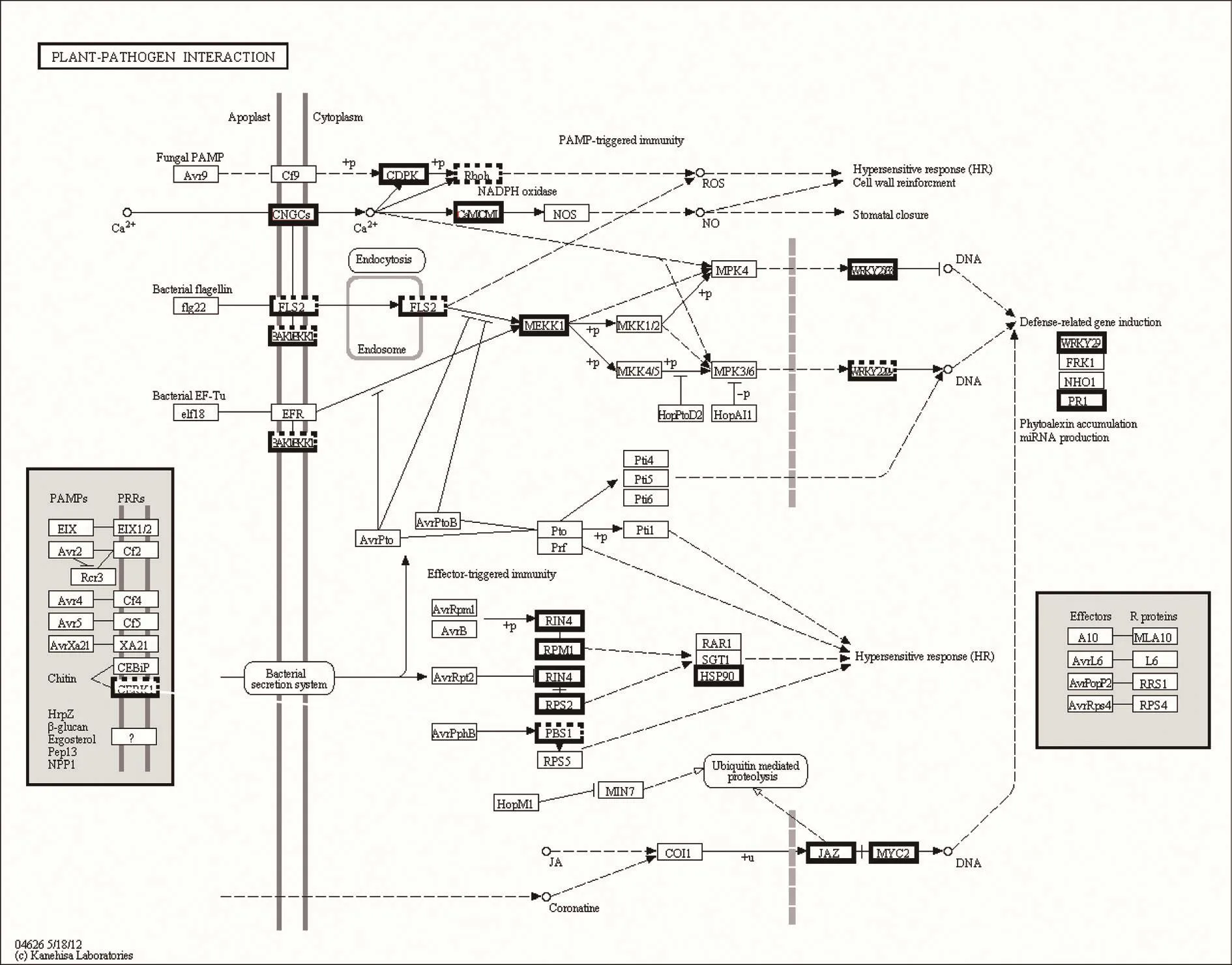
Fig. 7 Key enzymes and proteins regulating plant-pathogen interaction pathway. The genes encircled by bold solid line were upregulated. The genes encircled by bold dashed line were down-regulation. Other genes are no change in expression.
