Thermogenic protein UCP1 and UCP3 expression in nonsmall cell lung cancer: relation with glycolysis and anaerobic metabolism
Alexandra Giatromanolaki, Konstantina Balaska, Dimitra Kalamida, Christos Kakouratos, Efthimios Sivridis,Michael I. KoukourakisDepartment of Pathology, Department of Radiotherapy/Oncology, Democritus University of Thrace, University Hospital of Alexandroupolis, Alexandroupolis 6800, Greece
ORIGINAL ARTICLE
Thermogenic protein UCP1 and UCP3 expression in nonsmall cell lung cancer: relation with glycolysis and anaerobic metabolism
Alexandra Giatromanolaki1, Konstantina Balaska1, Dimitra Kalamida2, Christos Kakouratos2, Efthimios Sivridis1,Michael I. Koukourakis21Department of Pathology,2Department of Radiotherapy/Oncology, Democritus University of Thrace, University Hospital of Alexandroupolis, Alexandroupolis 68100, Greece
Uncoupling protein 1 (UCP1) is a proton transporter/channel residing on the inner mitochondrial membrane and is involved in cellular heat production. Using immunohistochemistry, we investigated the expression of UCP1 and UCP3 in a series of 98 patients with non-small cell lung cancer (NSCLC) treated with surgery. Expression patterns were correlated with histopathological variables, prognosis, and the expression of enzymes/proteins related to cell metabolism. Bronchial epithelium did not express UCP1 or UCP3, while alveolar cells strongly expressed UCP1. In tumors, strong expression of UCP1 and UCP3 was recorded in 43/98 (43.8%) and 27/98 (27.6%) cases, respectively. UCP1 was significantly associated with squamous cell histology (P = 0.05),whilst UCP3 was more frequently overexpressed in large cell carcinomas (P = 0.08), and was inversely related to necrosis (P =0.009). In linear regression analysis, UCP1 was directly related to markers of glycolysis [hexokinase (HXKII) and phosphofructokinase (PFK1)] and anaerobic glucose metabolism [pyruvate dehydrogenase kinase (PDK1) and lactate dehydrogenase (LDH5)]. UCP3 was directly linked with a glucose transporter (GLUT2), monocarboxylate transporter (MCT2),glycolysis markers (PFK1 and aldolase), and with the phosphorylation of pyruvate dehydrogenase (pPDH). Kaplan-Meier survival analysis showed that UCP3 was significantly related to poor prognosis in squamous cell carcinomas (P = 0.04). UCP1 and UCP3 are overexpressed in a large subgroup of non-small cell lung tumors and their expression coincides with increased glucose absorption, intensified glycolysis, and anaerobic glucose usage. Whether UCPs are targets for therapeutic interventions in lung cancer is a hypothesis that demands further investigation.
Lung cancer; thermogenesis; UCP1; UCP3; glycolysis; anaerobic metabolism
Introduction
Mitochondrial oxidative phosphorylation is the main source of ATP production in normal tissues. During this process, a proton gradient is generated across the mitochondrial inner membrane, which is essential for the formation of ATP by ATP synthase. Around 25% of the energy of this proton gradient is, however, consumed by pumping protons into the inner membrane space, which is a 'proton-leak' process called mitochondrial uncoupling. This process bypasses ATP synthase and produces heat1,2. The uncoupling protein,(UCP), is a specific proton transporter/channel residing onthe inner mitochondrial membrane that is responsible for some of this heat production. Indeed, five different UCPs have been identified in mammals: UCP1, thermogenin identified in brown adipose tissue; UCP2, widely expressed in tissues; UCP3, mainly located in muscle tissues, and SLC25A27/UCP4, and SLC25A14/BMCP1/UCP5,predominantly expressed in the central nervous system3.Several functions have been attributed to UCPs, including cold- and diet-related thermogenesis, repression of reactive oxygen species (ROS) formation, and glucose sensing4.
The role of UCPs in tumorigenesis and cancer growth has recently been a focus for many researchers. UCP1 for example has been shown to be expressed in prostate cancer and be related to a metastatic phenotype5. UCP2 has been identified to induce the Warburg effect and to promote tumor growth in breast cancer6. A recent study in skin cancer suggests that UCP3 increases lipid oxidation but limits tumorigenesis through the inhibition of Akt7. Alternatively,increased UCP3 expression has been reported in gastrointestinal adenocarcinomas8and in renal cell carcinoma9. Defects in UCP3 protein are speculated to be involved in obesity, a condition characterized by an excessive increase in body weight because of a massive accumulation of body fat.
In the current study, we investigated the expression of UCP1 and UCP3 in a series of non-small cell lung cancers(NSCLCs) and examined their association with histopathological variables and prognosis, as well as the expression of other proteins related to metabolism.
Materials and methods
Formalin-fixed paraffin-embedded (FFPE) material from 98 tissue samples, from patients treated with surgery for NSCLC, was retrieved from the archives of our Pathology Department. Patients were serial in terms of date of operation. The median age of patients was 68 years (range 32-81). Twelve out of 98 patients were female. Using the UICC staging system, the distribution of patients among stages was 46 in stage I, 22 in stage II, and 30 in stage III.Regarding histology, 58 were of the squamous type, 22 were adenocarcinomas, and 18 were large cell undifferentiated carcinomas. The time of follow-up ranged from 26-112 months, with a median of 46 months. This study was approved by the Research Ethics Committee of the Democritus University of Thrace, in the context of the ARISTEIA II project.
UCP immunohistochemistry
Immunohistochemistry was performed on FFPE tissue sections, 3 μm thick, that were placed on positively charged slides. The slides were then deparaffinized by xylene and rehydrated through graded ethanol solutions to water, so that aqueous antibodies, molecular probes, and detection reagents could penetrate and adhere to the tissue. The heat-induced epitope retrieval (HIER) process was performed in a microwave oven using Dako EnVision FLEX Target Retrieval Solution (pH 6.0). Slides were immersed into a pre-heated container following incubation at 97°C for 3 × 5 min. After the container with the slides was removed from the microwave oven, it was allowed to cool at 25–27°C for 20 min and then it was placed for 10 min in the wash buffer solution (Tris-buffered saline containing Tween 20, pH 7.6).For both antibodies (UCP1 and UCP3) tested in the current study, polymer detection methods were employed due to their increased sensitivity and detection simplicity. Omission of the primary antibody was used as a negative control. The anti-UCP antibodies were validated with Western blot detection of specific bands in cancer cell line extracts(prostate cancer 22RV1, glioblastoma U87, and lung cancer H1299). Indeed, specific protein bands at 32 and 46 kDa for UCP1 and UCP3, respectively, were identified(Supplemental Figure 1S-A).
