Expression of a novel Ceratopteris thalictroides-derived anti-HIV protein CVNH and its mutant in Pichia pastoris
CUI Limin, WANG Zhen, WANG Ting, SU Yingjuan, 4
(1. School of Life Sciences, Sun Yat-sen University, Guangzhou 510275, China; 2. College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, China; 3. College of Life Sciences, South China Agricultural University, Guangzhou 510642, China; 4. Shenzhen Research Institute of Sun Yat-sen, Shenzhen 518057, China)
Expression of a novel Ceratopteris thalictroides-derived anti-HIV protein CVNH and its mutant in Pichia pastoris
CUI Limin1, WANG Zhen2, WANG Ting3, SU Yingjuan1, 4
(1. School of Life Sciences, Sun Yat-sen University, Guangzhou 510275, China; 2. College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, China; 3. College of Life Sciences, South China Agricultural University, Guangzhou 510642, China; 4. Shenzhen Research Institute of Sun Yat-sen, Shenzhen 518057, China)
CVN (Cyanovirin-N) is a highly potent anti-HIV protein. Cyanovirin-N homology (CVNH) family is a homologous protein to CVN, which exhibits a common fold structure similar to CVN and possess a highly conserved anti-HIV CVN domain. In this study, DNA encoding sequences of the wild-type and mutant CtCVNH fromCeratopteristhalictroideswere separately cloned into eukaryotic expression vector pPICZαA. AllwCtCVNHand mutatedmCtCVNHgenes produced two bands corresponding to 22 000 and 24 000, which were validated by western blot. The proteins were purified by ultra-filtered centrifugation and Ni-NTA affinity chromatograph. Glycosylation assay further confirmed that no N-glycosylation site occurred in wCtCVNH protein. The possible reasons were ascribed to incomplete cleavage of the α-factor signal peptide sequence in vector pPICZαA or some other post-translational modifications rather than N-glycosylation. The study laid a solid foundation for further exploring the pharmacokinetics and pharmacodynamics of CtCVNH in future.
anti-HIV protein;Ceratopteristhalictroides; mutant;Pichiapastoris
AIDS (Acquired Immune Deficiency Syndrome) is a global concern epidemic caused by human immunodeficiency virus (HIV), which forces a great impact on society and economy. Globally, 36.9 million people are living with HIV until the end of 2014. Over the past three decades, more than 25 anti-HIV compounds are used for treatments of AIDS[1-2], such as reverse transcriptase inhibitors, protease inhibitors, integrase inhibitors, and cell entry inhibitors. Unfortunately, defects of these antiviral drugs in high costs, resistance development, serious side effects, or pharmacokinetic interactions greatly limit their use. Hence, it is top priority to develop safe, efficacious, and natural anti-HIV agents.
Cyanovirin-N (CVN) is a novel and structurally unique anti-HIV protein isolated from the cyanobacteriumNostocellipsosporum[3-4]. Eleven-kDa CVN exhibits very low homology to any other known protein and contains a two-fold pseudosymmetry[5-6]. Its antiviral activity is ascribed to specifically bind to HIV-1 envelope glycoproteins gp120 to prevent interaction of the virus with a host cell[7-8]. Potent activity of CVN reflected in irreversibly neutralizing HIV-1 at low nanomolar concentrations[3]. In addition, it is also active against other viruses such as SIV, Ebola, influenza, and hepatitis C[9-12]. CVN is a remarkably stable protein, which is insensitive to organic solvents, high concentrations of salt, strong detergents, continuous freeze-thaw cycles and high temperature[3, 5].
Cyanovirin-N homology (CVNH) family is a homologous protein to CVN, which exhibits a common fold structure similar to CVN[13]and possess a highly conserved anti-HIV CVN domain[14-15]. CVNHs are also potential anti-HIV agents. The protein is widely distributed in bacteria, fungi, and fern[13-15]. CtCVNH, which is from fernCeratopteristhalictroides[16], is expressed inEscherichiacoli[17-18]. However, due to lack of post-translational modifications and formation of inclusion body in prokaryotic systems, it is necessary to consider eukaryotic hosts for cost-efficient and mass functional production. In addition, because of the possible occurance of typical drawbacks such as short plasma half-life, proteolysis, and immunogenicity, it is also key to study expression of its mutant in eukaryotic systems.
Pichiapastorisis increasing popular as an exogenous gene expression system[19], which has been widely applied in drug and pharmaceutical industries[20]. Its advantages embody in easy to operate and obtain high density cell cultivation with low cost[21]. Furthermore,P.pastoriscan produce large quantities of foreign secretory proteins[21]. Most importantly,P.pastorissuccessfully performs protein post-translational modifications, such as phosphorylation or glycosylation[21-23]. Nowadays, more than 500 proteins have been successfully expressed inP.pastoris[24-25], such as antigen 5 (Ag5), bovine neutrophil, β-defensins, and human neuritin[26- 28].
