Inhibition of CYP3A4 by natural accurring flavonoids:A SAR and 3D-QSAR study
, , , ,ng , , , ,
(1.Key Laboratory for Basic Pharmacology of Ministry of Education,Zunyi Medical University,Zunyi Guizhou 563099,China; 2.Pharmaceutical Lab,School of Pharmacy,Zunyi Medical University,Zunyi Guizhou 563099,China; 3.School of Pharmaceutical Engineering,Shenyang Pharmaceutical University,Shenyang Liaoning 110000,China; 4.Department of Biotechnology,Dalian Institute of Chemical Physics,Chinese Academy of Sciences,Dalian Liaoning 116023,China)
基础医学研究
Inhibition of CYP3A4 by natural accurring flavonoids:A SAR and 3D-QSAR study
WangWei1,2,LiMing3,AiChunzhi4,DuYimei1,2,LingLei1,2,MaFeifei1,2,ZhouXumei2,LuYanliu1,HeYuqi1,2
(1.Key Laboratory for Basic Pharmacology of Ministry of Education,Zunyi Medical University,Zunyi Guizhou 563099,China; 2.Pharmaceutical Lab,School of Pharmacy,Zunyi Medical University,Zunyi Guizhou 563099,China; 3.School of Pharmaceutical Engineering,Shenyang Pharmaceutical University,Shenyang Liaoning 110000,China; 4.Department of Biotechnology,Dalian Institute of Chemical Physics,Chinese Academy of Sciences,Dalian Liaoning 116023,China)
Objective Mining the key structure feature of natural occurring flavonoids for inhibition on CYP3A4 via SAR and 3D-QSAR strategy.Methods Evaluate the activity of CYP3A4 in human liver microsome incubation system via checking the specific probe reaction:6β-hydroxylation of testosterone.Screen the inhibitory effect of 25 flavonoids at a relatively higher concentration.Check the IC50values of flavonoids with inhibitory rate more than 50%.Using the tested and reported IC50values to construct a 3D-QSAR model to predict the inhibitory potency of flavonoids other than the investigated 25 objects.Results Nine flavonoid glycones showed high inhibitory effect on CYP3A4.The number,type,and position of the substituted groups on flavonoid core structures are correlated with their inhibitory effect on CYP3A4.C1-C2 double bond promotes the inhibitory effect.A 3D-QSAR model was established and applied with good prediction.Conclusion Key structure features for CYP3A4 inhibition were extracted from flavonoids.A 3D-QSAR model for flavonoids related CYP3A4 inhibition prediction was established.Results of the present study would benefit the prediction of Food or Drug-Drug interaction mediated by flavonoids.
flavonoids; CYP3A4; inhibition; SAR; 3D-QSAR
Cytochrome P450 enzymes play an important role on the metabolism of endogenous and xenobiotic small molecules in animals and human.Usually,cytochrome P450 enzymes catalyze compounds to inactive them,however,sometimes,they also active non-toxic or non-active molecules to be toxic and active.Some of those P450-catalyzed metabolites even too active to attach biomacromoleculars such as DNA and protein[1].As the cytochrome P450-mediated metabolism would change the activity or toxicity of drugs,inhibition on P450 enzymes is extremely important to understand the potential metabolism-mediated food or drug-drug interaction,toxicity,and carcinogenicity[2-3].CYP3A4 is the most important cytochrome P450 isozyme.It occupies around 30% of total P450s and accounts for more than 50% of the metabolism of current drugs[1].Thus,inhibition of CYP3A4 is the prior choice while entering into inhibition studies for cytochrome P450 isozymes.
Flavonoids exist extensively in foods,herbs and drinks (tea,red wine etc.)[4].By the year 2000,more than 8000 flavonoids have been isolated from plants and over 5000 flavonoids have been described[5].They exhibit broad pharmacological activities such as antioxidant,anti-hypertension,anti-arrhythmia,and anti-tumor.In addition,clinical observations indicated that foods with high level of flavonoids showed significant effects on preventing cancer,thus theyhave been considered as the valuable chemo-preventive agent for anti-cancer[6].Recently Amin A and Buratovich M.illustrated the charm of flavonoids in the treatment of hormone-dependent cancers and they even proposed that “A Cup-of-Tea Will Do”[7].