Validation of the antibodies was further performed by examining the signal from the antibodies in tissues expected to express the mRNAs in the Human Protein Atlas https://www.proteinatlas.org/ENSG00000109424-UCP1/tissue and https://www.proteinatlas.org/ENSG00000175564-UCP3/tissue). Thus, the immunohistochemical signal obtained in skeletal muscles, breast tissue, adrenal tissue,colon (reported as negative), and bronchus (reported as negative) were performed before the immunostaining of lung cancer tissues. Images of this control immunostaining are shown in Supplemental Figure 1S-B.
With respect to the UCP1 primary antibody, the UltraVision Quanto Detection System was applied.Nonspecific background staining was blocked by preincubation with UltraVision Protein Block for 5 min,following 2 × 6 min PBS buffer washing. The slides were then incubated with the UCP1 primary antibody overnight at 4°C.The rabbit polyclonal anti-UCP1 antibody (ab10983, Abcam,Cambridge, UK) was used at a dilution of 1:500. This antibody was raised against a synthetic peptide expressed in brown adipose tissue and localized in the mitochondrial inner membrane. The peptide was conjugated to keyhole limpet hemocyanin (KLH) corresponding to amino acids 145–159 of human UCP1, with an N-terminal cysteine added(http://www.abcam.com/ucp1-antibody-ab10983.html).After an overnight incubation, tissue sections were washed in PBS 2 × 6 min and then UltraVision Hydrogen Peroxide Block was applied for 10 min, in order to neutralize endogenous peroxide activity. The slides were then washed with PBS buffer 2 × 6 min and then incubated with Primary Antibody Amplifier Quanto for 10 min, which is suitable for both mouse and rabbit primary antibodies. After 2 × 6 min PBS buffer washing, HRP Polymer Quanto was applied and slides were incubated for another 10 min. Subsequently,extensive PBS buffer washing was performed (3 × 6 min) and tissue sections were incubated with DAB Quanto Chromogen for 6 min. The slides were then PBS buffer washed,counterstained with hematoxylin OS (Vector H-3404),dehydrated through graded ethanol solutions as well as xylene, and mounted in a synthetic resin.
The detection of UCP3 was performed with the Dako EnVision FLEX detection system and tissue sections were incubated with primary antibody for 60 min at room temperature. The rabbit polyclonal anti-UCP3 antibody
(ab125830, Abcam, Cambridge, UK) was used at a dilution of 1:100. This antibody was raised against a synthetic peptide conjugated to KLH, derived from residues 100–200 of human UCP3 (http://www.abcam.com/ucp3-antibodyab125830.html). UCP3 is only expressed in skeletal muscle(more highly expressed in glycolytic rather than oxidative skeletal muscle) and heart tissue. Following primary antibody incubation, slides were washed with PBS buffer (2 × 6 min)and EnVision FLEX Peroxidase-Blocking Reagent was applied for 10 min, in order to block endogenous peroxidase activity. Slides were then washed with PBS buffer solution 2 ×6 min and then incubated with EnVision FLEX+ Rabbit Linker for 15 min in order to amplify the signal. Tissue sections were washed with buffer (2 × 6 min) and the Dako EnVision FLEX HRP detection reagent was applied for 30 min. This reagent consists of a dextran backbone onto which a large number of peroxidase (HRP) molecules and secondary antibody molecules have been coupled. Sections were then washed in buffer (3 × 6 min) and EnVision FLEX DAB+ Chromogen was applied to the sections for 6 min. The final step of the procedure involved washing the sections with buffer (2 × 6 min), counterstaining with hematoxylin OS(Vector H-3404), dehydrating through graded ethanol solutions as well as xylene, and mounting of tissue sections in a synthetic resin.
Metabolism-related immunohistochemistry
Our UCP data collection was correlated with data previously obtained on glucose transport, glycolysis, and anaerobic metabolism. The details on the antibodies, the methodology of immunostaining, and the assessment of the results have been previously published (Table 1)10. The membrane expression of the glucose transporters GLUT1 and GLUT2 were studied to assess glucose absorption. For monocarboxylate (e.g., lactate) transport, the expression of ΜCT1 and MCT2 was studied. For glycolysis, we examined the expression of hexokinase II (HXKII, known to play an important role in cancer cell metabolism, tumor initiation,and is a putative target for cancer therapy11,12),phosphofructokinase 1 (PFK1) and fructose bisphosphate aldolase (aldolase; defects in aldolase lead to glycogen storage disease type 12 or red cell aldolase deficiency). For anaerobic pyruvate metabolism, we examined the expression of the lactate dehydrogenase isozyme, LDH5 (composed by 4 M-subunits, tetramer encoded entirely by the LDHA gene). For aerobic usage of pyruvate through oxidative phosphorylation, we studied the expression of pyruvate dehydrogenase (PDH) and its phosphorylated inactive form pPDH, as well as the expression of PDH-kinase, PDK1, that phosphorylates/inactivates PDH.
Cell lines
A549 (human alveolar basal epithelial adenocarcinoma cells)and NCI-H1299 (human NSCLC cells) were purchased from ATCC (ATCC, USA), (ATCC® CCL-185TMand ATCC®CRL-5803TM, respectively).
Cells were cultured under aseptic conditions using DMEM basal medium (31885-023, Gibco, Thermo Fisher Scientific,USA) supplemented with 10% fetal bovine serum (FB-1000/500, Biosera, France), 100 units/ml penicillin and 100 μg/mL streptomycin (15140-122, Gibco, Thermo Fisher Scientific, USA), and 2 mM L-glutamine (25030, Gibco,Thermo Fisher Scientific, USA). Cells were maintained under standard conditions (37°C, 5% CO2) in a humidified atmosphere and were used after reaching 70%–90%confluency.