In this study, we obtained wild-type and mutated CtCVNH genes. They were successfully expressed inP.pastoris, respectively. The present study lays a solid foundation for further pharmaceutical investigation on CtCVNHs.
1 Materials and methods
1.1 Materials
Oligonucleotide primers were synthesized by BGI (Guangzhou, China).TaqDNA polymerase, DNA marker, protein marker were purchased from Takara (Dalian, China). Restriction enzymes (EcoRⅠ、NotⅠ、SacⅠ) and T4 DNA ligase were from New England Biolabs (MA, USA). ZeocinTMwere obtained from Invitrogen (Carlsbad, CA, USA). Ni-NTA His Bind Reisn was acquired from Novagen (San Diego, CA, USA). Amicon Ultra-15 centrifugal filter units (10 000) were purchased from Millipore (Billerica, MA, USA). All chemicals used in this study were of analytical grade.
1.2 Methods
1.2.1 Strains, plasmid and media TheP.pastorisstrain GS115 (Invitrogen) was used to express proteins. Plasmid pPICZαA (Invitrogen) was served as an expression vector.E.coliDH5α (Tiangen) was used as the host for gene manipulation and plasmid amplification.E.coliwas maintained in low-salt LB medium (1 % tryptone, 0.5% yeast extract, 0.5% sodium chloride, and 25 μg/mL Zeocin). TheP.pastorisstrain GS115 was grown in different media including YPD medium (2% peptone, 1% yeast extract, 2%D-glucose), YPDS plates (2% peptone, 1% yeast extract, 18.22% sorbitol, 2%D-glucose, and 2% agar), MD/MM plates (1.34% YNB, 4×(10%~5%) biotin, 2 %D-glucose or 0.5% methanol, and 2% agar), and BMGY/BMMY medium (2% peptone, 1% yeast extract, 100 mmol/L potassium phosphate buffer (pH 6.0), 1.34% yeast nitrogen base, 4×(10%~4%) biotin, and 1% glycerol or 0.5% methanol).
1.2.2 Amplification and synthesis of theCtCVNHgenes The coding sequence of wild-typeCtCVNH(wCtCVNH) was amplified from previous pBluescript-wCtCVNH in our lab. The primers were 5′-CCGGAATTCATGGAGTACTTTACACCGC-3′ (forward primer), which contains anEcoR Ⅰ restriction site (underlined), and 5′-ATTTGCGGCCGCTCCGCTGCTTGCTTCTTC-3′(reverse primer), which contains aNotⅠ site (underlined). The PCR conditions were set as follows: 94 ℃ for 5 min, 30 cycles of 94 ℃ for 45 s, 57.5 ℃ for 45 s, 72 ℃ for 1 min, and a final extension at 72 ℃ for 7 min. The PCR products were separated on 1.5% agarose gel.
The potential N-glycosylation site of wCtCVNH was predicted using NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/). Three amino acids, Asn (23), Pro (76), and Pro (108), were mutated by Ala, Gly, and Gly, respectively. An Asn23Ala mutation was performed to eliminate the N-glycosylation motif, whereas a Pro76Gly mutation was used to avoid protein misfolding and conformational heterogeneity. The key physicochemical profiles of the mutant, positive charge, isoelectric point, and instability index were analyzed by ProtParam (http://web.expasy.org/protparam/). To achieve high-level expression of CtCVNH inP.pastoris, thewCtCVNHgene was optimized according to Codon Usage Database (http://www.jcat.de/). Finally, the mutatedCtCVNHgene (mCtCVNH) also designed with anEcoR Ⅰ restriction site and aNotⅠ restriction site was synthesized by BGI (Guangzhou, China).
1.2.3 Construction of the expression vectors The resulting PCR product ofwCtCVNHandmCtCVNHwere digested withEcoR Ⅰ andNotⅠ and ligated to the corresponding site of the expression vector pPICZαA, respectively (Fig.1). The recombinant plasmids were transformed intoE.coliDH5α competent cells, and cultured on low-salt LB agar plates containing 25 μg/mL Zeocin. Transformants were confirmed by restriction endonuclease digestion and DNA sequencing.
