石油烃厌氧降解产甲烷研究进展
胡恒宇,韦安培,刘少梅,李 静,赵东风
(1.临沂大学资源环境学院,山东 临沂 276000;2.中国石油大学(华东)化学工程学院,山东 青岛 266580)
石油烃厌氧降解产甲烷研究进展
胡恒宇1,韦安培1,刘少梅1,李 静1,赵东风2
(1.临沂大学资源环境学院,山东 临沂 276000;2.中国石油大学(华东)化学工程学院,山东 青岛 266580)
随着油藏的开采大量残余原油留存地下,通过微生物作用使残余原油(石油烃)降解产甲烷已成为油藏深度开发利用的新方法。油藏微生物由诸多菌群组成,这些菌群协同作用实现石油烃的厌氧降解。为进一步提高石油烃的降解效率,总结了降解石油烃的厌氧微生物菌群及其代谢特性,并对微生物厌氧降解石油烃产甲烷的代谢途径进行了比较。
残余原油;石油烃;厌氧降解;甲烷
数百万年来油藏内部进行着原油厌氧降解产甲烷过程,而随着油藏的开采大量残余原油留存地下,通过微生物作用使残余原油(石油烃)降解产甲烷已成为油藏深度开发利用的新方法。油藏微生物由诸多菌群组成,这些菌群协同作用实现石油烃的降解,石油烃降解过程中涉及到的反应步骤多、反应速率影响因素多,代谢方式也是多种多样,而残余原油中可以供微生物利用的营养物质和厌氧产生的毒害物质的数量是影响降解效率的关键因素[1]。目前,研究者多通过外加碳源、氮源、磷源、微量元素、维生素、络合剂等来激活微生物,从而提高石油烃的降解效率。石油烃降解的终端产物是甲烷,而电子受体(硝酸盐、四价锰、三价铁、硫酸盐等)的耗尽是甲烷产生的基础。产甲烷古菌会和其它菌群形成互营共生关系,最终使石油烃降解并接受末端电子产生甲烷[2]。为进一步提高石油烃的降解效率,作者总结了降解石油烃的厌氧微生物菌群及其代谢特性,并对微生物厌氧降解石油烃产甲烷的代谢途径进行了比较。
1 降解石油烃的厌氧微生物菌群
高温、高压、高地层水矿化度的厌氧环境使得微生物的生存环境异常苛刻,就在人们认为在这种环境下微生物很难生长时,不同功能的厌氧微生物菌群相继被发现和分离[3]。80年代后期,Vogel等证明了石油烃的厌氧降解[4];Lovley等[5]分离得到了高效降解甲苯的GS-15菌株,是第一株以三价铁为电子受体的菌株;2年后,Aeckersberg等[6]首次分离到以硫酸盐为电子受体的还原菌Hxd3,它们能够利用长链饱和烃;随后,多种厌氧降解电子受体(硝酸盐、硫酸盐、三价铁、二氧化碳、锰离子等)被大量发现[7-12]。近年来,人们越来越认识到石油烃降解产甲烷过程是多种菌群协同作用的结果,涉及到的菌群有发酵菌、硫酸盐/硝酸盐还原菌、厌氧产甲烷菌等[13],降解过程主要涉及4种反应:第一,在各种还原菌群的共同作用下石油烃失去电子发生氧化反应产甲烷;第二,乙酸失去电子发生氧化反应产甲烷;第三,乙酸分解产甲烷;第四,二氧化碳接受电子还原产甲烷。其中,二氧化碳接受电子还原是产甲烷的主要反应[14],长链烷烃通过复合菌群的共同作用产生乙酸、二氧化碳、氢气,最终在产甲烷古菌和其它硫酸盐/硝酸盐还原菌的协同作用下产生甲烷和其它气体[15-16]。