MitoTracker staining
The integrity and functionality of mitochondria can be assessed with confocal microscopy using specific staining compounds that diffuse passively across mitochondrial membranes and accumulate in active mitochondria.MitoTracker is the most common dye of this group.MitoTracker® Deep Red FM (Invitrogen, Thermo Fisher Scientific, USA) staining was performed as per the manufacturer’s instructions, MitoTracker (250 nM) for 30 min. MitoTracker enters mitochondria through passive diffusion (independent of mitochondria membrane potential) and links to thiol groups becoming permanently bound to the mitochondria. Thus, it remains after the cell dies or is fixed. The fluorescence of MitoTracker® Deep Red FM can be monitored by confocal microscopy at 644 nm excitation wavelength and 665 nm emission wavelength.
Confocal immunofluorescence and image analysis
For immunofluorescence staining, cells were grown on No.1.5 glass coverslips, treated with MitoTracker dye if required,fixed in 3.7% paraformaldehyde/phosphate-buffered saline(PBS; pH 7.4) for 20 min at 37°C, and then permeabilized in PBS/0.1% v/v Triton X-100 (pH 7.4) for 5 min at room temperature. In addition, cells were blocked in PBS/5% w/v BSA (pH 7.4) for 20 min and stained with various primary antibodies (i.e., anti-UCP1 rabbit polyclonal antibody (1:200;Abcam, UK), anti-UCP3 rabbit polyclonal antibody (1:200;Abcam, UK)) for 1 h at room temperature. Cells were then washed in PBS (pH 7.4), incubated with an appropriate CF 488 secondary antibody at RT, and counterstained with Hoechst 33342 (1 μg/mL; Sigma-Aldrich, USA). After the final washes, coverslips were mounted in homemade Mowiol mounting medium. Imaging was performed on a customized Andor Revolution Spinning Disk Confocal System built around a stand (IX81; Olympus) with a 60× lens and a digital camera (Andor Ixon+885) (CIBIT Facility, MBG-DUTH,GREECE). Image acquisition was performed in Andor IQ 2 software. Optical sections were recorded every 0.3 μm. All confocal microscopy images presented in this work are 2D maximum intensity projections of z-stack images, and image analysis for the obtained data sets has been performed using ImageJ 1.47v (National Institute of Health, USA).Colocalization images and calculation of the Pearson’s coefficient for the analyzed images were performed using a combination of the Colocalization Finder and the Coloc 2 plugins in ImageJ 1.47v (National Institute of Health, USA).
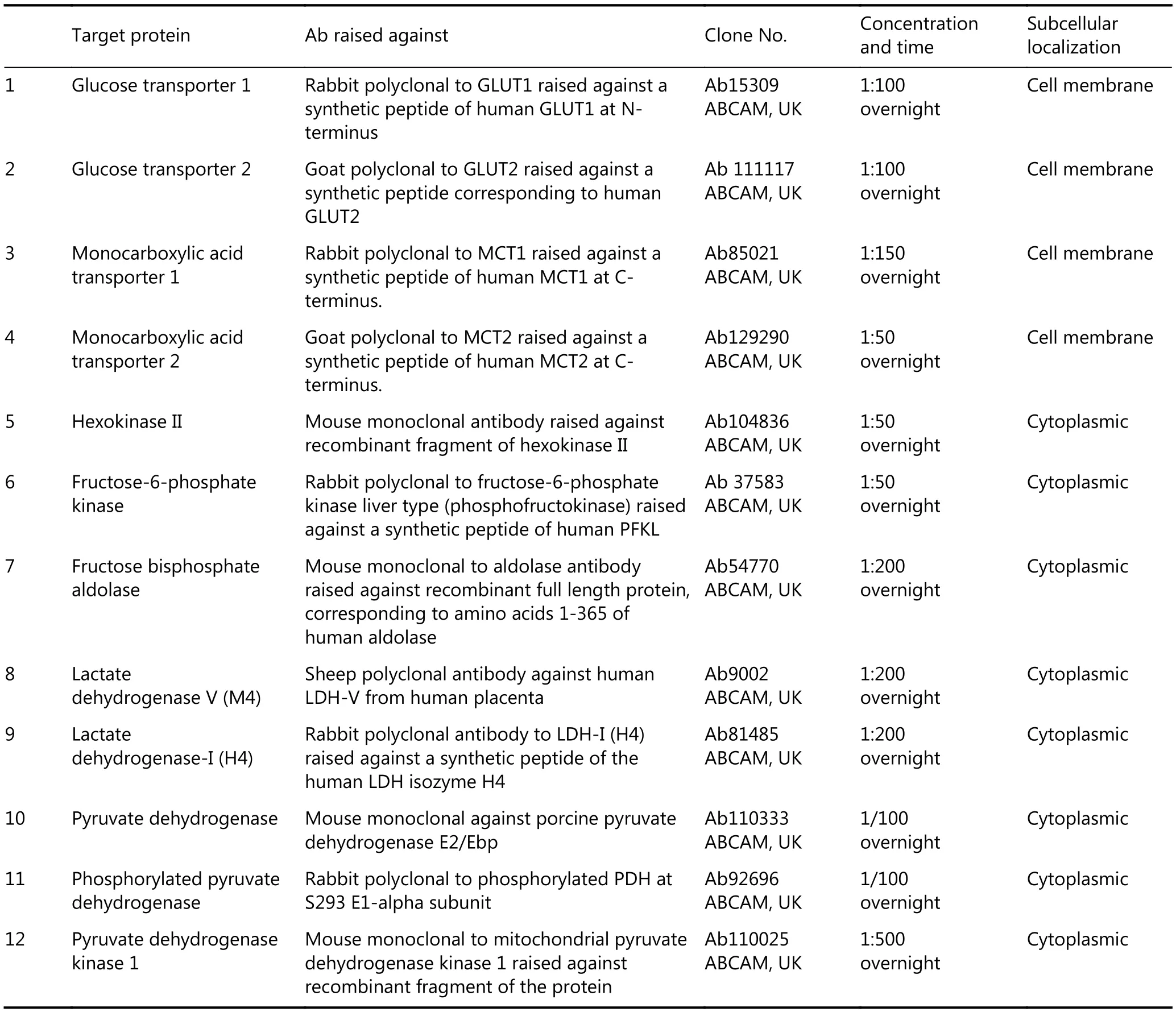
Table 1 Characteristics of the metabolism-related antibodies used for immunohistochemistry.