4. Discussion
Salinity threatens approximately 20% of arable soil and 50% of irrigated land worldwide (Caruso et al. 2008). Salt stress is caused by excessive concentrations of salt, mainly in the form of NaCl, in the soil. In this study, eno2–mutant plants exposed to a high salt concentration displayed several prominent phenotypes (Fig. 1), which indicates that AtENO2 is active in signal transduction pathways that are triggered by NaCl stress. Although research underlying the mechanism of ENO2 tolerance in plant species has largely progressed in recent studies (Kang et al. 2013), it is still unclear how ENO2 has led to the evolution of pathways that enables plants to withstand stress challenges in many biological environments. In the present study, the RT-qPCR and Western blotting results (Fig. 2) showed that AtENO2 responded rapidly to 300 mmol L–1NaCl stress. Namely,AtENO2 played a more important role than AtENO1 or AtENO3. AtENO2 also played a greater role at high NaCl concentrations compared to other abiotic stresses, such as the cold, heat, and drought stress. The 454 GS FLX sequencing was used to investigate the causes of the different phenotypes and the multiple processes that were affected in the WT and eno2–Arabidopsis plants. Pathways,such as biosynthesis of secondary metabolites, plantpathogen interactions (Fig. 7), and plant hormone signal transduction (Fig. 8), were highly in fluenced by high salinity.We also analyzed key enzymes and proteins that regulate glycolysis because ENO2 catalyzes a key step in glycolysis(Fig. 9). Based on our GO analysis, we realize that ENO2 play a role in metabolic processes, response to stimuli;particularly the cellular processes, cellular development,membrane binding, and catalytic activity, among others.Previous studies have shown that cell wall components and cytoskeleton organization maintained cell turgor by transforming the cell size (Ndimba et al. 2005; Li et al.2011). ENO2-GFP localizes in the nucleus as well as in the cytoplasm (Lee et al. 2002). The ENO2 promoter region was fused to the α-glucuronidase reporter gene (GUS),GUS expression was detected in all plant tissues (Lee et al.2002). Analysis of fluorescence using confocal microscopy indicated that AtENO2-YFP was localized in both cytosolic and nuclear cellular compartments, while AtMBP-1-YFP was preferentially localized in nuclei (Kang et al. 2013;Eremina et al. 2015). DEGs with pathway annotations contributed to 115 pathways in the KEGG mapping analyses.The expression levels of the four genes validated using RT-qPCR con firmed the accuracy of the 454 sequencing results (Fig. 10).

Fig. 8 Key enzymes and proteins regulating the plant hormone signal transduction pathway. The genes encircled by bold solid line were up-regulated. The genes encircled by bold dashed line were down-regulation and the genes encircled conjointly by bold solid and other genes are no change in expression.

Fig. 9 Key enzymes and proteins regulating glycolysis. The number in the square represents the ID of the enzyme. The genes encircled by bold solid line were up-regulated. The genes encircled by bold dashed line were down-regulation. Other genes are no change in expression.
Because multi-genes regulate stress response, it is dif ficult to signi ficantly enhance plant stress tolerance using a single functional gene approach (Mittler and Blumwald 2010). A large array of stress regulatory genes, such as TFs and protein kinases, have been authenticated in many plants (Shinozaki et al. 2003). Progressively more studies have found that some TFs or protein kinases have vital roles in multiple abiotic stress tolerances (Banerjee and Roychoudhury 2015). The calcium-dependent protein kinase (CDPK) (Schaller et al. 2008) pathways and mitogen-activated protein kinase (MAPK) (Huang et al.2012) involved in some plant stress responses. Three genes encoding the CDPK among the DEGs, namely,AT5G66210.4, AT4G04700.1 and AT4G04695.1, were up-regulated. In addition, another three genes encoding the MAPK among the DEGs, namely, AT1G01560.1,AT1G01560.2 and AT5G67080.1 were also up-regulated.This shows that ENO2 is very important in stress responses.
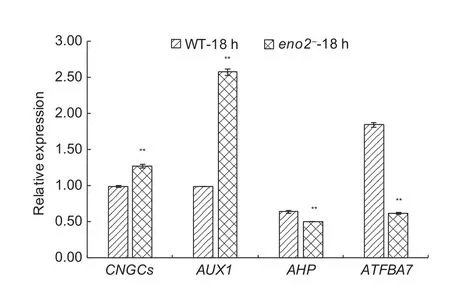
Fig. 10 The relative expression levels of CNGCs, AUX1,AHP, and ATFBA7. CNGCs, AUX1, AHP, and ATFBA7 were randomly selected from differentially expressed genes (DEGs)in response to 300 mmol L–1 NaCl stress, as revealed by quantitative real-time RT-PCR (RT-qPCR) analysis. 30-d-old wild type (WT) and ENO2 T-DNA insertion mutant (eno2–)plants were treated with 300 mmol L–1 NaCl for 18 h. * and**, signi ficances at P<0.05 and P<0.01, respectively. Bars represent the values of stand error.