To data,flavonoids inhibit cytochrome P450 sextensively[6,8].As the significant correlation between flavonoids treatment and cancer prevention,most studies were focused on the inhibition effect of flavonoids to CYP1A1/1A2[9-11].Regarding CYP3A4,the most important cytochrome P450 isozyme,strong inhibition by flavonoids were also observed[12-14].Naringenin inhibited CYP3A1/2 and increased the AUC of paclitaxel (substrate of CYP 3A) in rats[15].The grapefruit juice (with kinds of flavonoids)[16],increased the bioavailability of drugs metabolized by CYP3A4[17].Quercetin and kaempferol[18]showed a dose-dependent and time-dependent inhibition to CYP3A4.Isoliquiritigenin,licopyranocoumarin,4-hydroxyguaiacol apioglucoside,liquiritin,liquiritigenin 7,4-diglucoside,liquiritinapioside,silybin,silydianin,silycristin,and dehyfrosilybin[14,19]show strong inhibition to CYP3A4 with IC50values of less than 100 μmol/l.Quercetin and baicalein inhibited CYP3A4 with IC50values as low as less than 20 μmol/L[16,18,20].Glabridin,an isoflavone,showed mechanism-based inactivation to CYP3A4 with a KIvalue (inhibition constant) as low as 7 μmol/l[21].A few structure features of flavonoids were noted for CYP3A4 inhibition.For example,hydroxylation on C-7of flavonoids is considered to promote the inhibition on CYP3A4.In addition,the number of substituted hydroxyl radical is also important for CYP3A4 inhibition.However,these data can’t be used for SAR analysis because they were generated by different experimental platforms.Until now,a strictly speaking SAR study between flavonoid structures and CYP3A4 inhibitory activity is still deficient.
In this study,twenty five flavonoids were collected.They covered five subgroup of flavonoids (Fig 1).Their inhibitory effect on CYP3A4 activity would be check in human liver microsome incubation system by using testosterone as probe.SAR would be done while a 3D-QSAR model would be established.The present study provided not only a descriptive model but also a prediction model for CYP3A4 inhibition by flavonoids.
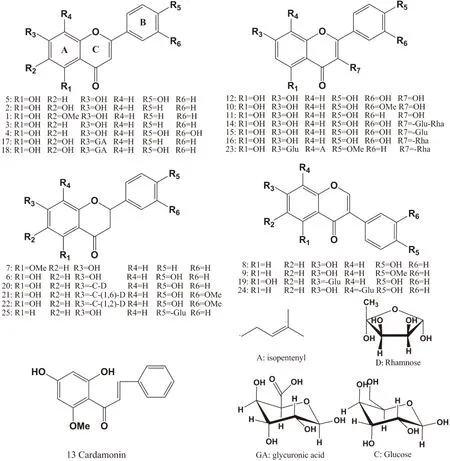
Series numbers represent compounds as follows:1 oroxylin A,2 baicalein,3 chrysin,4 luteolin,5 apigenin,6 naringenin,7 alpinetin,8 daidzein,9 formononetin,10 isorhamnetin,11 kaempferol,12 quercetin,13 cardamonin,14 rutin,15 hyperoside,16 quercitrin,17 baicalin,18 scutellarin,19 genistin,20 naringin,21 hesperidin,22 neohespeidin,23 icariin,24 puerarin,25 liquritin.Fig 1 Structures of investigated flavonoids
1 Materials and methods
1.1 Chemicals Flavonoids were isolated by our lab and the purities are more than 98% analyzed by HPLC.Testosterone,D-glucose-6-phosphate,glucose-6-phosphate dehydrogenase,NADP+,methanol and acetonitrile were purchased from Sigma-Aldrich (St.Louis,MO,USA).Ketoconazole (KTZ) was obtained from ICN Biomedicals Inc.(Aurora,Ohio,USA).All other reagents were of HPLC grade or of the highest grade commercially available.
1.2 Preparation and characterization of liver microsomes Human livers were obtained from autopsy samples (n=5,male Chinese,ages from 27 to 48) from Dalian Medical University,with the approval of the ethics committee of Dalian Medical University.The medication history of the donors was not known.Research involving human subjects was done under full compliance with government policies and the Helsinki Declaration.Liver specimens were stored in liquid nitrogen until preparation of microsomes.Microsomes were prepared from liver tissue by differential ultra centrifugation as described previously[22-23].
1.3 Assay of the CYP3A4 activity A standard incubation system included:human liver microsome (HLM,0.5 mg/ml),D-glucose-6-phosephate (1 mmol/l),D-glucose-6-phosephate dehydrogenase (1 U/ml),NADPNa (1 mmol/l),and substrate.In case of CYP3A4 activity probing,testosterone (50 μmol/l,1 μl for each reaction system) was used as the specific substrate[24].Total volume of incubation system was 200 μl and total organic volume was less than 1% of the reaction system.After incubation for 10 min at 37 ℃,100 μl acetonitrile with corticosterone (20 μmol/l,used as internal standard) was added to quench the reaction.Then the quenched reaction system was centrifuged at 14 300 rpm for 10 min.The supernatant was injected to HPLC to determine the generation of 6β-hydroxylated testosterone (product of testosterone catalyzed by CYP3A4).All reaction parameters followed the reported methods[24].
1.4 HPLC analysis CYP3A4-specific metabolite of testosterone was analyzed on a Shimadzu LC-10A HPLC system with an Agilent Zorbax C18column (150×4.6 mm,5 μm).An aliquot of 20 μl samples were separated at 25oC by ddH2O and methanol with elution gradient as follow:0-15 min,52% to 70% of methanol; 15-22 min,70% to 80% of methanol.Testosterone,cortisone (internal standard),and 6β-hydroxylated testosterone was determined at 254 nm.
