Natural and artificial small RNAs: a promising avenue of nucleic acid therapeutics for cancer
Sunny Yadav, Mamta Shekhawat, Devashree Jahagirdar, Nilesh Kumar Sharma
Cancer and Translational Research Lab, Dr. D.Y Patil Biotechnology & Bioinformatics Institute, Dr. D. Y. Patil Vidyapeeth, Pune 411033, Maharashtra, India
Natural and artificial small RNAs: a promising avenue of nucleic acid therapeutics for cancer
Sunny Yadav, Mamta Shekhawat, Devashree Jahagirdar, Nilesh Kumar Sharma
Cancer and Translational Research Lab, Dr. D.Y Patil Biotechnology & Bioinformatics Institute, Dr. D. Y. Patil Vidyapeeth, Pune 411033, Maharashtra, India
Since the failure of traditional therapy, gene therapy using functional DNA sequence and small RNA/DNA molecules (oligonucleotide) has become a promising avenue for cancer treatment. The discovery of RNA molecules has impelled researchers to investigate small regulatory RNA from various natural and artificial sources and determine a cogent target for controlling tumor progression. Small regulatory RNAs are used for therapeutic silencing of oncogenes and aberrant DNA repair response genes. Despite their advantages, therapies based on small RNAs exhibit limitations in terms of stability of therapeutic drugs, precisionbased delivery in tissues, precision-based intercellular and intracellular targeting, and tumor heterogeneity-based responses. In this study, we summarize the potential and drawbacks of small RNAs in nucleic acid therapeutics for cancer.
Cancer; drug targeting; gene; intracellular; therapeutics; tumor heterogeneity
Introduction
Cancer is a group of diseases involving uncontrolled and abnormal cell proliferation and can potentially invade or spread to other parts of the body. To date, cancer leads to 168.1 million deaths, of which 3% and 12% of the cases are due to cervical cancer and breast carcinoma, respectively. The number of estimated cancer cases worldwide until 2012 reached 14.1 million, which involved 7.4 million men and 6.7 million women1.
Traditional therapies for treatment of cancer include surgery, radiation, and chemotherapy2-4. Radiation and chemotherapy use non-selective agents, which may cause toxicity to normal tissues5,6. In this regard, scholars have developed a promising technology, namely, gene therapy or nucleic acid therapy (NAT), which uses functional genes and small genetic materials, such as RNAs and DNAs7-13. In this process, DNA or RNA as coding gene or small non-coding genetic materials is transferred into different cellular compartments of the host (human or animal) to alleviate various pathophysiological conditions, including cancer12,13. In addition to their roles in functional gene therapy, small RNAs have been widely investigated as a tumor suppressor agent to suppress the aberrant expression of oncogenes and DNA repair response genes and control the growth and proliferation of cells14-20. Therefore, small RNAs obtained from natural sources and artificial mimetic agents have been increasingly studied to explore new class of anticancer drugs. In this paper, we present the role of small RNAs in cancer therapeutics in terms of pre-clinical and clinical perspectives and the concomitant challenges.
Small RNAs from natural sources
Gene silencing and small RNAs were first studied in the early 1990s21-23. The discovery of small ~20–30 nucleotide RNA molecules is an outstanding discovery in biology because of their distinct role in the expression and function of eukaryotic genomes14-20. The predominant small RNA classes include short-interfering RNAs (siRNAs) and microRNAs (miRNAs), which function in somatic and germline lineages in a broad range of eukaryotic species17,19,20,24. Small RNAs exhibit potential regulatory roles at various cellular strata, including in chromatin structure, chromosome segregation, transcription, and RNA, stability, and translation17,24. The regulatory mechanisms of small RNAs in the intra-cellular, intercellular, and extracellular levels are generally inhibitory and interpreted as RNA silencing22,23. Small RNAs can also activate gene transcription and are classified as small-activating RNAs. Natural endogenous small RNAs are found in various organisms including humans, plants, mouse, fungi, bacteria, flies, and worms17-20. In addition to their versatile roles in intra-cellular signaling, intercellular communication, and cell growth and development, small RNAs, particularly miRNAs, which are found in biological fluids, such as serum and plasma, function as potential biomarkers for cancer prognosis and detection25-27. Various types of small RNAs found in several species of plants and microorganisms exhibit potential as therapeutic agents (Figure 1)17-20.
Small RNAs from plants as anti-cancer NAT
The plant genome encodes numerous small RNAs that are involved in genetic and epigenetic silencing pathways. The abundance and diversity of small RNA classes differ among plant species20. The diversity of small RNAs from plants could be related to coevolution between environmental adaptations and influences from other organisms. Hence, the possibility of cross-kingdom transfer of these small RNAs can be predicted. Small RNAs are commonly found in staple foods, such as rice (Oryza sativa) and corn (Zea mays). A previous study showed that miRNA plays an important role in cross-kingdom gene regulation, where exogenous plant miRNAs are found in human serum and possibly acquired through food intake. According to the dietary xenomiRNA hypothesis, miRNAs present in food stuffs may regulate the gene expression in cross-kingdom species28-30. Philip et al.29demonstrated that plant miRNAs remain intact even after storage, processing, and cooking. Common food materials, such as rice and soybean, contain miRNA that will remain intact after ingestion. Moreover, miRNAs that underwent genetic modifications can be incorporated into the food to protect it from degradation. The regulatory ability of these molecules must be further explored for cancer therapeutics.
MiRNA-168a from rice (Oryza sativa) has been found in human serum in its stable form; this miRNA can bind to mRNA encoding for low-density lipoprotein receptor adaptor protein1 (LDLRAP1) and inhibit the protein expression28,30. Plant miRNA159, which is abundant in mammalian serum because these organisms consume a variety of food, is inversely correlated to the incidence and progression of breast cancer31. Chin et al.31reported that miRNA159, encoding for the Wnt signaling transcription factor, can bind to and inhibit the proliferation of TCF7, resulting in reduced MYC protein level.