硫酸盐还原菌是一个复杂的生物菌群,存在于大部分油藏中。硫酸盐还原菌以硫酸盐作为电子受体进行无氧呼吸[17]。根据rRNA序列分析,可将其分为4类:革兰氏阴性嗜温菌、革兰氏阳性芽孢菌、嗜热细菌、嗜热古细菌。有学者研究革兰氏阴性嗜温菌的演化历史时发现,硫酸盐还原菌的共同祖先是一些光养生物,在漫长的演变过程中,这些光养生物逐渐失去光合能力演变成异氧细菌[18]。研究表明,Desulfovibrionaceae 和Desulfobacteriaceae 已经出现很大分化,Desulfobacteriaceae是原始的细菌,包括1个δ-Proteobacteria,而Desulfovibrionaceae却没有δ-Proteobacteria[19-20]。部分厌氧降解石油烃的硫酸盐还原菌[21]列于表1。
产甲烷古菌因天然气方面的巨大作用而受到研究者的广泛关注,加上近年来石油烃厌氧降解产甲烷成为研究热点,人们开始通过研究产甲烷古菌来揭示石油烃厌氧降解产甲烷过程。研究发现,产甲烷古菌和其它菌群通过协同作用经厌氧发酵将长链有机物降解成短链的无机物或有机物,然后再降解产生甲烷[41-42],这期间伴随着其它降解产物的生成,如氢气和二氧化碳;产甲烷古菌处在厌氧降解产甲烷的最后环节[43-44]。
表1 部分厌氧降解石油烃的硫酸盐还原菌

Tab.1 Some sulfate-reducing bacteria capable of anaerobic degradation of petroleum hydrocarbon
产甲烷古菌广泛存在于土壤、底泥、地热环境、油井、海底沉积物中[45],有3种类型:(1)氢营养型:伊万诺夫甲烷杆菌[46]、热自养甲烷杆菌[47]、布氏甲烷杆菌和嗜热嗜碱甲烷杆菌[48]、热自养甲烷球菌[49]、石油甲烷盘菌属[50]、耐盐甲烷卵圆形菌[51];(2)甲基营养型:盐水甲烷嗜盐菌[52-53]、斯氏甲烷八叠球菌[54-55];(3)乙酸营养型:马氏八叠球菌[56]。从产甲烷古菌的系统分类和降解产物可以判断其具体生活方式和属性情况[22],产甲烷古菌的系统分类和降解产物如表2所示。
表2 产甲烷古菌系统分类及其降解产物

Tab.2 System classification and degradation product of Methanogenus
研究表明,当缺乏硫酸盐和硝酸盐电子受体时,烷烃仍然可以通过富集培养的菌群厌氧降解生成二氧化碳和甲烷。Foght[57]认为芳烃厌氧降解产甲烷是以氯酸盐、高氯酸盐或锰离子为电子受体的,如图1所示。
从图1可以看出,芳烃厌氧降解产甲烷过程基本上涉及到了石油烃厌氧降解产甲烷的主要过程,如延胡索酸加成反应、羟基化反应、甲基化反应、羧基化反应等[57]。
2 微生物厌氧降解石油烃产甲烷的代谢途径
2.1 延胡索酸加成反应
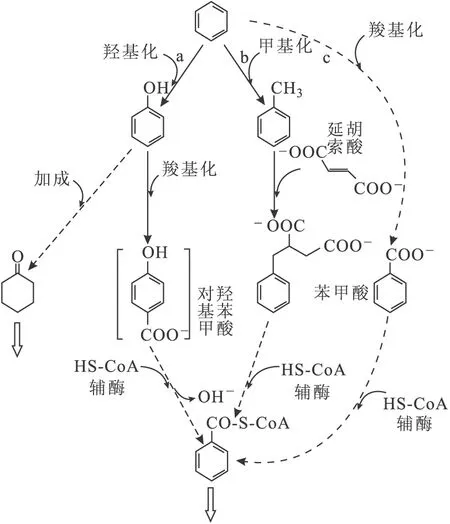
图1 芳烃厌氧降解产甲烷过程