1.5 Screening of the inhibitory effect of flavonoids on CYP3A4 A relatively high concentration was used to screen flavonoids with potential inhibitory effect on CYP3A4.For baicalein,kaempferol,luteolin,quercetin,rutin,quercitrin,naringenin,alpinetin,naringin,cardamonin,puerarin,and liquiritin,the screening concentrations were set at 100 μmol/l.Saturated concentrations of other tested compounds were used in screening assay.Details were shown below:40 μmol/l for oroxylin A,15 μmol/l for isorhamnetin,33.3 μmol/l for chrysin,16 μmol/l for apigenin,25 μmol/l for hyperoside,33.35μmol/l for baicalin,50 μmol/l for scutellarin,40 μmol/l for daidzein,50 μmol/l for formononetin,8.9 μmol/l for genistin,5 μmol/l for hesperidin,12.5 μmol/l for neohesperidin,and 20 μmol/l for icariin.KTZ (1 μmol/l) was used as the positive inhibitor.For test flavonoids and the positive inhibitor,an aliquot of 1 μl was added into the incubation system.Incubation without test flavonoids and KTZ was used as control.The remaining enzyme activity (REA) was calculated based on the HPLC peak area of 6β-hydroxylated testosterone.
1.6 Determination of IC50values Nine compounds with the inhibition rate more than 50% were subjected IC50value tests.Concentrations of flavonoids were set as follows:15,5,3,0.75 and 0.375 μmol/l for chrysin,5,2.5,1.25,0.625 and 0.312 5 μmol/l for apigenin,15,5,3,1.5 and 0.75 μmol/l for cardamonin,6,3,2,1 and 0.5 for quercetin,15,5,3,1.5 and 0.75 μmol/l for luteolin,15,5,3,1.5 and 0.75 μmol/l for kaempferol,90,40,20,10 and 5 μmol/l for naringenin,15,10,5,3 and 1.5 μmol/l for baicalein,15.45,10,5,2.5 and 1.25 μmol/l for isorhamnetin.According to the remaining enzyme activity (REA),the IC50value was calculated using the formula as follows:
lgIC50=Xm-I(P-(3-Pm-Pn)/4)
(1)
WhereXmrepresents the logarithm value of the maximal concentration,Irepresents the logarithm value of the ratio that the maximal concentration to the second highest concentration,Pindicates the sum of the inhibition rate (1-REA) at all five concentrations,Pmrepresents the inhibition rate of flavonoids at the highest concentration,andPnrepresents the inhibition rate of flavonoids at the lowest concentration.
Regarding investigated compounds other than the aforementioned nine flavonoids,IC50values were calculated by two formulas as follows:
(2)
(3)
WhereKmis Michaelis constant of CYP3A4-catalyzed testosterone 6β-hydroxylation,[S] represents the concentration of substrates,Control% represents the REA,[I] is the concentration of inhibitor,Kiis the inhibition constant.
1.7 Computationalmethods All the molecular modeling and computational analyses were conducted and viewed on LINUX RedHat 8.0 operating system employing SYBYL6.9 package on a Dell precision 650 workstation.
1.7.1 Structure generation,conformational analysis,and alignment Energy minimization and conformational search were performed using Tripos force field (SYBYL6.9.Tripos Inc.).The convergence criterion was set at 0.001 kcal/mol.In order to obtain the most stable conformation,the energy gradient limit was set at 0.05 kcal/mol.Å and the partial atomic charges were calculated with Gasteiger-Huckel Charge method.Then the dataset was aligned to the template.Apigenin was randomly chosen as the initial template for congruence.Sixteen of the investigated flavonoids were thrown into the training set while the other 9 flavonoids were used as test set.
1.7.2 CoMFA Comparative molecular field analysis (CoMFA) module in Sybyl 6.9 is used to develop 3D-QSAR model[25].In CoMFA,two parameters,the steric and electrostatic potential fields,were used to construct a mathematic model against the IC50values of each flavonoids.A sp3carbon atom with a charge of +1.0 was used as the probe atom to calculate steric and electrostatic fields,and a cutoff value of 30 kcal/mol with smooth transition was used.
1.7.3 Partial Least-Squares (PLS) analysis The conventional CoMFA descriptors derived above served as explanatory variables,while the pIC50represented the target variable in PLS[26]regression analyses to generate 3D-QSAR models implemented in the SYBYL package.The optimal number of components were obtained by running the cross validation in leave-one-out (LOO) option,where the lowest standard error of prediction corresponds toq2coefficient.The cross-validated coefficient,q2,was calculated using the following equation:
(4)
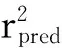
(5)
WhereSDisthesumofsquareddeviationsbetweenthebiologicalactivitiesofthetestsetandmeanactivityofthetrainingsetmolecules,andPRESSisthesumofsquareddeviationbetweenactualandpredictedactivitiesofthetestsetmolecules.
2 Results
2.1ScreeningofinhibitoryeffectonCYP3A4by25flavonoidsAttheconcentrationsusedforscreening(100μmol/lorsaturatedconcentration),flavonoidaglyconesshowedmuchstrongerinhibitionthanglycosides(Fig2).Amongtheseaglycones,baicalein,isorhamnetin,kaempferol,chrysin,luteolin,apigenin,quercetin,naringenin,andcardamonininhibitedmorethan90%oftheactivityofCYP3A4.Isorhamnetinshowedinhibitionratesofmorethan50%.