Many abiotic stresses can weaken plant defense mechanisms (Amtmann et al. 2008; Mittler and Blumwald 2010; Atkinson and Urwin 2012). Because NaCl is known as an antifungal agent (Blomberg and Adler 1993), salt stresspathogen interactions may be highly affected by stress concentrations (Souman and Kostandi 1998; Poschenrieder et al. 2006; Fones et al. 2010). Our results (Fig. 7) were consistent with previous researches.

Fig. 11 Interactions of ENO2 with PGK, PGK1, TPI, and PK under salt stress. A, yeast cells were co-transformed with the PGK,PGK1, TPI, and PK, pGBKT7-ENO2 (bait) constructs and the pGADT7 empty vector, pGBKT7-ENO2 proteins (bait) and all four pGADT7-PGK, pGADT7-PGK1, pGADT7-TPI, and pGADT7-PK proteins (prey) were grown on yeast SD drop-out medium that lacks Leu and Trp (–2) and Ade, His, Leu, and Trp (–4). B–E, the relative expression levels of PGK, PGK1, TPI, and PK in the wild type (WT) and ENO2 T-DNA insertion mutant (eno2–) plants. * and **, signi ficances at P<0.05 and P<0.01, respectively. Bars represent the values of stand error.
Additionally, hormones play a role in the integration of environmental stimuli under stress conditions (Kissoudis et al. 2014). The growth hormones, gibberellin, cytokinin,auxin, and brassinosteroid are active in response to adverse situations and pathogen attack (Robert-Seilaniantz et al.2011). Abscisic acid produced under high salinity conditions regulates stomatal closure and activates many stress-related genes to increase tolerance of plant to stresses (Nakashima et al. 2012). Ethylene participates in BR-induced AOX activity, which is important in the tolerance to abiotic stresses(Wei et al. 2015). Jasmonic acid (JA) is ubiquitous in plants (Schaller 2011; Pirbalouti et al. 2014). Under certain abiotic stresses, in particular salt stress, the application of exogenous JA was effective to improve tolerance of plants to stresses (Ahmad et al. 2016). Salicylic acid (SA) is a phenolic compound that triggers hypersensitive response to defense biotrophic pathogens (Eremina et al. 2016). It has also been reported that SA induces salinity tolerance by eventually activating the photosynthetic process and alleviating oxidative stress (Li et al. 2014). Thus, our study further con firms and supplements previous research investigating the relationship between abiotic stress and plant hormones.
Interestingly, only one protein family in the glycolysis pathway was up-regulated. This finding suggests that the molecular responses of ENO2 to abiotic stresses involve interactions among many pathways in addition to the glycolysis pathway. Therefore, it is very dif ficult to increase salt tolerance in complex organisms such as plants using only one approach.
PGK is an ATP-generating enzyme, a part of the glycolytic,photosynthetic, and gluconeogenic pathways (Banks et al.1979; McHarg et al. 1999; Lin et al. 2007). Ectopic expression of Pokkali phosphoglycerate kinase-2 (OsPGK2-P) improved yield in tobacco (Nicotiana tabaccum L.) plants under salinity stress (Joshi et al. 2016). TPI involved in response to a variety of abiotic stress in rice (Oryza sativa L.) (Sharma et al. 2012). PK was induced in soybean under flooding conditions and induced metabolic changes that promoted survival and recovery from flooding-induced damage (Khan et al. 2015). Results from the yeast two-hybrid assay showed that ENO2 interacts with other glycolysis enzymes such as PGK, PGK1, TPI, and PK (Fig. 11-A). Most importantly, the expression of PGK and PGK1 had an apparent reduction in the eno2–plants (Fig. 11-B–C), and ENO2 may regulate the expression of PGK, PGK1, TPI, and PK under salt stress(Fig. 11-B–E). The exceptions of these differences from the sequencing data in expression may result from the different criteria. For RT-qPCR analysis, it is relative expression level,but for sequencing, it is RPKM (reads per kiobase million reads). This result suggested that ENO2 had an important role in the plant growth and response to the high salt stress in some ways.