1.2.4 Transformation ofP.pastorisand target transformants screening After the two plasmids constructed above were linearized withSacⅠ, they were transformed intoP.pastorisGS115 competent cells by elec troporation at 1.5 kV, 25 μF, 200 Ω and 5 ms, respectively. Empty vector pPICZαA was used in the same conditions as a negative control. Transformed cells were plated onto MD agar plates for selection of His+ transformants at 28 ℃ for 2~3 days. Some His+ transformants were selected on YPDS plates with different concentrations of Zeocin (100, 200, 500, 800, 1 000 μg/mL)[29]. For Mut+colonies, Zeocin-resistant clones were picked on both selective MM and MD plates. The yeast transformants were selected and inoculated into YPD medium at 28 ℃ with shaking at 250 r/min. The “boiling-freezing-boiling” method was used to extract recombinant yeast genomic DNA[30]. The recombinant yeast strain was identified by PCR with the primers specific for the target gene and 5′α-Factor/3′AOX1 for the recombinant vector (Table 1).
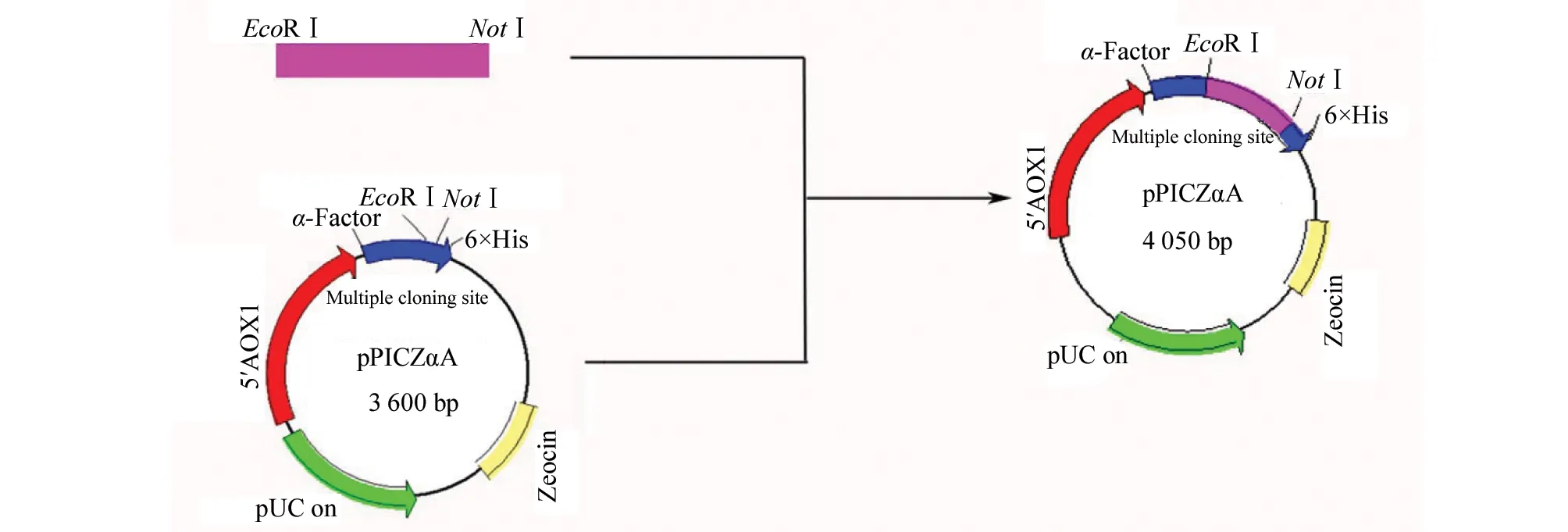
Fig.1 Construction strategy of expression vector pPICZαA. α-factor: native Saccharomyces cerevisiae α-mating factor secretion signal AOX1: methanol-inducible alcohol oxidase promoter; pUC ori: replication and maintenance of the plasmid in Escherichia coli
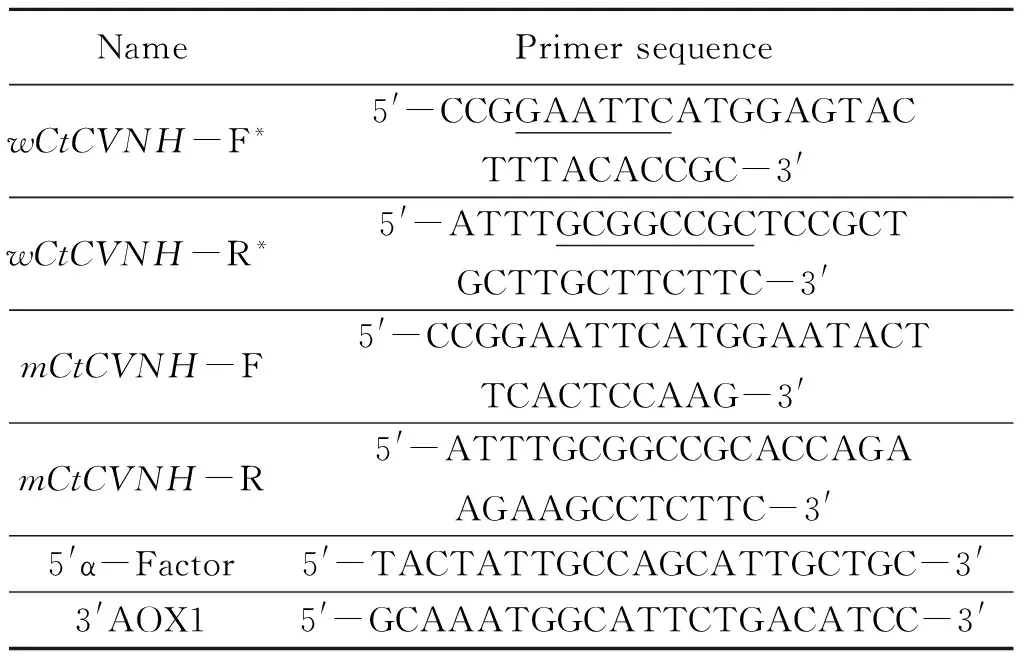
Table 1 Sequences of Primers
* Restriction sites are underlined
1.2.5 Expression of the CtCVNH proteins inP.pastorisTheP.pastoristransformants were inoculated into 9 mL YPD at 28 ℃ with 250 r/min for 16 h. Then 250 μL culture were transferred to 25 mL BMGY medium in a 250 mL baffled flasks. The transformants were grown in a shaking incubator at 28 ℃ with 250 rpm of agitation for 16~24 h until theA600was equal to 2~6. After the yeast cells were harvested by centrifuging at 4 000gfor 5 min at room temperature, the pellets were suspended in 100 mL BMMY medium for cultivation tillA600reachedto1.Methanol(100%)wasaddedtotheculturemediumatanevery24hintervaltothefinalconcentrationof1.0%tomaintaintheinduction.Theculturemediumwascollectedat24, 48, 72,and96h.Thesupernatantswereharvestedbycentrifugationat12000gfor 10 min.