Statistical analysis
Statistical analysis was performed by GraphPad Prism 5.0 and SPSS (v14.0, SPSS Inc.) statistical packages. The Fisher’s exact t-test or the unpaired two-tailed t-test was used to compare categorical variables, as appropriate. Linear regression analysis was used to test correlations between continuous variables. The Kaplan-Meier survival curves were used to assess the impact of various variables on the overall survival of patients. For survival analysis, disease-specific death events were used. A Cox proportional hazard model was used to assess the independent effect of variables on death events. A P-value of < 0.05 was used for significance.
Results
UCP subcellular localization
Using confocal microscopy and double immunostaining with the antibodies recognizing UCPs with MitoTracker, we found a Pearson’s correlation coefficient of 0.61 and 0.72 for the colocalization of UCP1 in the mitochondria of A549 and H1299 NSCLC cell lines, respectively. The coefficients for UCP3 were 0.60 and 0.53 for the A549 and H1299 NSCLC cell lines, respectively. This confirms that the antibodies do indeed identify mitochondrial proteins (Figure 1). The MitoTracker colocalization coefficient with UCPs was > 0.90.However, both UCPs showed also a diffuse localization in the cytoplasm and, particularly in the H1299 cell line, in the nuclei of cells within non-nucleolar areas.
UCP expression in normal lung and cancer tissues
Figure 2 shows the typical immunohistochemical images of normal lung structures. Bronchial epithelium did not express UCP1 or UCP3. Alveolar cells strongly expressed UCP1, but were unreactive to the UCP3 antibody. On the contrary,mucinous glands expressed UCP3 but not UCP1. The normal lung endothelium exclusively expressed UCP1. Cartilage expressed only UCP1. Alveolar lymphocytes did not express any of the UCPs.
In cancer cells, both UCPs showed a cytoplasmic granular localization in a varying percentage of cells (Figure 2). For UCP1, the absence of expression (i.e., normal bronchial epithelium pattern) was noted in 37/98 (37.8%), weak expression in a varying percentage of cells (10%–100%) in 18/98 (18.4%), and strong expression in 43/98 (43.8%) of cases. The percentage of cells with strong expression ranged from 10%–90% (with a median of 30%).
For UCP2, lack of expression was noted in 41/98 (41.8%),weak expression in 30/98 (30.6%), and strong expression in 27/98 (27.6%) of cases. The percentage of cells with strong expression ranged from 10%–100% (with a median of 50%).
A significant, but weak, association between UCP1 and UCP3 expression was noted, with r = 0.20 following linear regression analysis (P = 0.04). Absent/weak expression of both UCPs was noted in 44/98 (44.9%) of cases. Strong coexpression of the UCP proteins was noted in 16/98 (16.3%)of cases. In these cases, a strong association between UCP1 and UCP3 was noted, with r = 0.90 following linear regression analysis (P < 0.0001) (Figure 3A, B).
UCPs relation with patient and histopathological variables
Table 2 shows the association between UCPs and patient/disease characteristics. UCP1 was significantly associated with squamous cell histology (P = 0.05), male gender (P = 0.008), and younger age (P = 0.01). UCP3 was more frequently overexpressed in large cell carcinomas (P =0.08), and inversely related to necrosis (P = 0.009).
UCPs relation to glucose metabolism