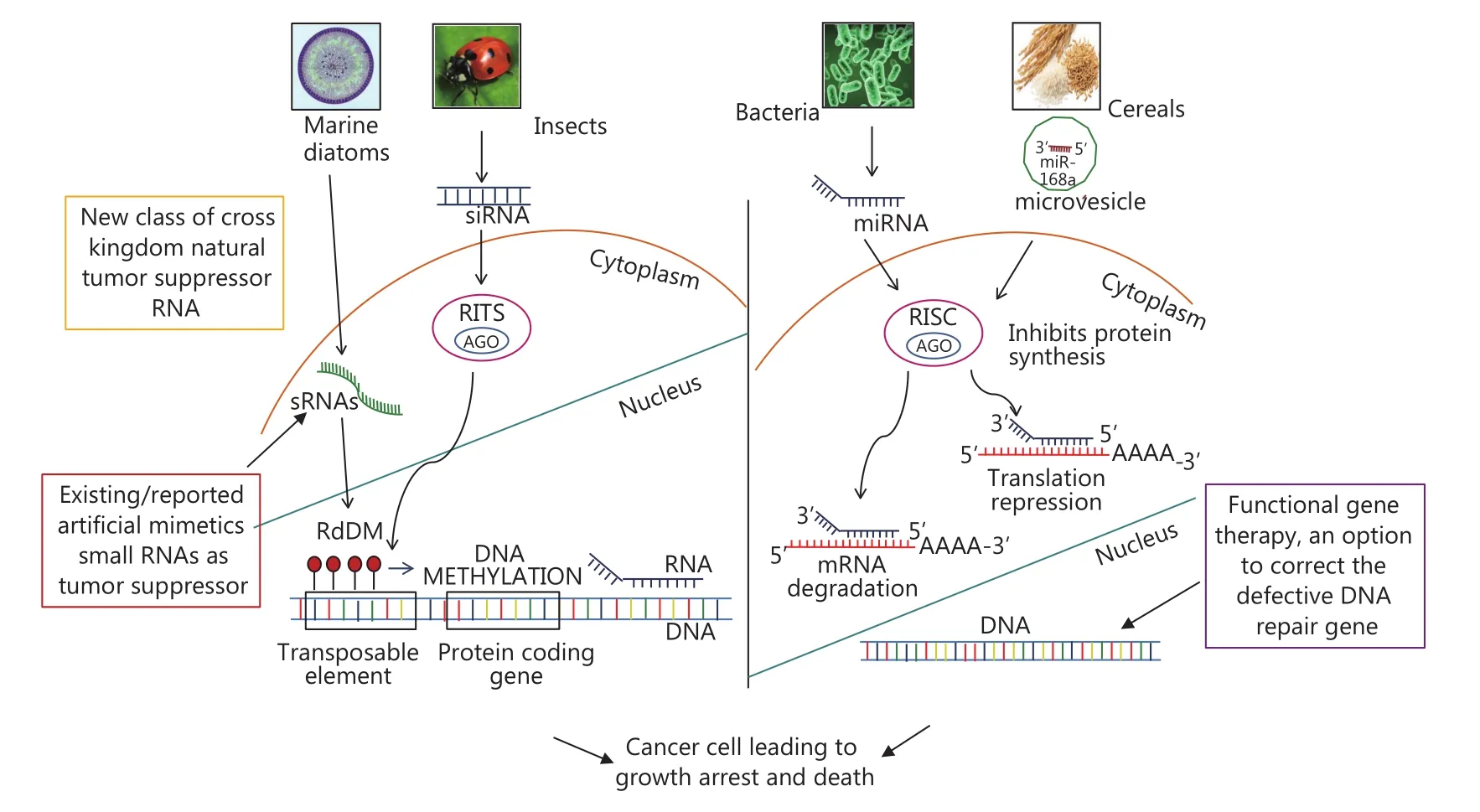
Figure 1 Mechanism of actions of cross kingdom natural small RNAs in cancer. This figure illustrates the mechanism of actions of cross kingdom natural small RNAs to bring cancer cell cycle arrest and death. Further, the mode of growth arrest in cancer cells via translational repression or mRNA degradation has been shown. Small molecular weight RNAs from various natural sources such as marine diatoms, insects, bacteria and cereals RNA have been shown to play a role in cancer. Additionally, pathway has been depicted for diatom encoded RNA directed DNA methylation (RdDM), which methylates the DNA leading to stall in transcription.
The potential of plant-based dietary small RNAs in cancer therapeutics have been investigated. Yang et al.32reported the presence of plant-specific small RNA MIR2911 in the serumof mice fed with diet rich in vegetables. The findings confirm the intrinsic stability of plant-based RNAs for future cancer NAT. The ingested miRNA persists in the gastrointestinal (GI) tract, are packaged into extracellular vesicles (EVs), and are released into circulation to regulate gene expression in animals31.
Small RNAs from bacteria and other lower organisms
Small, regulatory RNAs are present in bacteria. These sRNAs may base pair with target mRNAs by modulating protein activity and function as nucleic acid mimetics18. In prokaryotes, such as bacteria, clustered regularly interspersed short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) are analogous to small RNAs33.
In addition to various natural sources of small RNAs and other ncRNAs, the former has been identified in marine diatoms and function in gene-silencing activity. Most sRNAs possess length of 25 to 30 nt and target DNA-methylated protein-coding genes; hence, gene-driven methylation in diatoms may be mediated by sRNAs. Moreover, the majority of sRNAs comprise non-coding RNAs, tRNAs, and U2 snRNA, which play important roles in stressful environments like starvation, oxidative stress, etc. Furthermore, miRNAs have been found in two marine diatom species, namely, Thalassiosira pseudonana and P. tricornutum. Nevertheless, none of these species have been experimentally studied34.
Types of small RNAs in anticancer NAT
Earlier discoveries in molecular biology have resulted in a paradigm shift. Lines of evidence showed that in addition to its function as regulator between DNA and protein, RNA regulates gene expression and genome organization18-20. Studies show that the number of genes encoding for RNA is higher than that of genes encoding for proteins19. Evolutionary RNA is considered the ancestor of deoxyribonucleic acid (DNA). DNA and RNA exhibit minimal structural differences, but the hydroxyl group at 2′of the pentose ring significantly affects the interaction and folding properties of RNA. This 2′ hydroxyl allows the interactions between RNA fragments and RNAs and other biomolecules35,36.
The completion of the human genome project confirmed that all plant and animal species require the same number of genes to produce proteins. Many of these species also produce non-coding regulatory products, such non-proteincoding RNA (npc RNA), which are genes that encode small RNA molecules18-20. Small RNAs constitute different classes of non-protein-coding RNA molecules, such as miRNAs and small interfering RNAs (siRNAs), which function in many physiological and pathological processes. Scholars have revolutionized the understanding of gene performance and the physiology of cells by introducing RNAi37-39. With increased knowledge on molecular mechanisms through small-sized RNAs function in normal and malignant cells, researchers can elucidate tumor biology and discover novel therapeutic markers for cancer treatment10,12,40-46. In the human genome, many genes produce non-coding regulatory products instead of having 97% of non-coding DNA. Genes that encode for small or short length RNA are members of the non-protein-coding RNA (npcRNA) class, in which miRNAs constitute an important group37,47.
MiRNAs are small non-protein-coding (~22 nucleotide) RNAs that function in several physiological and pathological processes, such as cell development, evolution, cellular differentiation, proliferation, embryogenesis, cell death, and gene expression17,19,20,24. MiRNAs bind to the 3′untranslated regions (UTRs) of mRNA molecules, causing either mRNA degradation or translation repression and eventually resulting in silencing of unwanted genes47,48. The suppressed degradation and translation of mRNA could be attributed to imperfect complementary and perfect complementary between miRNA and mRNA, respectively17,19,20,49. MiRNAs do not encode any proteins but interfere with protein production by targeting specific mRNA.
Small temporal RNA (stRNA) and siRNAs are categorized as two classes of miRNA. Only 1% of human genes encode for miRNAs. The first miRNA discovered in Caenorhabditis elegans in 1993 is Lin-4, which controls the development of the organism15,50-52. Thereafter, let-7 was discovered in 2000 and found to have similar effects to those of Lin-451. Researchers have found the homologs of let-7 in many species, such as frog, mice, and humans, indicating the sequence similarity of let-7 among these species15. Tumor suppressor miRNAs, such as let-753and miR-3454, exhibit low expression levels in many cancers, leading to enhanced cell proliferation. Overexpressing or repressing miRNA expression, depending on the type of disease, and suppressing or replacing miRNA are promising areas for study in therapeutics. The overexpression of miRNA can be observed in many diseases. For example, miR-21 is overexpressed in many cancers, leading to the progression of cell cycle and enhanced proliferation49,55. Studies have mainly focused on the inhibition and replacement of miRNA for therapeutic purposes49. SiRNAs, which are a group of small dsRNA consisting of 21–23 nucleotides, can cleave the RNAmolecule as mediated by RNA inducing silencing complex (RISC), ultimately resulting in the disruption of translation48.