Kniemeyer等[34]提出烃降解的一个主要途径是生成芳香基或烷基琥珀酸盐,是通过延胡索酸盐上的双键碳被烃上的碳原子加成来实现的。例如,在己烷降解过程中,延胡索酸盐双键碳原子上被己烷的第二个碳原子结合,生成了(1-甲基戊基)琥珀酸盐(MPS);通过氢同位素标记方法证实了延胡索酸加成反应,并检验到了琥珀酸盐的生成。在间二甲苯的降解过程中,延胡索酸和苯环上的甲基发生加成反应,生成苯甲基琥珀酸这一短暂性的中间产物,然后脱氢产生中间产物苯甲酰辅酶A(图2)。在多环芳烃的降解过程中,由于多环芳烃2-甲基萘的甲基C-H解离能约为359.2 kJ·mol-1,2-甲基萘的甲基加到延胡索酸的双键碳上产生琥珀酸,经过β-氧化产生中间代谢产物2-萘酸,再经过一系列苯环还原反应,最终产生二氧化碳[8,22,58]。
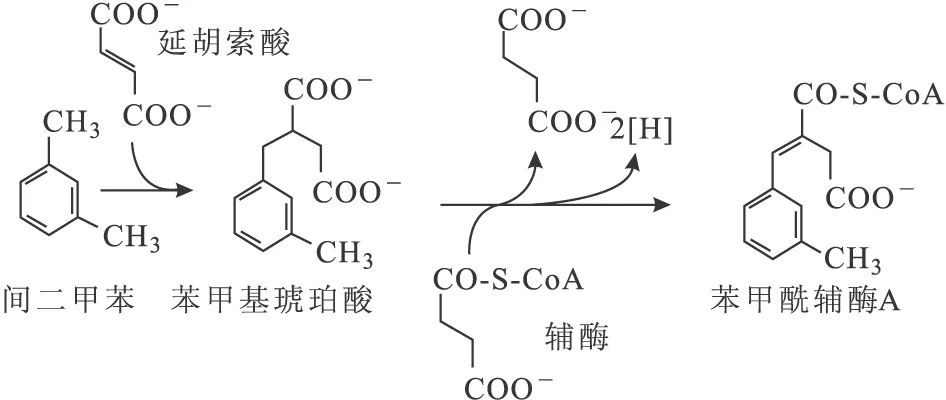
图2 延胡索酸加成反应代谢途径
2.2 羟基化反应
芳烃的另一种主要代谢途径为羟基化反应。有学者在研究乙苯降解时提出,羟基化反应发生在乙苯的亚甲基上,生成1-苯基乙醇,是由于羟基结合于芳烃的烷基链或苯环上。还有学者认为苯氧化为苯酚就是在脱氢酶的作用下,羟基进攻苯环发生羟基化反应的过程[59-62]。
2.3 甲基化反应和羧基化反应
有学者在研究萘的代谢反应时提出了甲基化反应代谢途径,在添加碳酸盐激活剂的情况下,实现了萘的甲基化作用生成2-甲基萘,而这一作用途径与一氧化碳脱氢酶催化的反应途径不同,从而加快了萘的代谢[63]。还有学者提出了可能的羧基化代谢途径(图3)[64]。
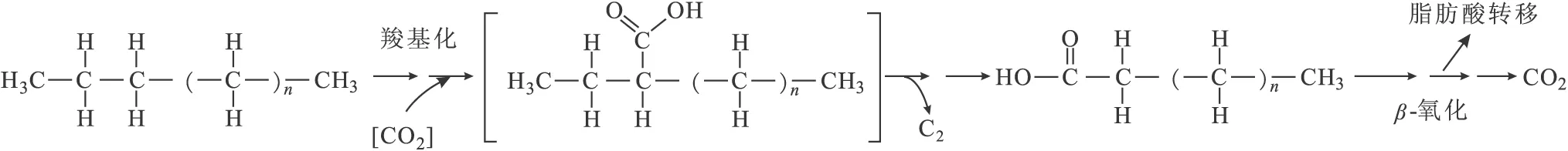
图3 羧基化代谢途径
3 展望
石油烃厌氧降解产甲烷过程涉及到的反应步骤很多,影响反应速率的因素也非常多,代谢途径也多种多样,为提高降解效率,研究者通过加入碳源、氮源、磷源、微量元素、维生素、络合剂等来激活微生物,这已是目前的研究热点。今后,还应在如下方面进行深入研究,探讨石油烃厌氧降解产甲烷过程,深度开发利用油藏。(1)通过油藏物性的分析、地层条件的分析、地层水分析,探讨环境条件如压力、温度、pH值和矿化度对微生物的代谢和生长的影响。(2)特定油藏条件下,厌氧降解产甲烷混合菌的筛选,建立生物反应器模拟地层环境,以石油烃为唯一碳源,通过富集培养石油烃降解产甲烷微生物,采用新技术确定微生物混合菌的组成及特征并且构建克隆文库。(3)通过研究营养激活剂(如碳源、氮源、磷源、微量元素)来提高石油烃厌氧降解产甲烷的效率,尤其是表面活性剂、络合剂之间的协同作用。(4)利用气相色谱、液相色谱分析石油烃的组分,考察石油烃的利用情况,利用GC-MS、NMR、IR分析石油烃厌氧降解产甲烷过程中的代谢中间产物的变化,同时用高级烷烃同位素示踪的方法确定石油烃厌氧降解机理。(5)考察石油烃在降解反应前后的性质变化,尤其是运动黏性、密度及界面张力,提出能解决实际问题的工艺途径。
[1] 汪卫东,王静,耿雪丽,等.储层残余油生物气化技术现状与展望[J].石油地质与工程,2012,26(1):78-81.