Following the linear regression analysis of the percentage of cells with strong UCP expression with enzymes/proteins involved in glucose metabolism, UCP1 was directly related to several markers of glycolysis such as HXKII (P=0.02, r=0.22),PFK1 (P < 0.0001, r = 0.42). UCP was also directly related to those proteins involved in anaerobic glucose metabolism such as PDK1 (P=0.04; r=0.20) and LDH5 (P=0.02, r=0.22).UCP3 was directly linked to glucose transporter, GLUT2(P=0.002, r=0.28), and to monocarboxylate transporter,MCT2 (P=0.02, r = 0.14) overexpression. UCP3 was also related to glycolytic markers, such as PFK1 (P=0.01, r=0.24)and aldolase (P=0.001, r=0.31), and with markers of Kreb’s cycle suppression, like phosphorylated PDH (P=0.01, r=0.25)and PDK1 (P < 0.0001, r=0.42). Supplemental Figure 2S shows the linear regression analysis plots.
Survival analysis
Kaplan-Meier survival analysis of UCP3 expression showed that UCP3 had a trend to define poor prognosis, which reached statistical significance in squamous cell carcinomas(P = 0.04; Figure 3C, D). No association between UCP1 and prognosis was noted. In multivariate analysis including histopathological features only advanced stage and poor differentiation were independent prognostic variables (P =0.0001 and P = 0.03, respectively).
Discussion
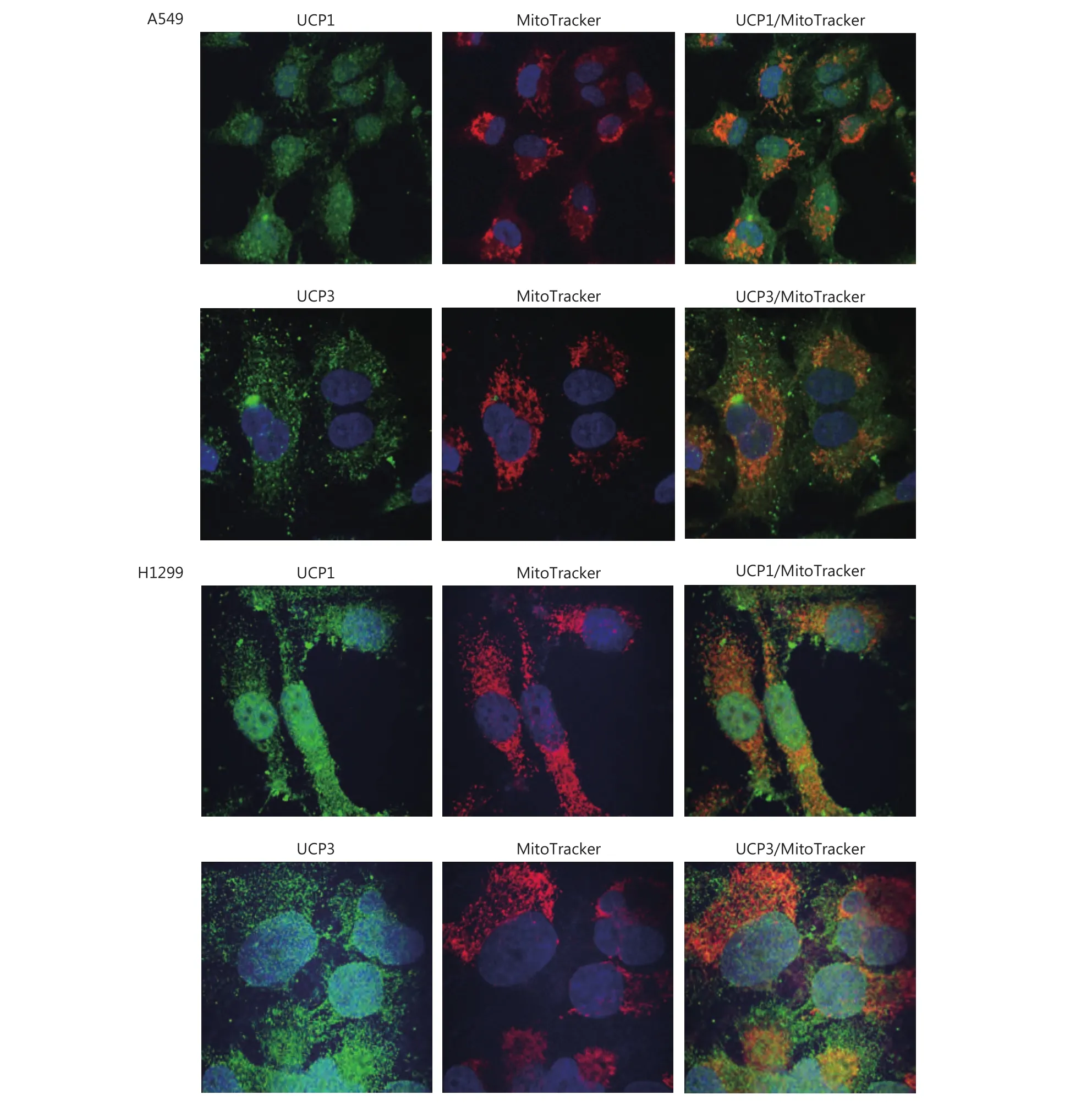
Figure 1 Confocal microscopy of A549 and H1299 lung cancer cell lines after double staining with MitoTracker and with UCP1 or UCP3(green: UCP1 and UCP3, red: MitoTracker, and orange: colocalization). The colocalization Pearson’s correlation coefficients for A549 were UCP1/MitoTracker 0.61, UCP3/MitoTracker 0.60 and MitoTracker/UCPs >0.90; for H1299 these were UCP1/MitoTracker 0.72,UCP3/MitoTracker 0.53 and MitoTracker/UCPs > 0.90).
Uncoupling proteins have been recently under intense investigation in multiple medical fields, including obesity,diabetes, immunology, aging, and cancer. The leak of protons across the mitochondrial inner membranes, which is mediated by UCP1, bypasses ATP synthase and allows for fast oxygen consumption and heat production. Aside from their thermogenic activity, UCPs also have important functions in controlling ROS generation by mitochondria, as studies show that activation of UCP2 and UCP3 attenuate ROS production and enhance fatty acid oxidation13,14.
The role of UCPs in carcinogenesis and cancer growth remains unclear. In 2005, Collins et al.15reported that UCP2 is overexpressed in the human hepatoma cell line HepG2 and that this protected cells against oxidative stress. Similarly,Horimoto et al.16reported a significant up-regulation of UCP2 in colon adenocarcinoma as compared to pre-neoplastic lesions. More recently, Kuai et al.17confirmed the frequent overexpression of UCP2 in colon cancer and suggested that this feature may be related to increased metastatic ability. UCP2 has also been reported to be overexpressed in breast cancer18,19and osteosarcomas20.
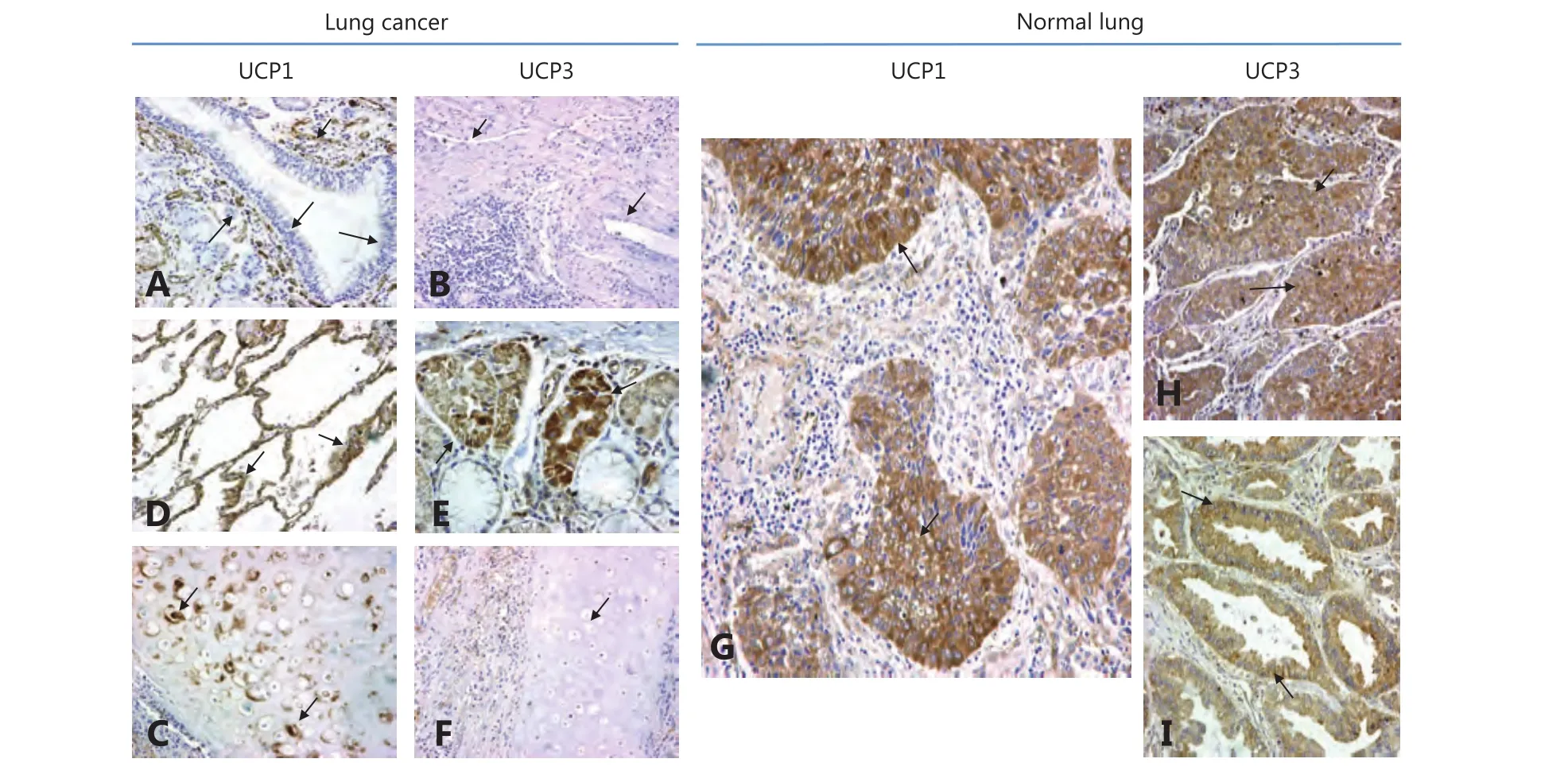
Figure 2 Immunohistochemical images of normal lung and lung cancer stained for UCP1 and UCP3 (IHC staining, 40×). (A) Normal bronchial epithelium is unreactive to the UCP1 antibody, while normal peribronchial vessels strongly express the protein. (B) Alveolar cells were strongly stained for UCP1. (C) Bronchial cartilage with intense staining of chondrocytes for UCP1. (D) Normal lung vessels were unreactive to the UCP3 antibody. (E) Peribronchial mucous glands express UCP3. (F) Bronchial cartilage does not express UCP3. (G) Strong cytoplasmic expression of UCP1 in squamous cell cancer. (H) Overexpression of UCP3 in a squamous cell carcinoma. (I) Overexpression of UCP3 in a lung adenocarcinoma.