Small nuclear RNAs (snRNAs), which are members of small RNAs, play an important role in splicing and are also known as spliceosomal RNA. As another class of small RNAs, small nucleolar RNAs (snoRNAs), which are found in the nucleolus, are involved in methylation and pseudouridylation of snRNAs, tRNAs, and rRNAs37,56. Cajal bodies, also called small cajal body-specific RNA (ScaRNAs) are small RNAs found in the subnuclear region19. Recently, two other classes of small RNAs have been identified in animals19; these classes include transcription initiation RNAs (tiRNAs)57, which initiate the RNA transcription and splice-site RNAs (spliRNAs)58. These small RNAs are involved in nucleosome positioning and chromatin organization. Scholars have also reported the presence of less distinct classes, namely, promoter-associated RNAs (PASRs)59, transcription start site-associated RNAs (TSSa-RNAs), and promoter upstream transcripts (PROMPTS)60. A summary of the information on different types of small RNAs is given in Table 1.
Obstacles and bottlenecks encountered in RNA delivery
One of the major challenges in small RNA therapy (RNAi technology) is the delivery of these molecules into the cell7-13. The delivery of nucleic acids (i.e., DNA, RNA, siRNA, shRNA, and antisense oligonucleotides) can down-regulate and silence unwanted gene expression and thus suppress tumor growth and invasion3,48,77. However, the delivery of these molecules remains challenging because of their large size and negative charges; the main obstacles are related to stability of small RNAs as therapeutic drug, controlled intracellular and inter-cellular release, unwanted inflammation due to immune responses, and precision to the target genes7-13. Therefore, the delivery system must be modified to achieve stability against serum nucleases, evade the immune system, avoid non-specific interactions with serum proteins and non-target cells, prevent renal clearance, and minimize off target effects41,78-80.
Synthetic small RNAs as mimetic agent
The two major types of small dsRNAs, namely, siRNAs and miRNAs, participate in RNA interference (RNAi), which involves several mechanisms, including gene silencing by remodeling chromatin to suppress transcription; degrading complementary mRNA; or blocking protein translation12,44-46. The development and design of synthetic small RNAs in cancer therapeutics have gained increasing research attention. Considering the different methods available for synthetic development of small RNAs, Yoo et al.81reported that synthetic small RNAs can be constructed through conventional gene cloning. The available different therapeutic options using drugs containing small RNAs are listed in Figure 218-20.
Chemically synthesized siRNAs have been developed to target mammalian cells without any innate immune responses to improve their inherent properties, such as in vivo instability, off-target effects, and immunogenicity. The most widely used chemically synthesized siRNA is (19+2traditional siRNA), which is similar to the nature’s dicer product13,82. Chemically synthesized siRNAs with two nucleotides overhanging at 3’ can efficiently cleave siRNA, and the cleavage site is located near the region covered by the guiding siRNA83. Major challenges associated with RNAi mechanism exist due to the naked RNA structure of naturally available small RNAs; such challenges include rapid RNA degradation in biological fluids, poor cellular uptake, and offtarget effects. In this regard, synthetic RNA mimetics have been developed for RNAi- based medicine11-13.
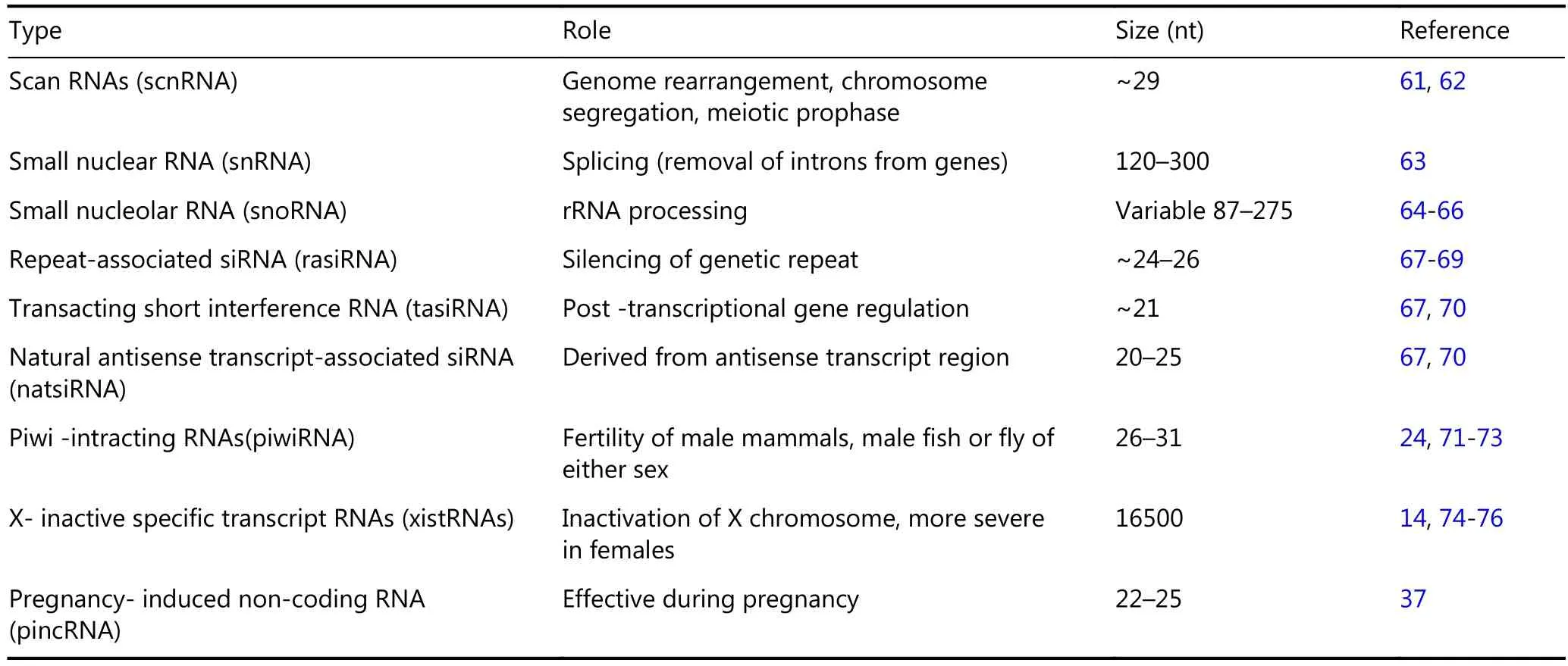
Table 1 List of small RNAs and their function
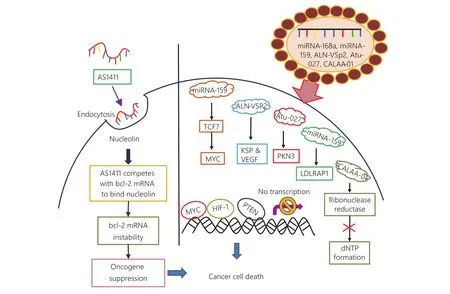
Figure 2 Various RNA based drugs to target cancer cell. This figure depicts the newly synthesized RNA and drugs, which targets the progression of cancer cell by inhibiting the transition from epithelial to mesenchymal cells and also by preventing angiogenesis. An aptamer drug AS1411 has been depicted to block the nucleolin receptor based signaling and inhibit the oncogene expression for cancer survival. On the other hand, this diagram shows the use of small RNAs as a cancer therapeutic approach including ALN-VSP2, miRNA-159, CALAA-01 etc.
Synthetic gRNAs induce a site-specific mutation in the target sequence. The site-specific mutation mediated by gRNA modifies pre-miRNA without synthesizing particular miRNAs84. Cancer cells possess the ability to modify premiRNA alternative splicing85. The gRNA lentiviral CRISPR/Cas9 vector can interrupt miR-21, which inhibits epithelial-to-mesenchymal transition (EMT)84.