5. Conclusion
In summary, we sequenced the WT and eno2–mutant Arabidopsis lines that were stressed with 300 mmol L–1NaCl for 18 h using the 454 GS FLX platform and identi fied a large number of DEGs. Our EST database can serve as a valuable resource for genetic and genomic studies of Arabidopsis mutants in response to salt stress. The present findings highlight the diverse modes of adaptation to salt stress, and additional research such as proteomics studies and determination of enzyme activities will be essential to elucidate the related regulatory mechanisms.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (31470399 and 31270365). We thank BGI-Tech Solutions Co., Ltd. (BGI-Tech, China) for performing the transcriptome sequencing.
Appendixassociated with this paper can be available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
Ahmad P, Prasad M N. 2012. Abiotic Stress Responses in Plants: Metabolism, Productivity and Sustainability.Springer-Verlag, New York.
工会是在党的领导下独立自主开展工作的群众性组织。作为国有特大型企业的工会组织,在维护职工权益、协调劳动关系、促进和谐创建等方面具有其他组织不可替代的优势。当前,企业的用工制度、工作环境、管理重点、员工队伍构成、需求层次、意识形态等各方面都发生了显著的变化,因而工会权益维护、参政议政、生产建设、教育引领的难度不断加大,工会工作面临着全新的选择和考验。在这种大环境下,如何把住时代的脉搏,高效规范地履行职责,使工会组织有作为、有地位、有影响,使工会干部有形象、有威信、有人气,笔者认为应当着重把握好以下四个方面:
Ahmad P, Rasool S, Gul A, Sheikh S A, Akram N A, Ashraf M, Kazi A M, Gucel S. 2016. Jasmonates: Multifunctional roles in stress tolerance. Frontiers in Plant Science,7, 813.
Altschul S F. 1993. A protein alignment scoring system sensitive at all evolutionary distances. Journal of Molecular Evolution,3, 290–300.
Amtmann A, Trouf flard S, Armengaud P. 2008. The effect of potassium nutrition on pest and disease resistance in plants.Plant Physiology,4, 682–691.
Andriotis V M, Kruger N J, Pike M J, Smith A M. 2010. Plastidial glycolysis in developing Arabidopsis embryos. New Phytologist,3, 649–662.
Atkinson N J, Urwin P E. 2012. The interaction of plant biotic and abiotic stresses: From genes to the field. Journal of Experimental Botany,10, 3523–3543.
Banerjee A, Roychoudhury A. 2015. WRKY proteins: Signaling and regulation of expression during abiotic stress responses. Scienti fic World Journal,2015, 807560.
Barkla B J, Vera-Estrella R, Hernández-Coronado M, Pantoja O. 2009. Quantitative proteomics of the tonoplast reveals a role for glycolytic enzymes in salt tolerance. The Plant Cell,12, 4044–4058.
Bent A F, Mackey D. 2007. Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annual Review of Phytopathology,45, 399–436.
Blakeley S D, Dekroon C, Cole K P, Kraml M, Dennis D T. 1994.Isolation of a full-length cDNA encoding cytosolic enolase from Ricinus communis. Plant Physiology,1, 455–456.
Blomberg A, Adler L. 1993. Tolerance of fungi to NaCl. In:Jennings D H, ed., Stress Tolerance of Fungi. Marcel Dekker, New York. pp. 209–232.
Bray A. 1997. Plant responses to water de ficit. Trends in Plant Science,2, 48–54.
Bray E A, Bailey-Serres J, Weretilnyk E. 2000. Responses to abiotic stresses. In: Gruissem W, Buchnnan B, Jones R, eds., Biochemistry and Molecular Biology of Plants.American Society of Plant Physiologists, Rockville, MD.pp. 1158–1249.
Caruso G, Cavaliere C, Guarino C, Gubbiotti R, Foglia P,Laganà A. 2008. Identi fication of changes in Triticum durum L. leaf proteome in response to salt stress by two-dimensional electrophoresis and MALDI-TOF mass spectrometry. Analytical and Bioanalytical Chemistry,1,381–390.
Conesa A, Götz S, García-Gómez J M, Terol J, Talón M,Robles M. 2005. Blast2GO: A universal tool for annotation,visualization and analysis in functional genomics research.Bioinformatics,18, 3674–3676.
Eremina M, Rozhon W, Poppenberger B. 2016. Hormonal control of cold stress responses in plants. Cellular and Molecular Life Sciences,4, 797–810.
Eremina M, Rozhon W, Yang S, Poppenberger B. 2015. ENO2 activity is required for the development and reproductive success of plants, and is feedback-repressed by AtMBP-1.The Plant Journal,6, 895–906.
Fones H, Davis C A, Rico A, Fang F, Smith J A, Preston G M. 2010. Metal hyperaccumulation armors plants against disease. PLoS Pathogens,9, e1001093.