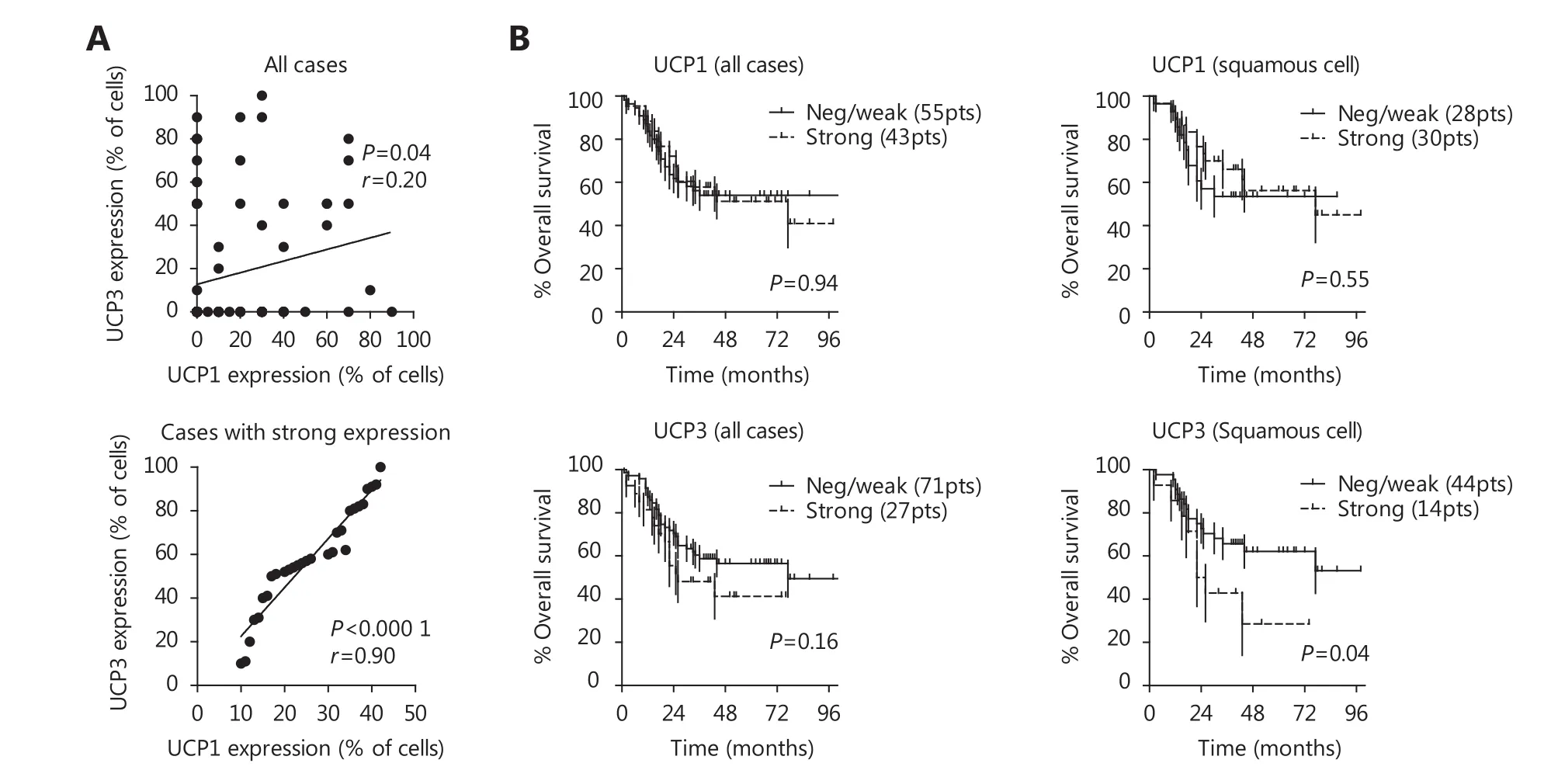
Figure 3 (A) Linear regression analysis of the percentage of cells with UCP1 vs. UCP3 expression, in all cases and in cases with strong expression. (B) Kaplan-Meier survival curves of patients stratified for UCP1 and UCP3 expression, in all cases and in cases with squamous cell carcinoma.