2.2IC50valuesAccordingtothescreeningofCYP3A4inhibitionby25investigatedflavonoids,chrysin,apigenin,cardamonin,quercetin,luteolin,kaempferol,naringenin,baicalein,andisorhamnetinshowedinhibitionratemorethan50%.Here,wetestedtheinhibitionratesofthese9flavonoidsataserialofconcentrations.ThenIC50valuesoftheseflavonoidswerecalculatedfollowingequation1 (Fig3).Amongthese9compounds,apigeninshowedthestrongestinhibitoryactivitybasedonthetestedIC50value(1.34μmol/l).IC50valuesforalltestedcompoundsarelessthan25μmol/l,suggestingthehighinhibitionpotencyofflavonoidaglycones.

1-25 represent the compounds as described in Fig 1.PC represents positive control (KTZ).Compounds 1-13 were flavonoid aglycones (black solid bar) and compounds 14-25 were flavonoid glycosides (white open bar).Fig 2 Inhibiton of CYP3A4 by investigated flavonoids at screening concentrations

The IC50 values of apigenin,baicalein,chrysin,isorhamnetin,luteolin,kaempferol,cardamonin,quercetin and naringenin were 1.34,3.16,3.27,11.93,3.31,2.85,3.25,1.75 and 23.08 μmol/l.Fig 3 Determination of IC50 values of 9 compounds showing high inhibition potency in screening test
For other investigated flavonoids,the IC50values were calculated according to Formula 2 and Formula 3.The calculated IC50values (Tab 1) were far higher than other 9 compounds described above.
Tab 1 Tested,calculated,and reported inhibition effect of flavonoids on CYP3A4
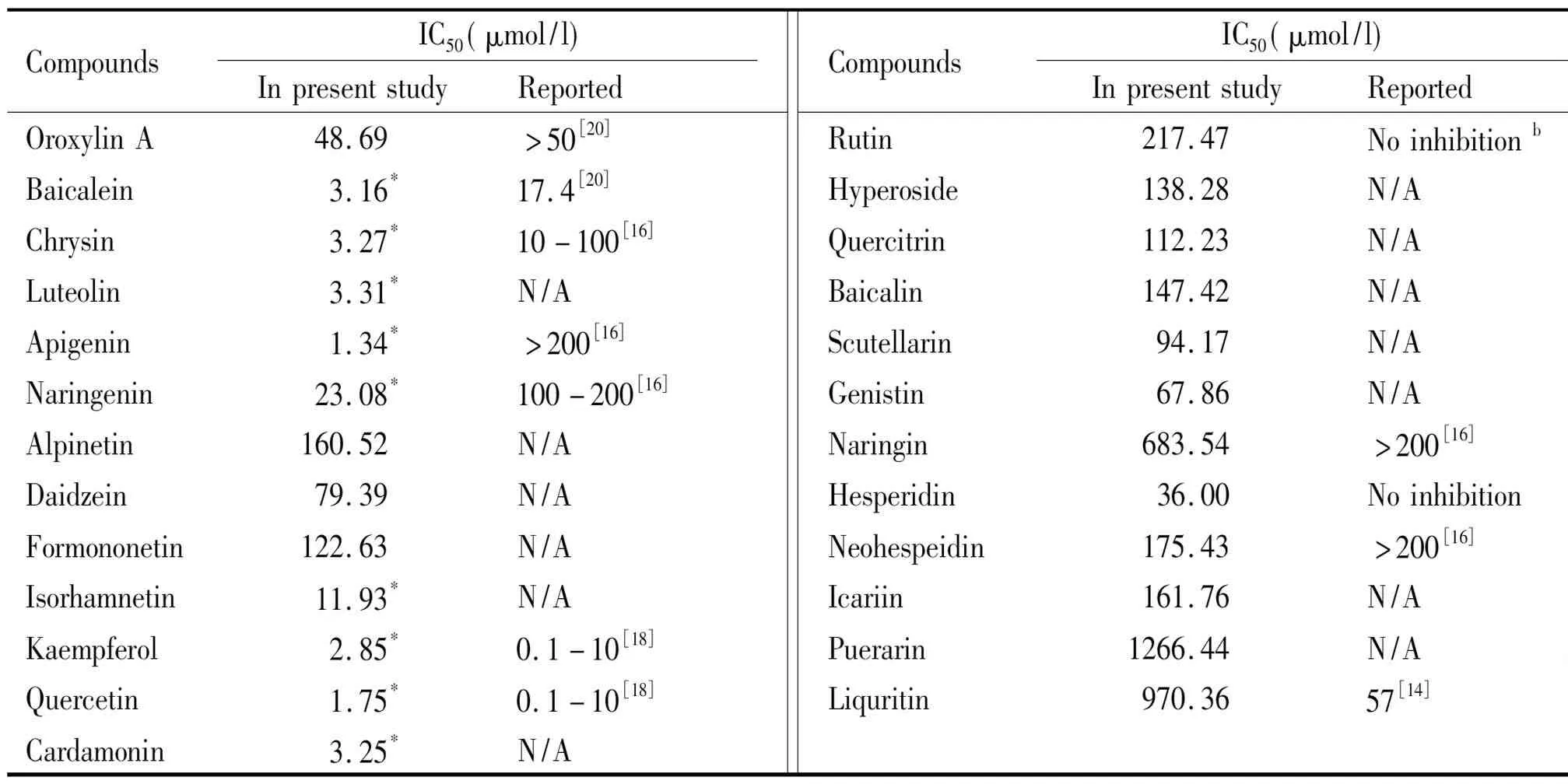
CompoundsIC50(μmol/l)InpresentstudyReportedCompoundsIC50(μmol/l)InpresentstudyRe-por-tedOroxylinA48.69>50[20]Rutin217.47Noin-hibitionbBaicalein3.16*17.4[20]Hyperoside138.28N/AChrysin3.27*10-100[16]Quercitrin112.23N/ALuteolin3.31*N/ABaicalin147.42N/AApigenin1.34*>200[16]Scutellarin94.17N/ANaringenin23.08*100-200[16]Genistin67.86N/AAlpinetin160.52N/ANaringin683.54>200[16]Daidzein79.39N/AHesperidin36.00Noin-hi-bi-tionFormonone-tin122.63N/ANeohespeidin175.43>200[16]Isorhamnetin11.93*N/AIcariin161.76N/AKaempferol2.85*0.1-10[18]Puerarin1266.44N/AQuercetin1.75*0.1-10[18]Liquritin970.3657[14]Cardamonin3.25*N/A
“*”,tested IC50values; “N/A”,reported IC50values are Not Avalable; “no inhibition”:this compound does not inhibit CYP3A4.
2.3 Apparent SAR analysis ①Flavones inhibited CYP3A4 stronger than flavanone.Apigenin and naringenin had extremely similar structures with each other.However,different connection of C2-C3 determined that apigenin was flavone while naringenin was flavonone.To table 1,apigenin (1.34 μmol/l) showed lower IC50value than naringenin (23.08 μmol/l),suggesting flavone favored CYP3A4 inhibition more than flavonone.