Synthetically created mRNAs exhibit strong therapeutic potential after its introduction into mammalian cells. These mRNAs are stable and can encode desired proteins86. Such mRNAs are known as naked mRNA because they elicit immune responses around the tumor cells. Naked mRNAs do not need any carriers for transport and require short period of time to show their effectiveness; hence, these mRNAs are important in the field of modern therapy87.
Nat delivery system
Considering the potential mechanisms of small RNAs in gene therapy against cancer, scholars have performed gene silencing by decreasing the expression of the target genes, which are mostly oncogenes, in cancer10,12,42,44-46. These interfering RNAs can be synthetic (i.e., oligonucleotide therapy) or encoded in novel genes; as such, the sequences are the inverse of the normal sequence (i.e., antisense) and can thus hybridize to the message and prevent its translation10,12,42,44-46. Systemic gene delivery is one of the major challenges in modern cancer gene therapy and is a limiting factor in experimental and clinical approaches9,40,88,89. Synthetic siRNAs are poly-anionic macromolecules that do not readily enter the cell but require a delivery system for effective gene silencing90,91. Previousstudies attempted the precise and effective delivery of these potential NAT agents by using several targeted delivery systems, such as exosomes, nanocarriers, and aptamers48,92. First, naked therapeutic DNA or RNA can be transferred into cells by using high voltage (i.e., electroporation), through uptake by invaginating vesicles (i.e., endocytosis), or by sheer mechanical force using a gene gun instrument. DNA or RNA can be packaged into liposomes (i.e., membrane bound vesicles) and naturally released exosomes, which can be more easily absorbed by the cells than naked DNA/RNA87,93,94. Different types of liposomes have been developed to preferentially bind to specific tissues and modify protein or RNA at different levels. Third, DNA or RNA can be packaged into virus-like particles by using a modified viral vector. Finally, DNA or RNA can be combined with cell therapy protocols95. Chemically synthesized siRNAs can be encapsulated within the nanocarriers and may be administered with chemotherapeutic drugs96. Figure 3 shows the schematic of therapeutic delivery options based on small RNAs for cancer treatment12,44-46.
With the advancements in nanotechnology, cancer therapy has considerably progressed. In nanomedicine, a wide variety of nano-carriers containing polysaccharides have been developed. Nanoparticle carriers, which are non-toxic, biocompatible, biodegradable, and immunoefficient, can be potentially used for cancer therapy97. Ideal polymers for preparing nano-systems include anionic polymers, such as hyaluronic acid, heparin, or alginate, which exhibit anticancer property98. Modified delivery systems include lipid carriers and polymers aptamers. Nanotechnology and modern cancer research have allowed the development of highly safe medicine with reduced toxicity and ability to carry large payload and multivalent ligand targeting and the improvements in cancer diagnostics7. Yuan et al.99showed that nanoparticles carrying three different siRNAs can be delivered to tumor xenografts. The simultaneous delivery of KRAS-, PIK3CA-, and PIK3CB-targeting siRNAs improved the therapeutic efficacy but did not increase the toxicity of the drug. Therefore, this approach can be used to develop nontoxic drugs for tumor suppression.
Exosomes as NAT cargo system
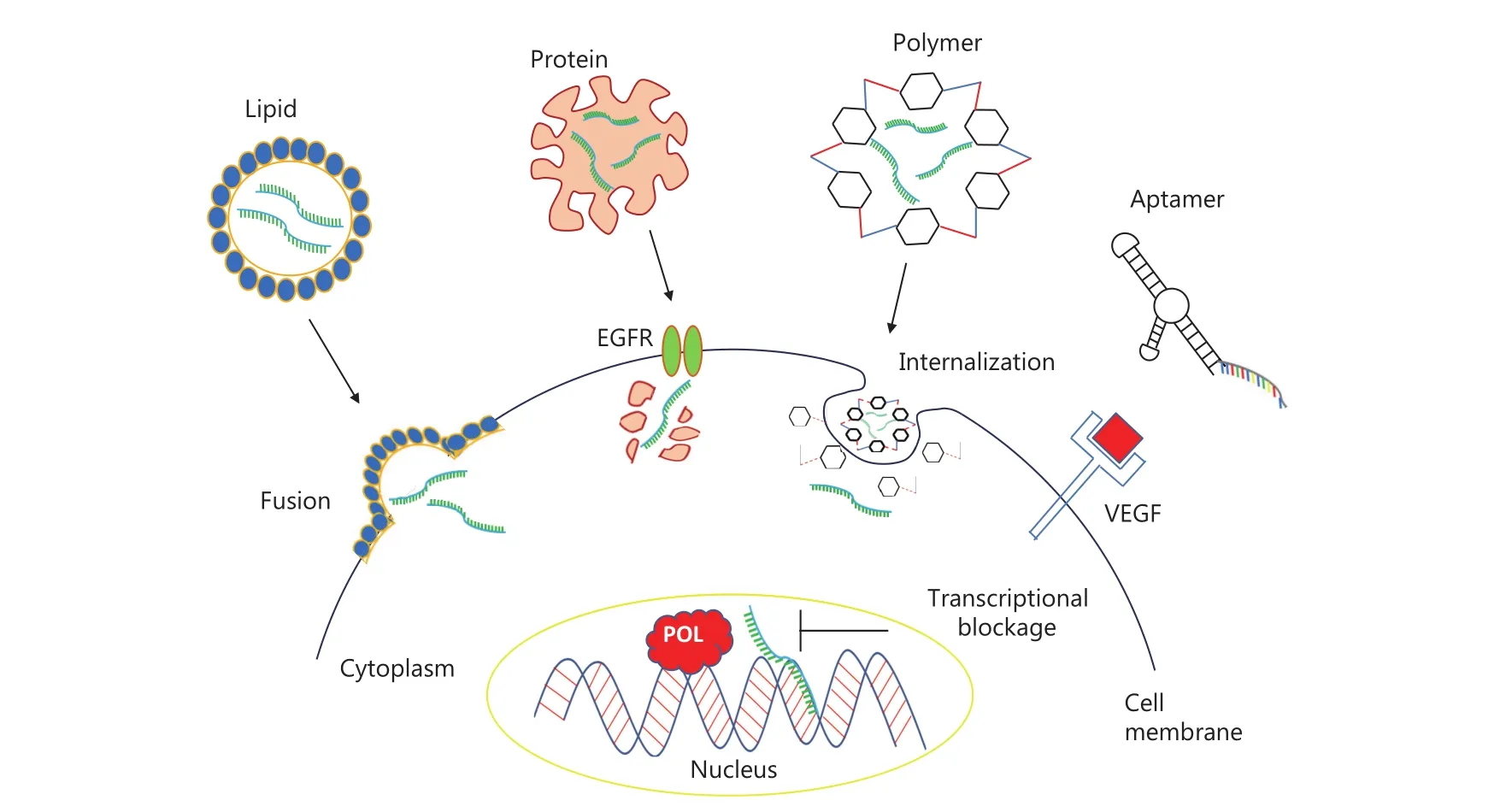
Figure 3 Import pathways of small molecular weight RNAs. This figure shows the diagrammatic representation of import of small molecular weight RNA through cellular or non-cellular contents. Small molecular weight RNAs have been shown to enter either through lipid vesicles, protein conjugates or through a polymer. These carriers are either embedded or engulfed by the cell membrane and RNA is released into the cells. After entry into cell, these small RNAs facilitate the blockage of transcription.