Holland M J, Holland J P. 1978. Isolation and identi fication of yeast messenger ribonucleic acids coding for enolase,glyceraldehyde-3-phosphate dehydrogenase, and phosphoglycerate kinase. Biochemistry,23, 4900–4907.
Huang G T, Ma S L, Bai L P, Zhang L, Ma H, Jia P, Liu J, Zhong M, Guo Z F. 2012. Signal transduction during cold, salt, and drought stresses in plants. Molecular Biology Reports,2,969–987.
Jiang Y, Yang B, Harris N S, Deyholos M K. 2007. Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. Journal of Experimental Botany,13,3591–3607.
Joshi R, Karan R, Singla-Pareek S L, Pareek A. 2016.Ectopic expression of Pokkali phosphoglycerate kinase-2(OsPGK2-P) improves yield in tobacco plants under salinity stress. Plant Cell Reports,1, 27–41.
Kang M, Abdelmageed H, Lee S, Reichert A, Mysore K S, Allen R D. 2013. AtMBP-1, an alternative translation product of LOS2, affects abscisic acid responses and is modulated by the E3 ubiquitin ligase AtSAP5. The Plant Journal,3,481–493.
Khan M N, Sakata K, Komatsu S. 2015. Proteomic analysis of soybean hypocotyl during recovery after flooding stress.Journal of Proteomics,121, 15–27.
Kissoudis C, van de Wiel C, Visser R G, van der Linden G. 2014.Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular crosstalk. Frontiers in Plant Science,5, 207.
Lal S K, Johnson S, Conway T, Kelley P M. 1991. Characterization of a maize cDNA that complements an enolase-de ficient mutant of Escherichia coli. Plant Molecular Biology,5,787–795.
Lee H, Guo Y, Ohta M, Xiong L, Stevenson B, Zhu K. 2002.LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional ENOLASE. EMBO Journal,11, 2692–2702.
Li T, Hu Y, Du X, Tang H, Shen C, Wu J. 2014. Salicylic acid alleviates the adverse effects of salt stress in Torreya grandis cv. Merrillii seedlings by activating photosynthesis and enhancing antioxidant systems. PLOS ONE,10,e109492.
Li W, Zhang C, Lu Q, Wen X, Lu C. 2011. The combined effect of salt stress and heat shock on proteome pro filing in Suaeda salsa. Journal of Plant Physiology,15, 1743–1752.
Lin J W, Ding M P, Hsu Y H, Tsai C H. 2007. Chloroplast phosphoglycerate kinase, a gluconeogenetic enzyme, is required for ef ficient accumulation of Bamboo mosaic virus.Nucleic Acids Research,35, 424–432.
Liu X, Zhang H, Zhao Y, Feng Z, Li Q, Yang HQ, Luan S, Li J, He Z H. 2013. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America,38, 15485–15490.
McHarg J, Kelly S M, Price N C, Cooper A, Littlechild J A. 1999.Site-directed mutagenesis of proline 204 in the ‘hinge’ region of yeast phosphoglycerate kinase. European Journal of Biochemistry,259, 939–945.
Mickelbart M V, Hasegawa P M, Bailey-Serres J. 2015. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nature Reviews Genetics,4, 237–251.
Mittler R. 2006. Abiotic stress, the field environment and stress combination. Trends in Plant Science,1, 15–19.
Mittler R, Blumwald E. 2010. Genetic engineering for modern agriculture: Challenges and perspectives. Annual Review of Plant Biology,61, 443–462.
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. 2012. NAC transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta,2, 97–103.
Ndimba B K, Chivasa S, Simon W J, Slabas A R. 2005.Identi fication of Arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics,16,4185–4196.
Pieterse C M, van der Does D, Zamioudis C, Leon-Reyes A, van Wees S C. 2012. Hormonal modulation of plant immunity.Annual Review of Cell and Developmental Biology,28,489–521.
Pirbalouti A G, Mirbagheri H, Hamedi B, Rahimi E. 2014.Antibacterial activity of the essential oils of myrtle leaves against Erysipelothrix rhusiopathiae. Asian Paci fic Journal of Tropical Biomedicine,4, 505–509.
Poschenrieder C, Tolrà R, Barceló J. 2006. Can metals defend plants against biotic stress? Trends in Plant Science,6,288–295.
Rangan P, Subramani R, Kumar R, Singh A K, Singh R. 2014.Recent advances in polyamine metabolism and abiotic stress tolerance. Biomed Research International,2014,239621.