②Flavones showed higher inhibitory potency than flavonol,which was supported by the comparison between IC50values of apigenin (1.34 μmol/l) and kaempferol (2.85 μmol/l).The only difference between these two compounds is R7.For apigenin,R7 is a hydrogen atom,determining it’s a flavone.For kaempferol,R7 is a hydroxyl,determining it’s a flavonol.As all left structures were the same as each other,data from apigenin and kaempferol indicated that flavones may inhibit CYP3A4 stronger than flavonol.With the same strategy,we got more information as follows:③Chalcone favored CYP3A4 inhibition more than flavonone,as cardamonin (3.25 μmol/l) showed higher inhibitory potency than alpinetin (160.52 μmol/l).④ Methoxyl substitution weakened the inhibitory effect of flavonoids.Methylation on R2 of baicalein formed oroxylin A,and significantly increased the IC50values from 3.16 to 48.69 μmol/l.The same phenomenon was also observed between daidzein (73.39 μmol/l) and formononetin (122.63 μmol/l),as well as quercetin (1.75 μmol/l) and isorhamnetin (11.93 μmol/l).⑤ C5-substitution promote the inhibition effect on CYP3A4.This is supported by tested high IC50values of daidzen and formononetin,as well as by data reported[16].⑥ Flavonols with two hydroxyls on ring B had stronger inhibitory activity than that with only one even no.This is supported by comparison between quercetinand kaempferol.⑦ However,flavones with two hydroxyls on ring B had weaker inhibitory activity than that with only one.This regularity is different from flavonols.For example,luteolin had weaker inhibitory activity than apigenin.
2.4 3D-QSAR analysis 3D-QSAR analysis was performed to connet the dimensionally structural features with the inhibition activities quantitatively.It would provide a predictive model for CYP3A4 inhibition by flavonoids.The aligned 16 compounds were included in the training set to establish the 3D-QSAR model while the other 9 inhibitors were used as test set to validate the model.The structures of molecules in test set are similar to the training set,including flavone (oroxylinA,luteolin),flavonol (isorhamnetin,icariin,quercitrin),isoflavone (liquiritin),flavanone (genistin),and chalcone (cardamonin) derivates.
Based on the molecular alignment (Fig 4),a predictive 3D-QSAR model was obtained with CoMFA default parameters.Aq2of 0.527 was yielded for the cross validated model.The optimal number of components was determined to be four to produce the conventional model using the SAMPLS method.Start from the structure,the electrostatic and steric fields of a flavonoid could be calculated and applied in the established 3D-QSAR model to predict the potential IC50value.
Anr2of 0.955 was finally determined for the non-cross-validated model with the standard estimation of error (SEE) 0.235 and theFratio of 57.969
(Tab 2).This model was used to predict the inhibitory bioactivities for the compounds in both training set and test set,see Table 3.