Exosomes are naturally occurring nano-sized vesicles released by all mammalian cells to facilitate cell-to-cell communication44. The delivery of exogenous siRNAs to the target cells poses many obstacles for effective gene therapy. The unmodified siRNA is unstable in the bloodstream, is immunogenic, and cannot easily cross the membrane and enter the cell. Therefore, a siRNA-based therapeutic approach requires a safe and reliable delivery method80. Hence, natural exosomes are used to deliver therapeuticsiRNAs to target cells42. O'Loughlin et al.46reported the use of extracellular vesicles to deliver small RNAs as therapeutic RNA cargo. Cholesterol-conjugated siRNAs (cc-siRNAs) should be used to develop EV-based therapeutics with increased loading of small RNAs to extracellular vesicles. Lunavat et al.44proposed a new class of extracellular vesicles to efficiently transport the RNA molecule into the cell cytoplasm. The authors provide evidence on the effective use of exosome-mimetic nanovesicles (NV) to reduce the expression of target genes, such as c-Myc, in cancer cells.
Aptamers and NAT
The conjugation of siRNA to delivery vehicles is a promising anticancer therapy. This single-component system uses equimolar concentration of the delivery system and siRNA. Several conjugate delivery systems have been developed by coupling siRNA to polymers, such as aptamers, peptides, and proteins80,100,101. Several researchers have transformed molecular recognition by fabricating synthetic RNA motifs that bind to specific targets. These molecules are called RNA aptamers102-104. These aptamers are selected through in vivo selection method called the systematic evolution of ligands by exponential enrichment (SELEX)103-105. These synthetically derived molecules are selective and demonstrate high affinity toward the target8,104. RNA aptamers are a special class of nucleic acids that can fold into composite structures106and possess pockets and hands for binding specific molecules. AS1411 is the first aptamer approved for clinical trial for treatment of different types of cancer. Wang et al.101demonstrated the application of aptamer AS1411-modified extracellular vesicles as RNAi nanoplatform for breast cancer therapeutics. Chen et al.107used a novel aptamer-siRNA chimera delivery system mediated by cationic Au-Fe3O4 nanoparticles (NPs) to reverse the multi-drug resistance (MDR) of ovarian cancer cells. Considering their development for systemic delivery, RNA aptamers have become active therapeutic agents particularly for blood stream and NAT104,108.
Liposome- and polymer-assisted delivery of NAT agents
Lipids are involved in the survival, proliferation, and death of cells and in cell-cell interaction45. Lipids, particularly phospholipids, function in cellular processes, such as signal transduction, post-translational modification, homeostasis, adhesion, migration, apoptosis, neurotransmission, vesicular trafficking, and energy storage109,110. A combination of cationic polymers, polyethyleneimines (PEIs), and liposomes can facilitate the formation of lipopolyplexes, which are used to deliver nucleic acids with improved efficacy and biocompatibility111. Many liposomes, such as cationic lipids, can be used to successfully deliver small RNAs. Positively charged lipids can improve the entrapment of small RNAs, increase the cellular uptake, and provide protection from endosomal escape. Examples of cationic lipids are Lipofectamine 2000 and RNAiMAX transfection reagent, which is a recently developed compound with high efficiency80,101. The modified cationic lipids do not act as carrier in the delivery of small RNA, but provide stability against serum and access to RNAi machinery101,112-114.
Polymer-assisted delivery vehicles comprise polymeric nanoparticles and are characterized as biodegradable; these vehicles prevent aggregation during storage, increase the circulation time, and reduce the off-target effects80,101,115. Cyclodextrin and polyethylenimine (PEI) are the most commonly used polymers for siRNA delivery101,116-118. Other polymers used for NAT delivery are polycaprolactone (PCL), poly (D, L-lactide) (PLA), poly (D, L-lactide-co-glycolide) (PLGA), chitosan, poly-L-lysine, dextran, polyglutamic acid, hyaluronic acid, and gelatin48,101,119. Magnetic molecularly imprinted polymers (MMIPs) are another form of delivery system and are synthesized through photo-polymerization of methacrylic acid and ethylene glycol dimethacrylate around the template molecule and in the presence of magnetite. These polymers exhibit controlled release and high magnetic properties120.
Clinical evaluation of therapeutic small RNAs
Various small RNAs have been reported by preclinical and clinical interventional studies7-13,46. Since the discovery of RNAi, more than 50 clinical trials involving 26 different siRNAs have been documented8-13. A previous study reported the use of siRNAs with liposome as a delivery vehicle to treat a patient with chronic myeloid leukemia121. Zou et al.122reported the potential of vascular endothelial growth factor siRNA (VEGF-siRNA) for treatment of hepatocellular carcinoma. RNAi is a potent tool in gene regulation, and the first in-man trial of ALN-VSP02, an RNAi therapy, has been reported in patients with advanced solid tumors123,124. Finally, preclinical and clinical trials and investigations have been conducted on the use of Atu027, a siRNA-lipoplex directed against protein kinase N3 (PKN3), for treatment of advanced solid cancers, including pancreatic cancer125,126.
CALAA-01, a type of siRNA targeted to curb theexpression of M2 sub-unit of ribonucleotide reductase (R2), can be used for siRNA-based therapies for cancer127. This drug is formulated in a stabilized nanoparticle to prevent nuclease-mediated degradation within tumor cells. The drug ALN-VSP02 is a lipid nanoparticle formulation containing two siRNAs targeting kinesin spindle protein (KSP) and VEGF with potential antitumor activity123. A phase 1 clinical study was conducted to investigate the use of TKM 080301, a type of lipid nanoparticles encapsulating siRNA targeted to the PLK1 gene for treatment of primary or secondary liver cancer128. Furthermore, the drug ALN-VSP02 demonstrated siRNA-mediated mRNA cleavage in the liver and exhibited antitumor activity123.
Another phase 1 interventional study evaluated the immunotherapy of melanoma by using tumor antigen RNA and small inhibitory RNA loaded into dendritic cells to target immunoproteasome beta subunits LMP2, LMP7, and MECL1129. Another clinical study evaluated the efficacy and safety of single-dose siG12D LODER administered with chemotherapy drugs to patients with unresectable locally advanced pancreatic cancer130. An ongoing clinical study reported on the use of siRNA-transfected peripheral blood mononuclear cells APN401 for treatment of pancreatic cancer, colorectal cancer, and other solid tumors that spread to other places in the body or have relapsed131.
Conclusions
Non-coding small RNAs have been increasingly investigated as NAT to repress pivotal specific genes related to tumor progression and drug resistance. These new class of drugs should be viewed from the perspective of combinatorial options as a cocktail of cancer drug therapy. Hence, the use of these small RNAs as NAT in cancer treatment should be encouraged in conjunction with other drug options, such as epigenetic, signaling, growth, and metastasis blockers. Such combinations may contribute to completely eradicate issues with regard to inherent tumor heterogeneity and drug resistance in clinical settings. In addition to new findings and the development of NAT based on small RNAs from natural and artificial sources, further studies should evaluate the stability and precision of NAT for targeting tumor tissues and analyze specific compartments of intercellular and intracellular locations. To achieve therapeutic approaches with effective targeting at the site of action, scientists, clinicians, and industry researchers should collaborate to develop an efficient drug delivery system and nano-imaging system for improved monitoring of cancer treatment. Overall, small-RNAs-based NAT is more effective and precise and poses less risks to healthy tissues than existing traditional therapeutic regimens for cancer.
Acknowledgements
The authors acknowledge the resources and facilities available at the Cancer and Translational Research Lab, Dr. D. Y Patil Biotechnology and Bioinformatics Institute, Dr. D. Y Patil Vidyapeeth, Pune, India. Grant No. DST-SERB, Government of India, New Delhi, India (SERB/LS-1028/2013) and Dr. D. Y Patil, Vidyapeeth, Pune, India (Grant No. DPU/05/01/2016).