Reed G H, Poyner R R, Larsen T M, Wedekind J E, Rayment I. 1996. Structural and mechanistic studies of enolase.Current Opinion in Structural Biology,6, 736–743.
Robert-Seilaniantz A, Grant M, Jones J D. 2011. Hormone crosstalk in plant disease and defense: More than just JASMONATE-SALICYLATE antagonism. Annual Review of Phytopathology,49, 317–343.
Rothberg J M, Leamon J H. 2008. The development and impact of 454 sequencing. Nature Biotechnology,26, 1117–1124.Sachs M M, Freeling M, Okimoto R. 1980. The anaerobic proteins of maize. Cell,3, 761–767.
Schaller G E, Kieber J J, Shiu S H. 2008. Two-component signaling elements and histidyl-aspartyl phosphorelays.Arabidopsis Book,6, e0112.
Schaller M. 2011. The behavioral immune system and the psychology of human sociality. Philosophical Transactions of the Royal Society of London Series B (Biological Sciences),1583, 3418–3426.
Sharma S, Musta fiz A, Singla-Pareek S L, Srivastava P S, Sopory S K. 2012. Characterization of stress and methylglyoxal inducible triose phosphate isomerase (OscTPI) from rice.Plant Signaling & Behavior,10, 1337–1345.
Shao H, Wang H, Tang X. 2015. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Frontiers in Plant Science,6, 902.
Shi H, Ye T, Chen F, Cheng Z, Wang Y, Yang P, Zhang Y, Chan Z. 2013. Manipulation of arginase expression modulates abiotic stress tolerance in Arabidopsis: Effect on arginine metabolism and ROS accumulation. Journal of Experimental Botany,5, 1367–1379.
Shinozaki K, Yamaguchi-Shinozaki K, Seki M. 2003. Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology,5, 410–417.
Souman M F, Kostandi S F. 1998. Effect of saline environment on yield and smut disease severity of different corn genotypes (Zea mays L.). Journal of Phytopathology,4,185–189.
van der Straeten D, Rodrigues-Pousada R A, Goodman H M, van Montagu M. 1991. Plant enolase: Gene structure,expression and evolution. The Plant Cell,7, 719–735.
Umeda M, Uchimiya H. 1994. Differential transcript levels of genes associated with glycolysis and alcohol fermentation in rice plants (Oryza sativa L.) under submergence stress.Plant Physiology,3, 1015–1022.
Voll L M, Hajirezaei M R, Czogalla-Peter C, Lein W, Stitt M,Sonnewald U, Börnke F. 2009. Antisense inhibition of enolase strongly limits the metabolism of aromatic amino acids, but has only minor effects on respiration in leaves of transgenic tobacco plants. New Phytologist,3, 607–618.
Wang H, Wang H, Shao H, Tang X. 2016. Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Frontiers in Plant Science,7, 67.
Wei L J, Deng X G, Zhu T, Zheng T, Li P X, Wu J Q, Zhang D W, Lin H H. 2015. Ethylene is involved in Brassinosteroids induced alternative respiratory pathway in cucumber(Cucumis sativus L.) seedlings response to abiotic stress.Frontiers in Plant Science,6, 982.
Yan S P, Tang Z, Su W, Sun W. 2005. Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics,1,235–244.
6 December, 2016 Accepted 5 May, 2017
Correspondence ZHANG Gen-fa, Tel: +86-10-58809453, E-mail:gfzh@bnu.edu.cn; LI Hong-jie, Tel: +86-10-82105321, E-mail:lihongjie@caas.cn
© 2018 CAAS. Publishing services by Elsevier B.V. All rights reserved.
10.1016/S2095-3119(17)61720-9
Section editor ZHANG Xue-yong
Managing editor WANG Ning
 Journal of Integrative Agriculture2018年1期
Journal of Integrative Agriculture2018年1期
- Journal of Integrative Agriculture的其它文章
- Climate change and agriculture: Impacts and adaptive responses in Iran
- Development of elite restoring lines by integrating blast resistance and low amylose content using MAS
- Evaluation of stability and yield potential of upland rice genotypes in North and Northeast Thailand
- ldenti fication of the resistance gene to powdery mildew in Chinese wheat landrace Baiyouyantiao
- New clues concerning pigment biosynthesis in green colored fiber provided by proteomics-based analysis
- Constitutive expression of feedback-insensitive cystathionine γ-synthase increases methionine levels in soybean leaves and seeds