Fig 4 Molecular alignment of flavonoid compounds as inhibitors of CYP3A4 based on the template of apigenin
Tab 2 Statistics of CoMFA models

q2NcSEPr2SEEFvalueContributionSEr2pred0.52740.7040.9550.23557.9690.4270.5730.632
Tab 3 Prediction of the inhibitory bioactivities of the flavonoid compounds in the training and test set

FlavonoidsActual(pIC50)Predicted(pIC50)Residual(pIC50)TrainingAlpinetin-2.21-2.280.07Apigenin-0.13-0.11-0.02Baicalein-0.50-1.000.50Baicalin-2.17-1.75-0.42Chrysin-0.52-0.45-0.07Daidzein-1.90-1.930.03Formonone-tin-2.09-1.94-0.15Hesperetine-1.56-1.610.05Hesperidin-2.14-2.01-0.13Kaempferol-0.45-0.560.11Naringenin-1.36-1.600.24Naringin-2.83-2.860.03Neohesperi-din-2.24-2.20-0.04Puerarin-3.10-3.200.10Quercetin-0.24-0.07-0.17Scutellarin-1.97-1.86-0.11TestCardamonin-0.51-1.320.81Genistin-1.83-1.50-0.33Icariin-2.21-1.59-0.62Isorhamnetin-1.08-1.150.07Liquiritin-2.99-1.41-1.58Luteolin-0.52-1.260.74Oroxylina-1.69-1.04-0.65Quercitrin-2.05-2.080.03Rutin-2.34-1.20-1.14
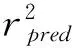

Circles represent compounds in training set and pentacle indicate test set compounds outside the training set. Fig 5 Correlation between CoMFA-predicted and actual inhibitory potencies of IC50
CoMFA contour maps were generated and interpolated upon the reference molecules to visualize the information content of the derived 3D-QSAR model.In steric (Fig 6A) and electrostatic contour (Fig 6B) maps,apigenin,hyperoside,neohesperidin and puerarin were superimposed to represent flavone,flavonol,flavanone and isoflavone analogues and illustrate the isocontour diagrams of the field contributions (“stdev*coeff”) for different properties calculated by the CoMFA analysis.Apigenin was shown in transparent stick-ball style,hyperoside in green transparent stick style,neohesperidin in red-orange stick style and puerarin in capped stick style.Puerarin was in a distinct coordinate from the other three molecules in the superimposition (Fig 6).The contour maps may be helpful to identify important regions related to CYP3A4 inhibition.
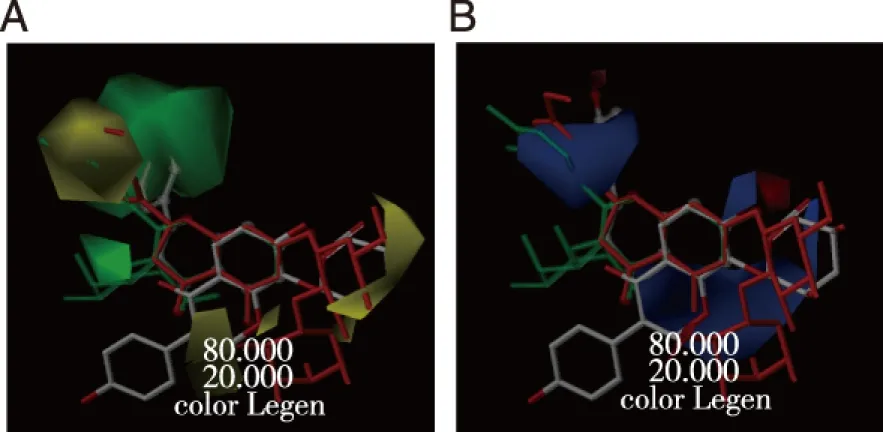
Apigenin was shown in transparent stick-ball style,hyperoside in green transparent stick style,neohesperidin in red-orange stick style and puerarin in capped stick style.Green contours indicate inhibitory efficacy increasing and yellow indicate efficacy decreasing upon addition of steric bulk.Blue contours indicate potency increasing upon addition of positive charge and red indicate potency increasing upon addition of negative charge. Fig 6 Steric (A),and electrostatic (B) contour maps of CoMFA shown for flavonoid inhibitors of CYP3A4 apigenin,hyperoside,neohesperidin and puerarin were superimposed to illustrate the isocontours
In Fig 6A,the green contours encode stericaly attractive regions and the yellow indicate repulsive areas.Several alkyl disfavored yellow plots were located around R1and R3position of apigenin,hyperoside,and neohesperidin,indicating that large substituted groups,such as -Glu and -Rha,would decrease the inhibitory effect of flavonoids.Whereas for puerarin these yellow contours were situated near R4area,which suggests isoflavone derivates are disfavored of bulky addition to R4.That’s why puerarin exhibits a lower bioactivity than daidzein and formononetin.Isorhamnetin shows a smaller inhibitory activity than kaempferol and quercetin.Perhaps,it could be explained that a relatively larger substructure of -OCH3 substitutes at R5region.Nevertheless flavone analogues wouldn’t repulse bulky substituents at R5and R6position because a bulky favored green contour was localized near the phenyl ring of apigenin (representative flavone).