[2] 王俊,俞理,黄立信.油藏生物气研究进展[J].特种油气藏,2010,17(5):8-12.
[3] ROLING W F,HEAD I M,LARTER S R.The microbiology of hydrocarbon degradation in subsurface petroleum reservoirs:perspectives and prospects[J].Research in Microbiology,2003,154(5):321-328.
[4] CHAKRABORTY R,COATES J D.Anaerobic degradation of monoaromatic hydrocarbons[J].Applied Microbiology and Biotechnology,2004,64(4):437-446.
[5] LOVLEY D R,BAEDECKER M J,LONERGAN D J,et al.Oxidation of aromatic contaminants coupled to microbial iron reduction[J].Nature,1989,339(6222):297-300.
[6] AECKERSBERG F,BAK F,WIDDEL F.Anaerobic oxidation of saturated hydrocarbons to CO2by a new type of sulfate-reducing bacterium[J].Archives of Microbiology,1991,156(1):5-14.
[7] HEIDER J,SPORMANN A M,BELLER H R,et al.Anaerobic bacterial metabolism of hydrocarbons[J].FEMS Microbiology Reviews,1998,22(5):459-473.
[8] SPORMANN A M,WIDDEL F.Metabolism of alkylbenzenes,alkanes,and other hydrocarbons in anaerobic bacteria[J].Biodegradation,2000,11(2):85-105.
[9] WIDDEL F,BOETIUS A,RABUS R.Anaerobic Biodegradation of Hydrocarbons Including Methane//The Prokaryotes[M].New York:Springer,2006:1028-1049.
[10] WIDDEL F,RABUS R.Anaerobic biodegradation of saturated and aromatic hydrocarbons[J].Current Opinion in Biotechnology,2001,12(3):259-276.
[11] GROSSI V,CRAVO-LAUREAU C,GUYONEAUD R,et al.Metabolism ofn-alkanes andn-alkenes by anaerobic bacteria:a summary[J].Organic Geochemistry,2008,39(8):1197-1203.
[12] MBADINGA S M,WANG L Y,ZHOU L,et al.Microbial communities involved in anaerobic degradation of alkanes[J].International Biodeterioration and Biodegradation,2011,65(1):1-13.
[13] GIEG L M,DUNCAN K E,SUFLITA J M.Bioenergy productionviamicrobial conversion of residual oil to natural gas[J].Applied and Environmental Microbiology,2008,74(10):3022-3029.
[14] JONES D M,HEAD I M,GRAY N D,et al.Crude-oil biodegradationviamethanogenesis in subsurface petroleum reservoirs[J].Nature,2008,451(7175):176-180.
[15] PARKES J.Cracking anaerobic bacteria[J].Nature,1999,401(6750):217-218.
[16] ZENGLER K,RIELMOW H H,ROSSELLO-MORA R,et al.Methane formation from long-chain alkanes by anaerobic microorganisms[J].Nature,1999,401(6750):266-269.
[17] CASTRO H F,WILLIAMS N H,OGRAM A.Phylogeny of sulfate-reducing bacteria(1)[J].FEMS Microbiology Ecology,2000,31(1):1-9.
[18] WOESE C R.Bacterial evolution backgroud[J].Microbiology,1987,51(2):221-271.