Conflict of interest statement
No potential conflicts of interest are disclosed.
1.Stewart BW, Wild CP. World cancer report 2014. World Health Organization: Geneva, 2014.
2.Tekade RK, Dutta T, Tyagi A, Bharti AC, Das BC, Jain NK. Surface-engineered dendrimers for dual drug delivery: a receptor up-regulation and enhanced cancer targeting strategy. J Drug Target. 2008; 16: 758-72.
3.Saraswathy M, Gong SQ. Recent developments in the co-delivery of siRNA and small molecule anticancer drugs for cancer treatment. Materialstoday. 2014; 17: 298-306.
4.Lage H. Gene therapeutic approaches to overcome ABCB1-mediated drug resistance. In: Walther W, editors. Current Strategies in Cancer Gene Therapy. Springer International Publishing: Switzerland, 2016. p.87-94.
5.Creixell M, Peppas NA. Co-delivery of siRNA and therapeutic agents using nanocarriers to overcome cancer resistance. NanoToday. 2012; 7: 367-79.
6.Gandhi NS, Tekade RK, Chougule MB. Nanocarrier mediated delivery of siRNA/miRNA in combination with chemotherapeutic agents for cancer therapy: current progress and advances. J Control Release. 2014; 194: 238-56.
7.Akhter S, Ahmad I, Ahmad MZ, Ramazani F, Singh A, Rahman Z, et al. Nanomedicines as cancer therapeutics: current status. Curr Cancer Drug Targets. 2013; 13: 362-78.
8.Germer K, Leonard M, Zhang XT. RNA aptamers and their therapeutic and diagnostic applications. Int J Biochem Mol Biol. 2013; 4: 27-40.
9.Walther W, Schlag PM. Current status of gene therapy for cancer. Curr Opin Oncol. 2013; 25: 659-64.
10.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genetics. 2014; 15: 541-55.
11.Ozcan G, Ozpolat B, Coleman RL, Sood AK, Lopez-Berestein G.Preclinical and clinical development of siRNA-based therapeutics. Adv Drug Delivery Rev. 2015; 87: 108-19.
12.Zuckerman JE, Davis ME. Clinical experiences with systemically administered siRNA-based therapeutics in cancer. Nat Rev Drug Discov. 2015; 14: 843-56.
13.Ku SH, Jo SD, Lee YK, Kim K, Kim SH. Chemical and structural modifications of RNAi therapeutics. Adv Drug Deliv Rev. 2016; 104: 16-28.
14.Hore TA, Koina E, Wakefield MJ, Graves JAM. The region homologous to the X-chromosome inactivation centre has been disrupted in marsupial and monotreme mammals. Chromosome Res. 2007; 15: 147
15.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001; 294: 862-864.
16.Mitrovich QM, Guthrie C. Evolution of small nuclear RNAs in S. cerevisiae, C. albicans, and other hemiascomycetous yeasts. RNA. 2007; 13: 2066-80.
17.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009; 136: 642-55.
18.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011; 43: 880-91.
19.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genetics. 2014; 15: 423-37.
20.Borges F, Martienssen RA. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol. 2015; 16: 727-41.
21.Guleria P, Mahajan M, Bhardwaj J, Yadav SK. Plant small RNAs: biogenesis, mode of action and their roles in abiotic stresses. Genom Proteom Bioinformat. 2011; 6: 183-99.
22.Budak H, Kantar M, Bulut R, Akpinar BA. Stress responsive miRNAs and isomiRs in cereals. Plant Sci. 2015; 235: 1-13.
23.Kamthan A, Chaudhuri A, Kamthan M, Datta A. Small RNAs in plants: recent development and application for crop improvement. Front Plant Sci. 2015; 6: 208
24.Axtell MJ, Snyder JA, Bartel DP. Common functions for diverse small RNAs of land plants. Plant Cell. 2007; 19: 1750-69.
25.Zhu WZ, Qin WY, Atasoy U, Sauter ER. Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes. 2009; 2: 89
26.Wu XW, Somlo G, Yu Y, Palomares MR, Li AX, Zhou WY, et al. De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J Transl Med. 2012; 10: 42
27.Jády BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body–specific localization signal. J Cell Biol. 2004; 164: 647-52.
28.Zhang L, Hou DX, Chen X, Li DH, Zhu LY, Zhang YJ, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012; 22: 107-26.
29.Philip A, Ferro VA, Tate RJ. Determination of the potential bioavailability of plant microRNAs using a simulated human digestion process. Mol Nutr Food Res. 2015; 59: 1962-72.
30.Lukasik A, Zielenkiewicz P. Plant MicroRNAs-novel players in natural medicine? Int J Mol Sci. 2016; 18: 9
31.Chin AR, Fong MY, Somlo G, Wu J, Swiderski P, Wu X, et al. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016; 26: 217-28.
32.Yang J, Hotz T, Broadnax L, Yarmarkovich M, Elbaz-Younes I, Hirschi KD. Anomalous uptake and circulatory characteristics of the plant-based small RNA MIR2911. Sci Rep. 2016; 6: 26834
33.Cooper EL, Overstreet N. Diversity, evolution, and therapeutic applications of small RNAs in prokaryotic and eukaryotic immune systems. Phys Life Rev. 2014; 11: 113-34.
34.Rogato A, Richard H, Sarazin A, Voss B, Navarro SC, Champeimont R, et al. The diversity of small non-coding RNAs in the diatom Phaeodactylum tricornutum. BMC Genomics. 2014; 15: 698
35.Westhof E, Fritsch V. RNA folding: beyond Watson-Crick pairs. Structure. 2000; 8: R55-65.
36.Jedrzejczyk D, Gendaszewska-Darmach E, Pawlowska R, Chworos A. Designing synthetic RNA for delivery by nanoparticles. J Phys Condens Matter. 2017; 29: 123001
37.Bahadori M. New advances in RNAs. Arch Iran Med. 2008; 11: 435-43.
38.Fire AZ. Gene silencing by double-stranded RNA. Cell Death Differ. 2007; 14: 1998-2012.
39.Mello CC. Return to the RNAi world: rethinking gene expression and evolution. Cell Death Differ. 2007; 14: 2013-20.
40.Walther W, Stein U. Viral vectors for gene transfer. Drugs. 2000; 60: 249-71.
41.Petros RA, DeSimone JM. Strategies in the design of nanoparticles for therapeutic applications. Nat Rev Drug Discov. 2010; 9: 615-27.
42.Shtam TA, Kovalev RA, Varfolomeeva EY, Makarov EM, Kil YV, Filatov MV. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun Signal. 2013; 11: 88
43.Martens-Uzunovaa ES, Böttchera R, Crocec CM, Jenstera G, Visakorpid T, Caline GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2013; 65: 1140-51.
44.Lunavat TR, Jang SC, Nilsson L, Park HT, Repiska G, Lässer C, et al. RNAi delivery by exosome-mimetic nanovesicles - Implications for targeting c-Myc in cancer. Biomaterials. 2016; 102: 231-8.
45.Bach DH, Hong JY, Park HJ, Lee SK. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int J Cancer. 2017; doi: 10.1002/ijc.30669.
46.O'Loughlin AJ, Mäger I, de Jong OG, Varela MA, Schiffelers RM, El Andaloussi S, et al. Functional delivery of lipid-conjugated siRNA by extracellular vesicles. Mol Ther. 2017; doi: 10.1016/j.ymthe.2017.03.021.
47.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001; 294: 858-62.