The electrostatic fields were demonstrated in Figure 6B.The blue and red contours represent the preference for electropositive and electronegative groups,respectively.Figure 7 showed the electro structure of flavonoid compounds.In flavonoids,π-πconjugation was formed between phenyl ring A and C3=OThus,the electron clouds tend to transfer from Ring A to =O,resulting in the positive-charged feature of C4 and C6.In case of flavone,flavonol and chalcone,C1 and C2 was connected by a double bond,π-πconjugation also exist between Ring B and C3=O.Substitution on C6 would enhance the conjugation while on C5 and C7 would weaken the conjugation.
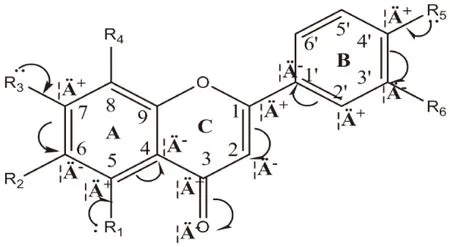
Fig 7 Electro structures of flavonoid derivates
In Fig 6,the large blue-colored block covering the region below Ring A and Ring C.Thus,positive charge at C3 and C4 would increase the bioactivities of flavonoid compounds.R1prefers to be substituted by groups like hydroxyl that would increase the positive charge at C4.This is supported by the inhibition effect of liquiritin with the lowest inhibition activity among all investigated flavonoids.Substitution of R2destroyed the conjugation status,decreasing the inhibitory potencies.Apigenin,chrysin and luteolin inhibited CYP3A4 sensitively.Once R2of them were replaced,they were converted to oroxylin A,scutellarin,and baicalin respectively.Along with the R2substitution,IC50values of aforementioned compounds significantly increased.The stronger inhibitory activity of flavone (apigenin) than corresponding flavonol also related to the distinct substitution at C5 site.Similarly,the blue plot at C6 position suggests that substitution at this site would enhance the conjugated effect.
On ring B of apigenin and hyperoside,a large blue contour were observed,indicating low electron density.Accordingly,C4’ prefers the addition of groups with lone electron pair rather than C3’.Thus,apigenin and quercetin showed higher inhibitory effect than luteolin and kaempferol,respectively.However,different from flavones,a hydroxylation at C3’ of flavonol enhances the bioactivities.Perhaps hydroxyl group at C3’ withdrawed electrons rather than strengthened the π-π conjugation.Although this blue block was also situated at Ring B of n eohesperidin,conjugated effect couldn’t be employed because no double carbon bond was formed between C1 and C2.For flavanone analogues,electro-withdrawn substituents would be favored to show positive charge characteristic.
3 Discussion
Hydroxylation was considered as the main factor influencing the inhibitory activity of flavonoids to CYP3A4.More hydroxylation were associated with higher CYP3A4 inhibition[16].However,the reversed effect of glucose and methoxyl substitution may completely cancel hydroxylation effect.For example,although isorhamnetin have more hydroxyls than chrysin,available methoxyl still make the inhibitory effect of isorhamnetin lower than chrysin.Moreover the double carbon bond of C1-C2 plays an important role on enhancing the inhibitory activity.To describe so many factors clearly,a 3D-QSAR analysis was performed using CoMFA approach.
We have correlated the inhibition of flavonoids to CYP3A4 with the steric and electrostatic fields calculated with CoMFA.Based on the training set with 16 flavonoid inhibitors,a predictive 3D-QSAR model have been establish.This model is successfully be validated by other 9 flavonoids.As the inhibition of CYP3A4 would induce potential drug drug interaction with agents metabolized by this enzyme,the prediction showed highly application value in both clinic and drug development.
Antioxidant activity is considered as one of the most important activities of flavonoids.In this paper we found that the CYP3A4-inhibition related SAR is similar to antioxidation-related SAR of flavonoids.For example,the more hydroxyls lead both strong CYP3A4 inhibition and antioxidation[27].Two hydroxyls at ring B and one oxygen atom at site 4 also increase the antioxidant activity of flavonoids[28-30].Some QSAR analysis between the flavonoids structures and the antioxidant activity suggested that more hydroxyl can increase the activity of flavonoids while 5-hydroxylation is the key factor.These data implied that the inhibition against CYPs may be one of the mechanisms of antioxidant activity of flavonoids.Cytochrome p450 enzymes generate reactive oxygen species (ROS), inhibition of oxidation enzymes may contribute to antioxidant acitivity of flavonoids.
In this study,by test inhibitory effect of 25 flavonoids in human liver microsome incubation system,we mined the key structure features benefit for CYP3A4 inhibition.A predictive 3D-QSAR model was established and validated.Result of the present study would be useful for new drug development as well as prevention of drug-drug interaction in clinic.
[1] Nebert D W,Dalton T P.The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis [J].Nature,2006,6:947-960.
[2] Tam L S,Chan T Y,Leung W K,et al.Warfarin interactions with Chinese traditional medicines:danshen and methyl salicylate medicated oil [J].Aust N Z J Med,1995,25(3):258.
[3] Patterson L H,Murray G I.Tumour cytochrome P450 and drug activation[J].Curr Pharm Des,2002,8(15):1335-1347.