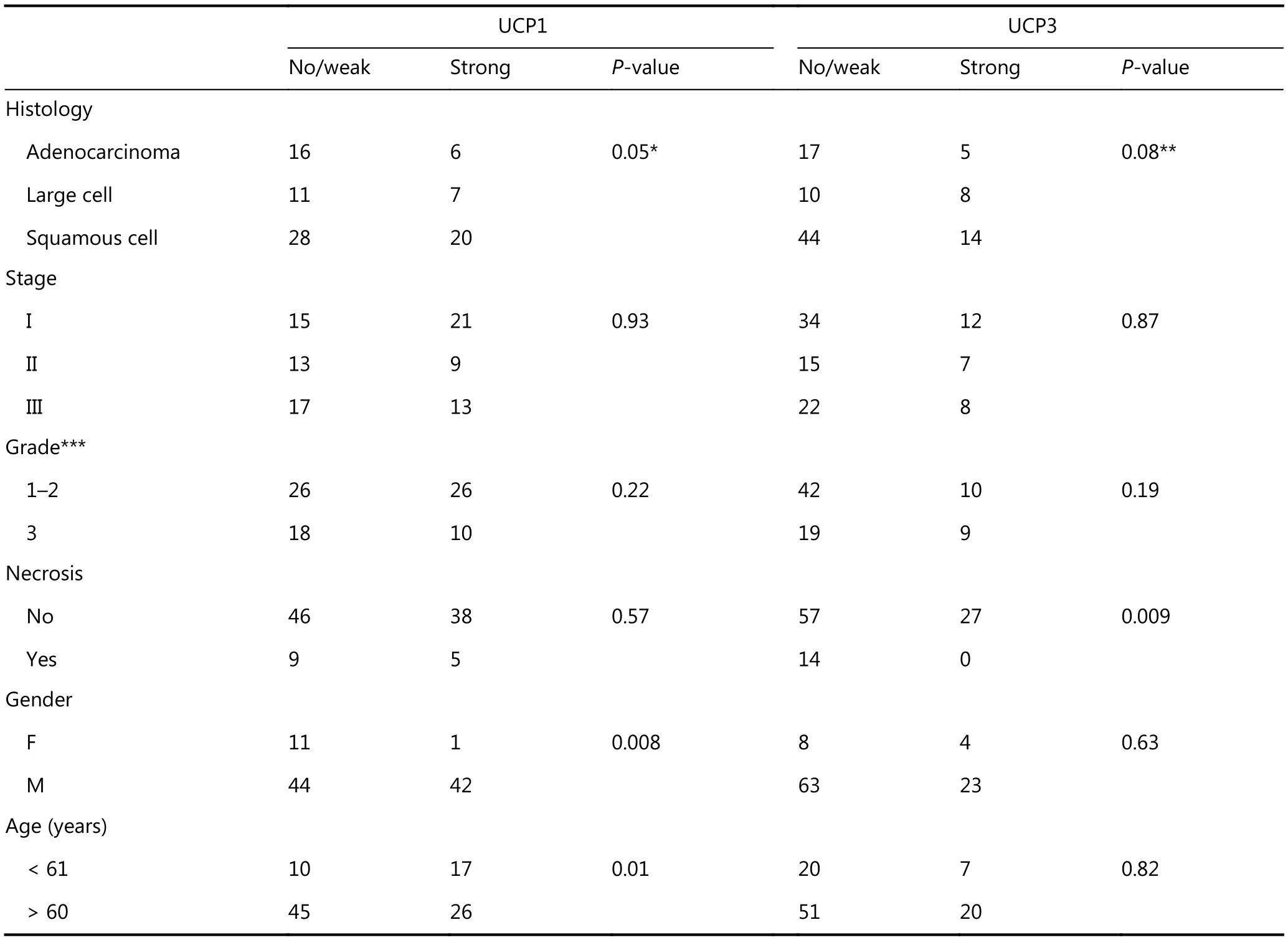
Table 2 Association of UCPs with patient and disease characteristics
The expression of UCP1 and UCP3 in human carcinomas and their biological and clinical role is unknown. In the current study, we investigated UCP1 and UCP3 expression in a series of NSCLC cases treated with surgery. In contrast to the bronchial epithelium, where UCP1 and UCP3 were not expressed, strong expression of UCP1, in a varying percentage of cancer cells, was noted in 43.8% of cases, while UCP3 was less frequently expressed (i.e., in 27.6% of cases).UCP1 was significantly overexpressed in squamous cell cancers, while UCP3 was more frequently noted in large cell carcinomas. In survival analysis, UCP3 was the uncoupling protein that was related to the worse prognosis, but this was statistically confirmed only in the squamous cell histology,probably owing to the higher number of cases in this histology subgroup.
An interesting observation from the current analysis was that both UCPs were significantly overexpressed in cases with intensified glycolytic activity, as assessed by the expression of HXKII, PFK1, and aldolase, as well as the anaerobic usage of pyruvate, as assessed by the overexpression of LDH5 and PDK1. UCP3 was also linked to the overexpression of glucose and monocarboxylate transporters. This is in accordance with a previous study where UCP2 expression was related to hypoxia markers in NSCLC21. This direct association of UCPs with anaerobic glycolysis may explain the inverse association found between UCP3 expression and necrosis,since intensification of glycolytic pathways, under poor blood flow and oxygen availability conditions, certainly offers a survival advantage to cancer cells.
The association of UCPs with anaerobic metabolism is not surprising, as several in vitro studies have reported a link between UCPs and the Warburg effect. Although the mechanism for this is obscure, it has been suggested that UCPs block the usage of pyruvate by the Kreb’s cycle by shifting mitochondrial function to thermogenesis and fatty acid or glutamate oxidation22. Brandi et al.23recently showed that UCP2 induces the expression of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2/B1), and regulates GLUT1 and LDH mRNA expression in pancreatic cancer cells, implying a direct involvement in anaerobic glucose usage. Interestingly, a UCP2-dependent decrease in mitochondria oxidative phosphorylation complexes was noted23.
In conclusion, UCP1 and UCP3 are overexpressed in a large subgroup of NSCLs and their expression coincides with an increase in glucose absorption, increased glycolytic rate,and anaerobic glucose usage. Whether UCPs are key elements in the induction of the Warburg effect in cancer, or whether interference with the function of these proteins has a therapeutic role in lung cancer requires further investigation.
Conflict of interest statement
No potential conflicts of interest are disclosed.
1.Rolfe DF, Newman JMB, Buckingham JA, Clark MG, Brand MD.Contribution of mitochondrial proton leak to respiration rate in working skeletal muscle and liver and to SMR. Am J Physiol. 1999;276: C692-9.