48.Mokhtarzadeh A, Alibakhshi A, Hashemi M, Hejazi M, Hosseini V, de la Guardia M, et al. Biodegradable nano-polymers as delivery vehicles for therapeutic small non-coding ribonucleic acids. J Control Release. 2017; 245: 116-26.
49.Simonson B, Das S. MicroRNA therapeutics: the next magicbullet? Mini Rev Med Chem. 2015; 15: 467-74.
50.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75: 843-54.
51.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000; 403: 901-6.
52.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001; 107: 823-6.
53.Eddy SR. Non–coding RNA genes and the modern RNA world. Nat Rev Genetics. 2001; 2: 919-29.
54.Craig VJ, Cogliatti SB, Imig J, Renner C, Neuenschwander S, Rehrauer H, et al. Myc-mediated repression of microRNA-34a promotes high-grade transformation of B-cell lymphoma by dysregulation of FoxP1. Blood. 2011; 117: 6227-36.
55.Sicard F, Gayral M, Lulka H, Buscail L, Cordelier P. Targeting miR-21 for the therapy of pancreatic cancer. Mol Ther. 2013; 21: 986-94.
56.Dreyfuss G, Philipson L, Mattaj IW. Ribonucleoprotein particles in cellular processes. J Cell Biol. 1988; 106: 1419-25.
57.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genetics. 2009; 41: 572-8.
58.Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat Struct Mol Biol. 2010; 17: 1030-1034.
59.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007; 316: 1484-8.
60.Hu CMJ, Zhang LF. Nanoparticle-based combination therapy toward overcoming drug resistance in cancer. Biochem Pharmacol. 2012; 83: 1104-11.
61.Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010; 17: F19-36.
62.Ahmed FE. Role of miRNA in carcinogenesis and biomarker selection: a methodological view. Expert Rev Mol Diagn. 2007; 7: 569-603.
63.Yang Z, Wu J. Small RNAs and development. Med Sci Monit. 2006; 12: RA125-9.
64.Mochizuki K, Gorovsky MA. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005; 19: 77-89.
65.Patel SB, Novikova N, Bellini M. Splicing-independent recruitment of spliceosomal small nuclear RNPs to nascent RNA polymerase II transcripts. J Cell Biol. 2007; 178: 937-49.
66.Li W, Jiang G, Zeng DB, Jin YX. Identification of six new box C/D snoRNA gene clusters from rice. IUBMB Life. 2007; 59: 664-74.
67.Barth S, Shalem B, Hury A, Tkacz ID, Liang XH, Uliel S, et al. Elucidating the role of C/D snoRNA in rRNA processing and modification in Trypanosoma brucei. Eukaryotic Cell. 2008; 7: 86-101.
68.Hertel J, Hofacker IL, Stadler PF. SnoReport: computational identification of snoRNAs with unknown targets. Bioinformatics. 2008; 24: 158-64.
69.Theurkauf WE, Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA. rasiRNAs, DNA damage, and embryonic axis specification. Cold Spring Harb Symp Quant Biol. 2006; 71: 171-80.
70.Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin HL. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 2007; 21: 3123-34.
71.Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, et al. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007; 35: 5430-8.
72.Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Zhu JK, et al. A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci USA. 2006; 103: 18002-7.
73.Lakshmi SS, Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008; 36: D173-7.
74.Sado T, Li E, Sasaki H. Effect of Tsix disruption on Xist expression in male ES cells. Cytogenet Genome Res. 2003; 99: 115-8.
75.Anguera MC, Sun BK, Xu N, Lee JT. X-chromosome kiss and tell: how the Xs go their separate ways. Cold Spring Harbor Symp Quant Biol. 2006; 71: 429-37.
76.Cohen HR, Panning B. XIST RNA exhibits nuclear retention and exhibits reduced association with the export factor TAP/NXF1. Chromosoma. 2007; 116: 373-83.
77.Liu X-Q, Sun C-Y, Yang X-Z, Wang J. Polymeric-micelle-based nanomedicine for siRNA delivery. Particle Particle Systems Characteriz. 2013; 30: 211-28.
78.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol Pharmaceutics. 2008; 5: 505-15.
79.Kanasty RL, Whitehead KA, Vegas AJ, Anderson DG. Action and reaction: the biological response to siRNA and its delivery vehicles. Mol Ther. 2012; 20: 513-24.
80.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013; 12: 967-77.
81.Yoo SM, Na D, Lee SY. Design and use of synthetic regulatory small RNAs to control gene expression in Escherichia coli. Nat Protoc. 2013; 8: 1694-707.
82.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001; 411: 494-8.
83.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001; 15: 188-200.
84.Huo WY, Zhao GN, Yin JG, Ouyang X, Wang YN, Yang CH, et al. Lentiviral CRISPR/Cas9 vector mediated miR-21 gene editing inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J Cancer. 2017; 8: 57-64.
85.Brosseau J-P, Lucier J-F, Nwilati H, Thibault P, Garneau D, Gendron D, et al. Tumor microenvironment–associated modifications of alternative splicing. RNA. 2014; 20: 189-201.
86.Rhoads RE. Synthetic mRNA: production, introduction into cells, and physiological consequences. In: Rhoads RE, editor. Synthetic mRNA: Production, Introduction Into Cells, and Physiological Consequences. Springer: New York, 2016; 1428: 3-27.
87.McNamara MA, Nair SK, Holl EK. RNA-based vaccines in cancer immunotherapy. J Immunol Res. 2015; 2015: 794528
88.Gillet JP, Macadangdang B, Fathke RL, Gottesman MM, Kimchi-Sarfaty C. The development of gene therapy: from monogenic recessive disorders to complex diseases such as cancer. In: Walther W, Stein US, editors. Gene Therapy of Cancer: Methods and Protocols. Humana Press: New York, 2009. p.5-54.
89.Ginn SL, Alexander IE, Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2012–an update. J Gene Med. 2013; 15: 65-77.
90.Farooqi AA, Rehman Z ur, Muntane J. Antisense therapeutics in oncology: current status. Onco Targets Ther. 2014; 7: 2035-42.
91.Gupta GJC, Pednekar PP, Jadhav KR, Chilajwar SV, Kadam VJ. Non viral synthetic siRNA delivery system an efficient tool for cancer treatment. World J Pharm Pharmaceut Sci. 2014; 3: 351-87.
92.Guo W, Chen WB, Yu WD, Huang WL, Deng WG. Small interfering RNA-based molecular therapy of cancers. Chin J Cancer. 2013; 32: 488-93.
93.Hoerr I, Obst R, Rammensee HG, Jung G. In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies. Eur J Immunol. 2000; 30: 1-7.
94.Steitz J, Britten CM, Wölfel T, Tüting T. Effective induction of anti-melanoma immunity following genetic vaccination with synthetic mRNA coding for the fusion protein EGFP.TRP2. Cancer Immunol Immunother. 2006; 55: 246-53.
95.Chen AM, Zhang M, Wei DG, Stueber D, Taratula O, Minko T, et al. Co-delivery of doxorubicin and Bcl-2 siRNA by mesoporous silica nanoparticles enhances the efficacy of chemotherapy in multidrug-resistant cancer cells. Small. 2009; 5: 2673-7.
96.Li JM, Wang YY, Xue SS, Sun JH, Zhang W, Wu P, et al. Effective combination treatment of lung cancer cells by single vehicular delivery of siRNA and different kinds of anticancer. Int J Nanomed. 2016; 11: 4609-24.
97.Gozuacik D, Yagci-Acar HF, Akkoç Y, Kosar A, Dogan-Ekici AI, Ekici S. Anticancer use of nanoparticles as nucleic acid carriers. J Biomed Nanotechnol. 2014; 10: 1751-83.