[19] DEVEREUX R,HE S H,DOYLE C L,et al.Diversity and origin ofDesulfovibriospecies:phylogenetic definition of a family[J].Journal Bacteriology,1990,172(7):3609-3619.
[20] WIDDEL F,BAK F.Gram-Negative Mesophilic Sulfate-Reducing Bacteria//The Prokaryotes[M].New York:Springer,1992:3352-3378. [21] 胡恒宇,顾贵洲,张强,等.残余油微生物气化产甲烷菌群的研究进展[J].化学与生物工程,2014,31(4):9-14.
[22] 周蕾.厌氧烃降解产甲烷菌系的组成及其代谢产物的特征[D].上海:华东理工大学,2012.
[23] RABUS R,NORDHAUS R,LUDWIG W,et al.Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium[J].Applied and Environmental Microbiology,1993,59(5):1444-1451.
[24] BELLER H R,SPORMANN A M,SHARMA P K,et al.Isolation and characterization of a novel toluene-degrading,sulfate-reducing bacterium[J].Applied and Environmental Microbiology,1996,62(4):1188-1196.
[25] HARMS G,ZENGLER K,RABUS R,et al.Anaerobic oxidation ofo-xylene,m-xylene,and homologous alkylbenzenes by new types of sulfate-reducing bacteria[J].Applied and Environmental Microbiology,1999,65(3):999-1004.
[26] KNIEMEYER O,FISCHER T,WILKES H,et al.Anaerobic degradation of ethylbenzene by a new type of marine sulfate-reducing bacterium[J].Applied and Environmental Microbiology,2003,69(2):760-768.
[27] MORASCH B,SCHINK B,TEBBE C C,et al.Degradation ofo-xylene andm-xylene by a novel sulfate-reducer belonging to the genusDesulfotomaculum[J].Archives of Microbiology,2004,181(6):407-417.
[28] OMMEDAL H,TORSVIK T.Desulfotignumtoluenicumsp. nov.,a novel toluene-degrading,sulphate-reducing bacterium isolated from an oil-reservoir model column[J].International Journal of Systematic and Evolutionary Microbiology,2008,57(Pt12):2865-2869.
[29] GALUSHKO A,MINZ D,SCHINK B,et al.Anaerobic degradation of naphthalene by a pure culture of a novel type of marine sulphate-reducing bacterium[J].Environmental Microbiology,1999,1(5):415-420.
[30] AECKERSBERG F,RAINEY F A,WIDDEL F.Growth,natural relationships,cellular fatty acids and metabolic adaptation of sulfate-reducing bacteria that utilize long-chain alkanes under anoxic conditions[J].Archives of Microbiology,1998,170(5):361-369.
[31] SO C M,YOUNG L Y.Isolation and characterization of a sulfate-reducing bacterium that anaerobically degrades alkanes[J].Applied and Environmental Microbiology,1999,65(7):2969-2976.
[32] CRAVO-LAUREAU C,GROSSI V,RAPHEL D,et al.Anaerobicn-alkane metabolism by a sulfate-reducing bacterium,Desulfatibacillumaliphaticivoransstrain CV2803T[J].Applied and Environmental Microbiology,2005,71(7):3458-3467.
[33] DAVIDOVA I A,DUNCAN K E,CHOI O K,et al.Desulfoglaebaalkanexedensgen. nov.,sp. nov.,ann-alkane-degrading,sulfate-reducing bacterium[J].International Journal of Systematic and Evolutionary Microbiology,2006,56(Pt12):2737-2742.
[34] KNIEMEYER O,MUSAT F,SIEVERT S M,et al.Anaerobic oxidation of short-chain hydrocarbons by marine sulphate-reducing bacteria[J].Nature,2007,449(7164):898-901.
[35] HIGASHIOKA Y,KOJIMA H,NAKAGAWA T,et al.A noveln-alkane-degrading bacterium as a minor member ofp-xylene-degrading sulfate-reducing consortium[J].Biodegradation,2009,20(3):383-390.
[36] RABUS R,FUKUI M,WILKES H,et al.Degradative capacities and 16S rRNA-targeted whole-cell hybridization of sulfate-reducing bacteria in an anaerobic enrichment culture utilizing alkylbenzenes from crude oil[J].Applied and Environmental Microbiology,1996,62(10):3605-3613.