1.2.6 SDS-PAGE analysis Supernatants were concentrated by TCA-acetone. The pellets were dried, dissolved in 20 μL distilled water, and separated on 15% SDS-PAGE gels. The protein bands were visualized by staining with Coomassie brilliant blue R-250.
1.2.7 Protein Purification The recombinant proteins wCtCVNH and mCtCVNH were purified by Ni-NTA affinity chromatography. After 96 h of methanol treatment, the cultures were collected by 12 000gcentrifugation for 10 min at 4 ℃ and filtered through a 0.2 μm filter. The supernatants were concentrated using a 10 000 MWCO ultrafiltration membrane. After mixed with 1 mL Ni-NTA resin for 3 h, the supernatant were applied to an affinity column. The column was first washed with 5~10 column volume of buffer A (50 mmol/L PBS, 500 mmol/L NaCl, 20 mmol/L imidazole, pH 7.4) to remove the unbinding proteins, and then eluted using buffer B (50 mmol/L PBS, 500 mmol/L NaCl, 200 mmol/L imidazole, pH 7.4). Eluted fractions were separated by 15% SDS-PAGE. The fractions containing wCtCVNH/mCtCVNH were collected, pooled, and desalted with an Ultra-3 000 device (Millipore, USA) using 10 mmol/L PBS buffer (pH 7.4), lyophilized, and stored at -80 ℃ for further analysis.
1.2.8 Western blot analysis After electrophoresis, the proteins were electrically transferred from the gel onto 0.2 μm nitrocellulose (NC) membrane. The membrane was blocked with TBST (20 mmol/L Tris-HCl, 150 mmol/L NaCl, 0.05% Tween 20) containing 5% skim milk for 2 h at room temperature. The NC membrane was first incubated with mouse anti-His monoclonal antibody (1∶3 000) at 4 ℃ overnight. After three times washing, the membrane was incubated with goat anti-mouse secondary antibody conjugated to horseradish peroxidase (1∶5 000) for 2 h at room temperature with gentle shaking. Target proteins were developed with ECL Chemiluminescent Kit and visualized using Syngene GBox-Chemi-XT4-E imaging system.
1.2.9 Determination of glycosylation sites In order to examine whether wCtCVNH was post-translationally glycosylated, we performed glycosylation assay. wCtCVNH was digested with Peptide N-glycosidase F (PNGase F, New England Biolabs, MA, USA) for 1 h at 37 ℃ according to manufacturer’s instructions. The products were separated on SDS-PAGE and analyzed by Western blot.