98.Martínez AM, Benito M, Pérez E, Teijón JM, Blanco MD. The role of anionic polysaccharides in the preparation of nanomedicines with anticancer applications. Curr Pharmaceut Design. 2016; 22: 3364-79.
99.Yuan TL, Fellmann C, Lee CS, Ritchie CD, Thapar V, Lee LC, et al. Development of siRNA payloads to target KRAS-mutant cancer. Cancer Discov. 2014; 4: 1182-97.
100.Jeong JH, Mok H, Oh YK, Park TG. siRNA conjugate delivery systems. Bioconjugate Chem. 2008; 20: 5-14.
101.Wang Y, Chen X, Tian B, Liu J, Yang L, Zeng L, et al. Nucleolintargeted extracellular vesicles as a versatile platform for biologics delivery to breast cancer. Theranostics. 2017; 7: 1360-72.
102.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990; 346: 818-22.
103.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990; 249: 505-10.
104.McKeague M, DeRosa MC. Challenges and opportunities for small molecule aptamer development. J Nucleic Acids. 2012; 2012: 748913
105.Darmostuk M, Rimpelova S, Gbelcova H, Ruml T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol Adv. 2015; 33: 1141-61.
106.Sundaram P, Kurniawan H, Byrne ME, Wower J. Therapeutic RNA aptamers in clinical trials. Eur J Pharm Sci. 2013; 48: 259-71.
107.Chen Y, Xu MJ, Guo Y, Tu KY, Wu WM, Wang JJ, et al. Targeted chimera delivery to ovarian cancer cells by heterogeneous gold magnetic nanoparticle. Nanotechnology. 2017; 28: 025101
108.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010; 9: 537-50.
109.Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010; 11: 593-8.
110.Zhao YY, Miao H, Cheng XL, Wei F. Lipidomics: Novel insight into the biochemical mechanism of lipid metabolism and dysregulation-associated disease. Chem Biol Interact. 2015; 240: 220-38.
111.Ewe A, Aigner A. Cationic lipid-coated polyplexes (Lipopolyplexes) for DNA and small RNA delivery. In: Candiani G, editor. Non-Viral Gene Delivery Vectors. Springer: New York, 2016; 1445: 187-200.
112.Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. In vivo activity of nuclease-resistant siRNAs. RNA. 2004; 10: 766-71.
113.Braasch DA, Jensen S, Liu YH, Kaur K, Arar K, White MA, et al. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003; 42: 7967-75.
114.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003; 9: 1034-48.
115.Bao YJ, Jin Y, Chivukula P, Zhang J, Liu Y, Liu J, et al. Effect of PEGylation on biodistribution and gene silencing of siRNA/lipid nanoparticle complexes. Pharm Res. 2013; 30: 342-51.
116.Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005; 12: 461-6.
117.Grayson ACR, Doody AM, Putnam D. Biophysical and structural characterization of polyethylenimine-mediated siRNA delivery in vitro. Pharm Res. 2006; 23: 1868-76.
118.Philipp A, Zhao XB, Tarcha P, Wagner E, Zintchenko A. Hydrophobically modified oligoethylenimines as highly efficient transfection agents for siRNA delivery. Bioconjugate Chem. 2009; 20: 2055-61.
119.Jiao J, Zou Q, Zou MH, Guo RM, Zhu S, Zhang Y. Aptamermodified PLGA nanoparticle delivery of triplex forming oligonucleotide for targeted prostate cancer therapy. Neoplasma. 2015; 63: 569-75.
120.Parisi OI, Morelli C, Puoci F, Saturnino C, Caruso A, Sisci D, et al. Magnetic molecularly imprinted polymers (MMIPs) for carbazole derivative release in targeted cancer therapy. J Mater Chem B. 2014; 2: 6619-25.
121.Koldehoff M, Steckel NK, Beelen DW, Elmaagacli AH. Therapeutic application of small interfering RNA directed against bcr-abl transcripts to a patient with imatinib-resistant chronic myeloid leukaemia. Clin Exp Med. 2007; 7: 47-55.
122.Zou Y, Guo CG, Yang ZG, Sun JH, Zhang MM, Fu CY. A small interfering RNA targeting vascular endothelial growth factor efficiently inhibits growth of VX2 cells and VX2 tumor model of hepatocellular carcinoma in rabbit by transarterial embolizationmediated siRNA delivery. Drug Des Devel Ther. 2016; 10: 1243-55.
123.Vaishnaw A. Dose escalation trial to evaluate the safety, tolerability, pharmacokinetics and pharmacodynamics of intravenous ALN-VSP02 in patients with advanced solid tumors with liver involvement. NCT00882180. NIH: USA, 2011.
124.Tabernero J, Shapiro GI, LoRusso PM, Cervantes A, Schwartz GK, Weiss GJ, et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013; 3: 406-17.
125.Aleku M, Schulz P, Keil O, Santel A, Schaeper U, Dieckhoff B, et al. Atu027, a liposomal small interfering RNA formulation targeting protein kinase N3, inhibits cancer progression. Cancer Res. 2008; 68: 9788-98.
126.Strumberg D. Atu027 plus gemcitabine in advanced or metastatic pancreatic cancer (Atu027-I-02) (Atu027-I-02). NCT01808638. NIH: USA, 2013.
127.Ribas A, Tolcher AW, Yen Y. Safety study of CALAA-01 to treat solid tumor cancers. NCT00689065. NIH: USA, 2008.
128.Rudloff U. A phase 1 dose escalation study of hepatic intra-arterial administration of TKM 080301 (lipid nanoparticles containing siRNA against the PLK1 gene product) in patients with colorectal, pancreas, gastric, breast, ovarian and esophageal cancers with hepatic. NCT01437007. NIH: USA, 2011.
129.Pruitt C. Immunotherapy of melanoma with tumor antigen rna and small inhibitory rna transfected autologous dendritic cells. NCT00672542. NIH: USA, 2014.
130.O'Reilly EM, Golan T. A phase II study of siG12D LODER in combination with chemotherapy in patients with unresectable locally advanced pancreatic cancer. NCT01676259. NIH: USA, 2017.
131.Triozzi P. APN401 in treating patients with recurrent or metastatic pancreatic cancer, colorectal cancer, or other solid tumors that cannot be removed by surgery. NCT03087591. NIH: USA, 2017.
Cite this article as: Yadav S, Shekhawat M, Jahagirdar D, Kumar Sharma N. Natural and artificial small RNAs: a promising avenue of nucleic acid therapeutics for cancer. Cancer Biol Med. 2017; 14: 242-53. doi: 10.20892/j.issn.2095-3941.2017.0038
Nilesh Kumar Sharma
E-mail: nilesh.sharma@dpu.edu.in
April 12, 2017; accepted May 22, 2017.
Available at www.cancerbiomed.org
Copyright © 2017 by Cancer Biology & Medicine
 Cancer Biology & Medicine2017年3期
Cancer Biology & Medicine2017年3期
- Cancer Biology & Medicine的其它文章
- Aberrant control of NF-κB in cancer permits transcriptional and phenotypic plasticity, to curtail dependence on host tissue: molecular mode
- 2016 Chinese expert consensus and guidelines for the diagnosis and treatment of papillary thyroid microcarcinoma
- Nanotechnology-based combination therapy for overcoming multidrug-resistant cancer
- BRAF mutation in colorectal carcinomas with signet ring cell component
- Recent progress on nanoparticle-based drug delivery systems for cancer therapy
- Preclinical and clinical applications of specific molecular imaging for HER2-positive breast cancer