[37] RUETER P,RABUS R,WILKES H,et al.Anaerobic oxidation of hydrocarbons in crude oil by new types of sulphate-reducing bacteria[J].Nature,1995,372(6505):455-458.
[38] SO C M,YOUNG L Y.Initial reactions in anaerobic alkane degradation by a sulfate reducer,strain AK-0l[J].Applied and Environmental Microbiology,1999,65(12):5532-5540.
[39] CRAVO-LAUREAU C,MATHERON R,JOULIAN C,et al.Desulfatibacillumalkenivoranssp. nov.,a noveln-alkene-degrading,sulfate-reducing bacterium,and emended description of the genusDesulfatibacillum[J].International Journal of Systematic and Evolutionary Microbiology,2004,54(Pt5):1639-1642.
[40] MUSAT F,GALUSHKO A,JACOB J,et al.Anaerobic degradation of naphthalene and 2-methylnaphthalene by strains of marine sulfate-reducing bacteria[J].Environmental Microbiology,2009,11(1):209-219.
[41] 程海鹰,肖生科,马光东,等.营养注入后油藏微生物群落16S rRNA基因的T-RFLP对比分析[J].石油勘探与开发,2006,33(3):356-359,373.
[42] FERRY J G.Methane:small molecule,big impact[J].Science,1997,278(5342):1413-1414.
[43] 龙胜祥,陈纯芳,李辛子,等.中国石化煤层气资源发展前景[J].石油与天然气地质,2011,32(3):481-488.
[44] LOWE D C.Global change:a green source of surprise[J].Nature,2006,439(7073):148-149.
[45] 傅霖,辛明秀.产甲烷菌的生态多样性及工业应用[J].应用与环境生物学报,2009,15(4):574-578.
[46] BELYAEV S S,WOLKIN R,KENEALY W R,et al.Methanogenic bacteria from the bondyuzhskoe oil field:general characterization and analysis of stable-carbon isotopic fractionation[J].Applied and Environmental Microbiology,1983,45(2):691-697.
[47] IVANOV M V,BELYAEV S S,ZYAKUN A M,et al.Microbiological methane formation in oil field development[J].Geokhimiya,1983,11:1647-1654.
[48] DAVYDOVA-CHARAKHCH′YAN I A,KUZNETSOVA V G,MITYUSHINA L L,et al.Methane-forming bacilli from oil fields of Tartaria and Western Siberia[J].Microbiology(English Translation of Mikrobiologiya),1993,61(2):202-207.
[49] NILSEN R K,TORSVIK T.Methanococcusthermolithotrophicusisolated from North Sea oil field reservoir water[J].Applied and Environmental Microbiology,1996,62(2):728-731.
[50] OLLIVIER B,CAYOL J L,PATEL B K,et al.Methanoplanuspetroleariussp. nov.,a novel methanogenic bacterium from an oil-producing well[J].FEMS Microbiology Letters,1997,147(1):51-56.
[51] OLLIVIER B,FARDEAU M L,CAYOL J L,et al.Methanocalculushalotoleransgen. nov.,sp. nov.,isolated from an oil-producing well[J].International Journal Systematic Bacteriology,1998,48(3):821-828.
[52] OBRAZTSOVA A Y,SHIPIN O V,BEZRUKOVA L V,et al.Properties of the coccoid methylotrophic methanogen,Methanococcoideseuhalobiussp. nov.[J].Mikrobiology (English Translation of Mikrobiologiya),1987,56:523-527.
[53] DAVIDOVA I A,HANNSEN H J M,STAMS A J M,et al.Taxonomic description ofMethanococcoideseuhalobiusand its transfer to theMethanohalophilusgenus[J].Antonie van Leeuwenhoek,1997,71(4):313-318.
[54] NI S S,BOONE D R.Isolation and characterization of a dimethyl sulfide-degrading methanogen,MethanolobussiciliaeHI350,from an oil well,characterization ofM.siciliaeT4/MT,and emendation ofM.siciliae[J].International Journal of Systematic Bacteriology,1991,41(3):410-416.