2 Results
2.1mCtCVNHdesign
Mutations did not change the isoelectric point (pI) and net charge of mCtCVNH. Compared to wide-type CtCVNH, the stability of mCtCVNH improved (22.69 and 24.21, respectively). In addition, codon optimization also increased the expression level of mCtCVNH inP.pastoris.
2.2 Construction of the expression plasmids, transformation ofP.pastoris, and screening of the positive transformants
ThewCtCVNHandmCtCVNHgene were confirmed to be successfully inserted into the vector pPICZαA, respectively. Both target genes were verified by restriction endonuclease and DNA sequencing. PCR was employed to detect the positive transformants with target genes (Fig.2a and Fig.2b). The 450 bp band was obtained using the gene-specific primer pairs, and the 650 bp product was amplified by the primer pair 5′α-Factor/3′AOX1, which suggested that the recombinant plasmids were successfully transformed into the host yeast.

Fig.2 Identification of Pichia integrant(a) PCR identification of positive transformants GS115-pPICZαA-wCtCVNH. M: 100 bp DNA marker; 1-3: negative control; 4-5: fragment from pPICZαA-wCtCVNH amplified by 5′α-Factor/3′AOXI primers; 6-7: fragment from pPICZαA-wCtCVNH amplified by the specific forward/reverse primers(b) PCR analysis of positive transformants GS115-pPICZαA-mCtCVNH. 1: negative control; 2-4: the transformed yeast cells containing pPICZαA-mCtCVNH with the specific forward/reverse primers; M: 200 bp DNA Marker; 5-8: the transformed yeast cells containing pPICZαA-mCtCVNH with 5′α-Factor/3′AOXI primers
2.3 Expression of recombinant proteins inP.pastoris
For wCtCVNH expression, two bands were detected corresponding to molecular mass approximately 22 000 and 24 000, which were higher than the predicted molecular weight (20 000) (Fig.3a). Similarly, mCtCVNH expressed protein also had two bands (24 000 and 26 000) (Fig.3b). The phenomena suggested that wCtCVNH synthesized by the yeast should be glycosylated. However, after wCtCVNH was treated with PNGase F for deglycosylation, two protein bands were apparent on the SDS-PAGE gel, which were confirmed by Western blot (Fig.4). The results indicated that no N-glycosylation site occurred in wCtCVNH protein.

Fig.3 SDS-PAGE analysis of the recombinant CtCVNHs using P. pastoris expression system (a) Recombinant wCtCVNH. 1: negative control; M: molecular protein marker; 2-5: culture supernatant (b) Recombinant mCtCVNH. M: molecular protein marker; 1: negative control; 2-4: culture supernatant.

Fig.4 Identification of N-glycosylation of wCtCVNH in yeast by western blot 1: two wCtCVNH bands after treated with PNGase F
2.4 Protein purification and Western blot analysis
Two protein bands from the supernatant were obtained by Ni-NTA resin purification (Fig.5a and Fig.6a). The purified proteins were further confirmed by Western blot with mouse anti-His tag monoclonal antibody and goat anti-mouse IgG antibody conjugated to horseradish peroxidase (Fig.5b and Fig.6b).

Fig.5 Electrophoresis and western blot after purification of recombinant wCtCVNH by affinity chromatography(a) SDS-PAGE. M: protein marker; 1: purified wCtCVNH; (b) Western blot. 1: negative control; 2: expressed wCtCVNH

Fig.6 SDS-PAGE and western blot analysis of purified mCtCVNH (a) SDS-PAGE. M: Protein marker; 1-2: purified mCtCVNH; (b) Western blot. M: Protein marker; 1-2: expressed mCtCVNH; 3: negative control
3 Discussion
Although CtCVNH has been successfully expressed inE.coli, it was expressed as inactive inclusion bodies[17], which is considered time-consuming, labor-intensive and not cost-effective[17]. Furthermore, recovery of protein fromE.coliinclusion bodies is very low. Currently, there has been no report on the expression of CVNH in a yeast expression system. Here,P.pastoriswas first selected to be the expression host of CtCVNH. As a eukaryotic host for the expression of recombinant proteins,P.pastorisis one of the most efficient cell factories for protein engineering and production[21]. Especially,P.pastorisexpression vector pPICZaA harbors alcohol oxidase 1 (AOX1) promoter, which is induced by methanol[31]. Methanol is sole source of carbon and inducer substrate[32]. Compared to other protein expression systems, it is easier to manipulate, culture, and undergone all the post-translational modifications[21]. Recombinant proteins are secreted into the culture medium, which do not need mechanical or chemical cell disruption and usually facilitate subsequent purification process. The purification of proteins was successfully achieved by affinity chromatography with a 6-His·tag. More importantly, free of endotoxins in yeast can reduce enormously the risk of side effects.