2.Brand MD, Chien LF, Ainscow EK, Rolfe DF, Porter RK. The causes and functions of mitochondrial proton leak. Biochim Biophys Acta. 1994; 1187: 132-9.
3.Azzu V, Brand MD. The on-off switches of the mitochondrial uncoupling proteins. Trends Biochem Sci. 2010; 35: 298-307.
4.Brand MD, Esteves TC. Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metab.2005; 2: 85-93.
5.Zhau HE, He H, Wang CY, Zayzafoon M, Morrissey C, Vessella RL,et al. Human prostate cancer harbors the stem cell properties of bone marrow mesenchymal stem cells. Clin Cancer Res. 2011; 17:2159-69.
6.Ayyasamy V, Owens KM, Desouki MM, Liang P, Bakin A,Thangaraj K, et al. Cellular model of Warburg effect identifies tumor promoting function of UCP2 in breast cancer and its suppression by genipin. PLoS One. 2011; 6: e24792
7.Nowinski SM, Solmonson A, Rundhaug JE, Rho O, Cho J, Lago CU, et al. Mitochondrial uncoupling links lipid catabolism to Akt inhibition and resistance to tumorigenesis. Nat Commun.2015; 6: 8137.
8.Collins P, Bing C, McCulloch P, Williams G. Muscle UCP-3 mRNA levels are elevated in weight loss associated with gastrointestinal adenocarcinoma in humans. Br J Cancer. 2002; 86: 372-5.
9.Braun N, Klumpp D, Hennenlotter J, Bedke J, Duranton C, Bleif M,et al. UCP-3 uncoupling protein confers hypoxia resistance to renal epithelial cells and is upregulated in renal cell carcinoma. Sci Rep.2015; 5: 13450
10.Giatromanolaki A, Sivridis E, Arelaki S, Koukourakis MI.Expression of enzymes related to glucose metabolism in non-small cell lung cancer and prognosis. Exp Lung Res. 2017; 43: 167-74.
11.Wu JY, Hu LR, Wu FP, Zou L, He TP. Poor prognosis of hexokinase 2 overexpression in solid tumors of digestive system: a meta-analysis. Oncotarget. 2017; 8: 32332-44.
12.Patra KC, Wang Q, Bhaskar PT, Miller L, Wang ZB, Wheaton W, et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer.Cancer Cell. 2013; 24: 213-28.
13.Bézaire V, Seifert EL, Harper ME. Uncoupling protein-3: clues in an ongoing mitochondrial mystery. FASEB J. 2007; 21: 312-24.
14.MacLellan JD, Gerrits MF, Gowing A, Smith PJS, Wheeler MB,Harper ME. Physiological increases in uncoupling protein 3 augment fatty acid oxidation and decrease reactive oxygen species production without uncoupling respiration in muscle cells.Diabetes. 2005; 54: 2343-50.
15.Collins P, Jones C, Choudhury S, Damelin L, Hodgson H.Increased expression of uncoupling protein 2 in HepG2 cells attenuates oxidative damage and apoptosis. Liver Int. 2005;25: 880-7.
16.Horimoto M, Resnick MB, Konkin TA, Routhier J, Wands JR, Baffy G. Expression of uncoupling protein-2 in human colon cancer. Clin Cancer Res. 2004; 10: 6203-7.
17.Kuai XY, Ji ZY, Zhang HJ. Mitochondrial uncoupling protein 2 expression in colon cancer and its clinical significance. World J Gastroenterol. 2010; 16: 5773-8.
18.Won KY, Kim GY, Kim YW, Lim SJ, Song JY. Uncoupling Protein 2 (UCP2) and p53 expression in invasive ductal carcinoma of breast. Korean J Pathol. 2010; 44: 565-70.
19.Sayeed A, Meng Z, Luciani G, Chen LC, Bennington JL, Dairkee SH. Negative regulation of UCP2 by TGFβ signaling characterizes low and intermediate-grade primary breast cancer. Cell Death Dis.2010; 1: e53.
20.Srivastava A, Rock C, Zhang KB, Ruan M, Bolander ME, Sarkar G.Expression of a novel alternatively spliced UCP-2 transcript in osteogenic sarcoma. J Orthop Sci. 2006; 11: 51-7.
21.Oleksiewicz U, Liloglou T, Tasopoulou KM, Daskoulidou N,Gosney JR, Field JK, et al. COL1A1, PRPF40A, and UCP2 correlate with hypoxia markers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2017; 143: 1133-41.
22.Samudio I, Fiegl M, Andreeff M. Mitochondrial uncoupling and the Warburg effect: molecular basis for the reprogramming of cancer cell metabolism. Cancer Res. 2009; 69: 2163-6.
23.Brandi J, Cecconi D, Cordani M, Torrens-Mas M, Pacchiana R,Dalla Pozza E, et al. The antioxidant uncoupling protein 2 stimulates hnRNPA2/B1, GLUT1 and PKM2 expression and sensitizes pancreas cancer cells to glycolysis inhibition. Free Radic Biol Med. 2016; 101: 305-16.
Cite this article as: Giatromanolaki A, Balaska K, Kalamida D, Kakouratos C,Sivridis E, Koukourakis MI, et al. Thermogenic protein UCP1 and UCP3 expression in non-small cell lung cancer: relation with glycolysis and anaerobic metabolism. Cancer Biol Med. 2017; 14: 396-404. doi:10.20892/j.issn.2095-3941.2017.0089
Michael I. Koukourakis
E-mail: targ@her.forthnet.gr
July 18, 2017; accepted November 13, 2017.
Available at www.cancerbiomed.org
Copyright © 2017 by Cancer Biology & Medicine
 Cancer Biology & Medicine2017年4期
Cancer Biology & Medicine2017年4期
- Cancer Biology & Medicine的其它文章
- Brain metastasis in non-small cell lung cancer (NSCLC)patients with uncommon EGFR mutations: a report of seven cases and literature review
- Complete pathologic response after chemoradiotherapy in a patient with rectal squamous cell carcinoma: a case report
- Increased expression of YAP1 in prostate cancer correlates with extraprostatic extension
- Profile of the breast cancer susceptibility marker rs4245739 identifies a role for miRNAs
- Promoter methylation of Wnt/β-Catenin signal inhibitor TMEM88 is associated with unfavorable prognosis of nonsmall cell lung cancer
- Brain metastasis in advanced colorectal cancer: results from the South Australian metastatic colorectal cancer (SAmCRC)registry