[55] NI S,WOESE C R,ALDRICH H C,et al.Transfer ofMethanolobussiciliaeto the genusMethanosarcina,naming itMethanosarcinasiciliae,and emendation of the genusMethanosarcina[J].Internationa Journal Systematic Bacteriology,1994,44(2):357-359.
[56] OBRAZTSOVA A Y,TSYBAN V E,LAUNNA-VICHUS K S,et al.Biological properties ofMethanosarcinanot utilizing carbonic acid and hydrogen[J].Microbiology(English Translation of Mikrobiologiya),1987,56:807-812.
[57] FOGHT J.Anaerobic biodegradation of aromatic hydrocarbons:pathways and prospects[J].Journal Molecular Microbiology and Biotechnology,2008,15(2/3):93-120.
[58] RABUS R,WILKES H,BEHRENDS A,et al.Anaerobic initial reaction ofn-alkanes in a denitrifying bacterium:evidence for (1-methylpentyl)succinate as initial product and for involvement of an organic radical inn-hexane metabolism[J].Journal of Bacteriology,2001,183(5):1707-1715.
[59] RABUS R,HEIDER J.Initial reactions of anaerobic metabolism of alkylbenzenes in denitrifying and sulfate-reducing bacteria[J].Archives of Microbiology,1998,170(5):377-384.
[60] CHAKRABORTY R,COATES J D.Hydroxylation and carboxylation——two crucial steps of anaerobic benzene degradation byDechloromonasstrain RCB[J].Applied and Environmental Microbiology,2005,71(9):5427-5432.
[61] JOHNSON H A,PELLETIER D A,SPORMANN A M.Isolation and characterization of anaerobic ethylbenzene dehydrogenase,a novel Mo-Fe-S enzyme[J].Journal of Bacteriology,2001,183(15):4536-4542.
[62] COATES J D,CHAKRABORTY R,McLNERNEY M J.Anaerobic benzene biodegradation——a new era[J].Research in Microbiology,2002,153(10):621-628.
[63] SAFINOWSKI M,MECKENSTOCK R U.Methylation is the initial reaction in anaerobic naphthalene degradation by a sulfate-reducing enrichment culture[J].Environmental Microbiology,2006,8(2):347-352.
[64] SO C M,PHELPS C D,YOUNG L Y.Anaerobic transformation of alkanes to fatty acids by a sulfate-reducing bacterium,strain Hxd3[J].Applied and Environmental Microbiology,2003,69(7): 3892-3900.
Research Progress in Methane-Producing byAnaerobic Degradation of Petroleum Hydrocarbon
HU Heng-yu1,WEI An-pei1,LIU Shao-mei1,LI Jing1,ZHAO Dong-feng2
(1.CollegeofResourcesandEnvironment,LinyiUniversity,Linyi276000,China;2.CollegeofChemicalEngineering,ChinaUniversityofPetroleum,Qingdao266580,China)
Withtheexploitationofoilreservoirs,alargeamountofresidualcrudeoilareleft.Residualcrudeoil(petroleumhydrocarbon)canbedegradedtoproducemethanebymicroorganisms,whichhasbecomeanovelapproachfordepthexploitationandutilizationofoilreservoirs.Oilreservoirmicroorganismsaremadeupofmanymicrobialcommunities,whichcanprovidesynergisticeffectstoachieveanaerobicdegradationofpetroleumhydrocarbon.Toimprovedegradationefficiency,wesummarizeanaerobicmicrobialcommunitieswithpetroleumhydrocarbonbiodegradabilityandtheirmetabolicproperties.Wealsocomparethemetabolicpathwaysofmethane-producingbyanaerobicdegradationofpetroleumhydrocarbon.
residualcrudeoil;petroleumhydrocarbon;anaerobicdegradation;methane
临沂大学校级博士科研项目(LYDX2016BS063)
2017-03-11
胡恒宇(1983-),男,黑龙江佳木斯人,博士,讲师,研究方向:石油烃的微生物降解,E-mail:hhyu01@163.com。
10.3969/j.issn.1672-5425.2017.08.004
TE357.4
A
1672-5425(2017)08-0016-06
胡恒宇,韦安培,刘少梅,等.石油烃厌氧降解产甲烷研究进展[J].化学与生物工程,2017,34(8):16-21.