In this study, we generated recombinant wCtCVNH and mCtCVNH by usingP.pastoris. InP.pastorisexpression system, the most common is N-glycosylation and post-translational modification[33]. If removal of N-glycan, proteins still possess extra molecular weight, which are possibly caused by post-translational modifications[33]. In this study, the wild-typeCtCVNHgene was expressed inP.pastoris, which produced two bands corresponding to 22 000 and 24 000, respectively. The two proteins were further identified by western blot. Similarly,mCtCVNHstill expressed two protein bands, which were also confirmed by western blot. The results were totally different from only one protein band expressed inE.coli. The possible reasons were ascribed to incomplete cleavage of the α-factor signal peptide sequence in vector pPICZαA or some other post-translational modifications rather than N-glycosylation. All of the cyanovirins were expressed as fusion proteins with α-factor in yeasts[34]. The phenomena of two bands occur in yeast expression of other proteins, such as FIP-fve from the mushroomFlammulinavelutipes[35]and interferon α from the bovine liver[36]. In addition, expression of CVN inP.pastorisindicates that CVN is N-glycosylated at Asn30, which is inactive against HIV[34]. On the contrary, there were no glycosylation sites in CtCVNH expressed inP.pastoris.
Mutation is one of the best ways to improve the activity of protein. Although CVN has been successfully expressed inE.coli, yeast, and transgenic plant, respectively[3, 34, 37-38], its derivatives, CVN2, LCVN, and 10 K PEG-ALD-LCVN[39-40], have displayed enhanced anti-HIV-1 activity. Hence, successful expression of wCtCVNH and mCtCVNH inP.pastoriswill contribute to investigate how to further improve anti-HIV activity of CtCVNH. The study also laid a solid foundation for further exploring the pharmacokinetics and pharmacodynamics of CtCVNH in future.
4 Conclusions
For the first time, we successfully expressed the wild-type and mutated CtCVNH genes inP.pastoris, respectively. To better understand CtCVNH, future research will emphasize on the biologically activity, large-scale preparation, and clinical trials of wCtCVNH and its mutant.
[1] de CLERCQ E.Anti-HIV drugs:25 compounds approved within 25 years after the discovery of HIV[J].Int J Antimicrob Agents,2009,33:307-320.
[2] ZHAN P,PANNECOUQUE C,de CLERCQ E,et al.Anti-HIV drug discovery and development:current innovations and future trends[J].J Med Chem,2016,59:2849-2878.
[3] BOYD M R,GUSTAFSON K R,MCMAHON J B,et al.Discovery of cyanovirin-N,a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120:potential applications to microbicide development[J]. Antimicrobial Agents Chemother,1997,41(7):1521-1530.
[4] GUSTAFSON K R,SOWDER R C,HENDERSON L E,et al.Isolation,primary sequence determination,and disulfide bond structure of cyanovirin-N,an anti-HIV ( human immunodeficiency virus) protein from the cyanobacteriumNostocellipsosporum[J].Biochem Biophys Res Commun,1997,238(1):223-228.
[5] BEWLEY C A,GUSTAFSON K R,BOYD M R,et al.Solution structure of cyanovirin-N,a potent HIV-inactivating protein[J].Nat Struct Biol,1998,5:571-578.
[6] ZAPPE H,SNELL M E,BOSSARD M J. PEGylation of cyanovirin-N, an entry inhibitor of HIV[J].Adv Drug Deliver Rev,2008,60:79-87.
[7] ESSER M T,MORI T,MONDOR I,et al.Cyanovirin-N binds to gp120 to interfere with CD4-dependent human immunodeficiency virus type 1 virion binding,fusion,and infectivity but does not affect the CD4 binding site on gp120 or soluble CD4-induced conformational changes in gp120[J].J Virol,1999,73:4360-4371.
[8] MORI T,BOYD M R.Cyanovirin-N, a potent human immunodeficiency virus-inactivating protein,blocks both CD4-dependent and CD4-independent binding of soluble gp120 (sgp120) to target cells,inhibits sCD4-induced binding of sgp120 to cell-associated CXCR4,and dissociates bound sgp120 from target cells[J].Antimicrob Agents Chemother,2001,45:664-672.
[9] BARRIENTOS L G,GRONENBORN A M.The highly specific carbohydrate-binding protein cyanovirin-N:Structure,anti-HIV/Ebola activity and possibilities for therapy[J].Mini-Rev Med Chem,2005,5:21-31.
[10] BARRIENTOS L G,O′KEEFE B R,BRAY M,et al.Cyanovirin-N binds to the viral surface glycoprotein,GP(1,2) and inhibits infectivity of Ebola virus[J].Antiviral Res,2003,58:47-56.
[11] O′KEEFE B R,SMEE D F,TURPIN J A,et al.Potent anti-influenza activity of cyanovirin-N and interactions with viral hemagglutinin[J].Antimicrob Agents Chemother,2003,47:2518-2525.
[12] DEY B,LERNER D L,LUSSO P,et al.Multiple antiviral activities of cyanovirin-N:Blocking of human immunodeficiency virus type 1 gp120 interaction with CD4 and coreceptor and inhibition of diverse enveloped viruses[J].J Virol,2000,74:4562-4569.
[13] PERCUDANI R,MONTANINI B,OTTONELLO S.The anti-HIV cyanovirin-N domain is evolutionarily conserved and occurs as a protein module in eukaryotes[J].Proteins:Struct Funct Bioinf,2005,60:670-678.
[14] KOHARUDIN L M I,VISCOMI A R,JEE J G,et al.The evolutionarily conserved family of cyanovirin-N homologs:structures and carbohydrate specificity[J].Structure,2008,16:570-584.
[15] MATEI E,LOUIS J M,JEE J G,et al.NMR solution structure of a cyanovirin homolog from wheat head blight fungus[J].Proteins:Struct Funct Bioinf,2011,79:1538-1549.
[16] QI X Q,YANG Y X,SU Y J,et al.Molecular cloning and sequence analysis of cyanovirin-N homology gene inCeratopteristhalictroides[J].Am Fern J,2009,99:78-92.
[17] SUN J B,SU Y J,WANG T.Expression,purification and identification of CtCVNH,a novel anti-HIV (human immunodeficiency virus) protein fromCeratopteristhalictroides[J].Int J Mol Sci,2013,14:7506-7514.
[18] 李宁,孙君波,韦剑,等.抗艾滋病病毒蛋白 CVNH 在大肠杆菌中表达条件的优化[J].中山大学学报(自然科学版),2013,52(2):90-96. LI N,SUN J B,WEI J,et al.Optimization for culture condition of CVNH inEscherichiacoli, a novel anti-HIV(human immunodeficiency virus) protein[J].Acta Sci Natur Univ Sunyatseni,2013,52(2):90-96.
[19] AHMAD M,HIRZ M,PICHLER H,et al.Protein expression inPichiapastoris:recent achievements and perspectives for heterologous protein production[J].Appl Microbiol Biotechnol,2014,98:5301-5317.
[20] VOGL T,HARTNER F S,GLIEDER A.New opportunities by synthetic biology for biopharmaceutical production inPichiapastoris[J].Curr Opin Biotechnol,2013,24:1094-1101.
[21] DALY R,HEARN M T.Expression of heterologous proteins inPichiapastoris:a useful experimental tool in protein engineering and production[J].J Mol Recognit,2005,18:119-138.
[22] CREGG J M,CEREGHINO J L,SHI J Y,et al.Recombinant protein expression inPichiapastoris[J].Mol Biotechnol,2000,16:23-52.
[23] de SCHUTTER K,LIN Y C,TIELS P,et al.Genome sequence of the recombinant protein production hostPichiapastoris[J].Nat Biotechnol,2009,27:561-566.
[24] WEINACKER D,RABERT C,ZEPEDA A B,et al.Applications of recombinantPichiapastorisin the healthcare industry[J].Braz J Microbiol,2013,44:1043-1048.
[25] RABERT C,WEINACKER D,PESSOA A,et al.Recombinants proteins for industrial uses:utilization ofPichiapastorisexpression system[J].Braz J Microbiol,2013,44:351-356.
[26] MONSALVE R I,LU G,KING T P.Expressions of recombinant venom allergen,antigen 5 of yellowjacket (Vespulavulgaris) and paper wasp (Polistesannularis),in bacteria or yeast[J].Protein Expr Purif,1999,16:410-416.
[27] KANG J,ZHAO D,LYU Y,et al.Antimycobacterial activity ofPichiapastoris-derived mature bovine neutrophil beta-defensins 5[J].Eur J Clin Microbiol,2014,33:1823-1834.
[28] ZHANG Y,ZHANG S,XIAN L,et al.Expression and purification of recombinant human neuritin fromPichiapastorisand a partial analysis of its neurobiological activity in vitro[J].Appl Microbiol Biotechnol,2015,99:8035-8043.
[29] HU H,GAO J,HE J,et al.Codon optimization significantly improves the expression level of a keratinase gene inPichiapastoris[J].PLoS One,2013,8:e58393.
[30] 剧海,梁东春,郭刚,等.用于PCR实验的毕赤酵母基因组DNA制备方法的比较[J].天津医药,2003,31:270-272. JU H,LIANG D,GUO G,et al.Comparison of four methods to preparePichiagenomic DNA for PCR[J]. Tianjin Med J,2003,31:270-272.
[31] CHA G H,LUO S W,QI Z H,et al.Optimal conditions for expressing a complement component 3b functional fragment (α2-macroglobulin receptor) gene fromEpinepheluscoioides, inPichiapastoris[J].Protein Expr Purif, 2015,109:23-28.
[32] BARRIGON J M,VALERO F,MONTESINOS J L,et al.A macrokinetic model-based comparative meta-analysis of recombinant protein production byPichiapastorisunder AOX1 promoter[J].Biotechnol Bioeng,2015,112:1132-1145.
[33] WANG C H,ZHANG J K,WANG Y,et al.Biochemical characterization of an acidophilic β-mannanase fromGloeophyllumtrabeumCBS900.73 with significant transglycosylation activity and feed digesting ability[J].Food Chem,2016,197:474-481.
[34] MORI T,BARRIENTOS L G,HAN Z Z,et al.Functional homologs of cyanovirin-N amenable to mass production in prokaryotic and eukaryotic hosts[J].Protein Expr Purif,2002,26:42-49.
[35] LIN J W,JIA J,SHEN Y H,et al.Functional expression of FIP-fve,a fungal immunomodulatory protein from the edible mushroomFlammulinavelutipesinPichiapastorisGS115[J].J Biotechnol,2013,168:527-533.
[36] SHAO J,CAO C,BAO J,et al.Characterization of bovine interferon α1:expression in yeastPichiapastoris,biological activities,and physicochemical characteristics[J].J Interferon Cytokine Res,2015,35:168-175.
[37] SEXTON A,DRAKE P M,MAHMOOD N,et al.Transgenic plant production of Cyanovirin-N,an HIV microbicide[J].Faseb J,2006,20:356-358.
[38] ELGHABI Z,KARCHER D,ZHOU F,et al.Optimization of the expression of the HIV fusion inhibitor cyanovirin-N from the tobacco plastid genome[J].Plant Biotechnol J,2011,9:599-608.
[39] KEEFFE J R,GNANAPRAGASAM P N P,GILLESPIE S K,et al.Designed oligomers of cyanovirin-N show enhanced HIV neutralization[J].Proc Natl Acad Sci USA,2011,108:14079-14084.
[40] CHEN J,HUANG D,CHEN W,et al.Linker-extended native cyanovirin-N facilitates PEGylation and potently inhibits HIV-1 by targeting the glycan ligand[J].PLoS One,2014,9:e86455.
2016-10-17 基金项目:国家自然科学基金(31370364, 31570652, 31670200); 深圳市科技创新委员会资助项目(JCYJ20160425165447211)
崔利敏(1990年生),女;研究方向:植物进化遗传学;E-mail: cuiliminly@163.com
苏应娟(1965年生),女;研究方向:植物进化遗传学;E-mail: suyj@mail.sysu.edu.cn
水蕨抗艾滋病病毒蛋白CVNH及其突变体在毕赤酵母中的表达
崔利敏1,王桢2,王艇3,苏应娟1, 4
(1. 中山大学生命科学学院,广东 广州 510275;2. 南京农业大学生命科学学院,江苏 南京 210095;3. 华南农业大学生命科学学院,广东 广州 510642;4. 中山大学深圳研究院,广东 深圳 518057)
CVN(Cyanovirin-N)是一种高效的抗HIV蛋白。 Cyanovirin-N(CVNH)家族是CVN的同源蛋白,具有与CVN的肽链折叠类型并且具有高度保守的抗HIV 的结构域。把水蕨Ceratopteristhalictroides野生型和突变型CtCVNH蛋白的编码序列分别克隆到真核表达载体pPICZαA,结果所有野生型wCtCVNH和突变型mCtCVNH基因分别产生两条带,Western blot确认大小分别为22 000和24 000。采用超滤离心和Ni-NTA亲和色谱法纯化蛋白质,糖基化检测证实wCtCVNH蛋白不存在N-糖基化位点,可能的原因是载体pPICZαA中α因子信号肽序列的不完全切割或存在一些非糖基化的其它翻译后修饰,为今后进一步研究CtCVNH的药代动力学和药效学奠定了坚实的基础。
抗HIV蛋白;水蕨;突变体;毕赤酵母
10.13471/j.cnki.acta.snus.2017.04.019
Q78 Document code:A Article ID:0529-6579(2017)04-0118-08