[4] Cook N C,Samman S.Flavonoids-Chemistry,metabolism,cardioprotective effects,and dietary sources[J].J Nutr Biochem,1996,7(2):66-76.
[5] Pietta P G.Flavonoids as antioxidants [J].J Nat Prod,2000,63(7):1035-1042.
[6] Hodek P,Trefil P,Stiborova M.Flavonoids-potent and versatile biologically active compounds interacting with cytochromes P450[J].Chem Biol Interact,2002,139(1):1-21.
[7] Amin A,Buratovich M.The anti-cancer charm of flavonoids:A cup-of-tea will do[J].Recent Patents on Anti-Cancer Drug Discovery,2007,2(2):109-117.
[8] Moon Y J,Wang X,Morris M E.Dietary flavonoids:effects on xenobiotic and carcinogen metabolism[J].Toxicol In Vitro,2006,20(2):187-210.
[9] Ishibe N,Hankinson S E,Colditz G A,et al.Cigarette smoking,cytochrome P450 1A1 polymorphisms,and breast cancer risk in the nurses' health study[J].Cancer Research 1998,15:667-671.
[10] Zhai S,Dai R,Friedman F K,et al.Comparative inhibition of human cytochromes P450 1A1 and 1A2 by flavonoids[J].Drug Metab Dispos,1998,26(10):989-992.
[11] Wen X,Walle U K,Wallet.5,7-Dimethoxyflavone downregulates CYP1A1 expression and benzo[a]pyrene-induced DNA binding in Hep G2 cells[J].Carcinogenesis,2005,26(4):803-809.
[12] Obermeier M T,White R E,Yang G C S.Effects of bioflavonoids on hepatic P450 activities [J].Xenobiotica,1995,25(6):575-584.
[13] Obach R S.Inhibition of human cytochrome P450 enzymes by constituents of St.John's Wort,an herbal preparation used in the treatment of depression[J].J Pharmacol Exp Ther,2000,294(1):88-95.
[14] Tsukamoto S,Aburatani M,Yoshida T,et al.CYP3A4 inhibitors isolated from Licorice[J].Biol Pharm Bull,2005,28(10):2000-2002.
[15] Lim S C,Choi J S.Effects of naringin on the pharmacokinetics of intravenous paclitaxel in rats[J].Biopharm Drug Dispos,2006,27(9):443-447.
[16] Ho P C,Saville D J,Wanwimlruk S.Inhibition of human CYP3A4 activity by grapefruit flavonoids,furanocoumarins and related compounds[J].J Pharm Pharm Sci,2001,4(3):217-227.
[17] Schmiedlin-Ren P,Edwards D J,Fitzsimmons M E,et al.Mechanisms of enhanced oral availability of CYP3A4 substrates by grapefruit constituents.Decreased enterocyte CYP3A4 concentration and mechanism-based inactivation by furanocoumarins[J].Drug Metab Dispos,1997,25(11):1228-1233.
[18] Zhang F F,Zheng Y F,Zhu H J,et al.Effects of kaempferol and quercetin on cytochrome 450 activities in primarily cultured rat hepatocytes[J].Journal of Zhejiang University,2006,35(1):18-22.
[19] Zuber R,Modriansky M,Dvorak Z,et al.Effect of silybin and its congeners on human liver microsomal cytochrome P450 activities[J].Phytother Res,2002,16(7):632-638.
[20] Kim J Y,Lee S,Kim D H,et al.Effects of flavonoids isolated from Scutellariae radix on cytochrome P-450 activities in human liver microsomes[J].J Toxicol Environ Health A,2002,65(5-6):373-381.
[21] Backman J T,Maenkpaa J,Belle D J,et al.Lack of correlation between in vitro and in vivo studies on the effects of tangeretin and tangerine juice on midazolam hydroxylation[J].Clin Pharmacol Ther,2000,67(4):382-390.
[22] Sanderink G,Bournique B,Stevens J,et al.Involvement of human CYP1A isoenzymes in the metabolism and drug interactions of riluzole in vitro[J].J Pharmacol Exp Ther,1997,282:1465-1472.
[23] Lowry O H,Rosebrough N J,Farr A L,et al.Protein measurement with the Folin phenol reagent [J].J Biol Chem,1951,193(1):265-275.
[24] Liu Y,Li W,Deng M C,et al.The inhibitory effect of intestinal bacterial metabolite of ginsenosides on CYP3A activity[J].Biol Pharm Bull,2004,27(10):1555-1560.
[25] Cramer R D,Patterson D E,Bunce J D.Comparative molecular field analysis (CoMFA).1.Effect of shape on binding of steroids to carrier proteins[J].J Am Chem Soc,1988,110(18):5959-5967.
[26] Wold S,Ruhe A,Wold H,et al.The collinearity problem in linear regression.the partial least squares (PLS) approach to generalized inverses[J].SIAM Journal on Scientific Computing,1984,5(3):735-743.
[27] Farkas O,Jakus J,Héberger K.Quantitative structure - antioxidant activity relationships of flavonoid compounds[J].Molecules,2004,9(12):1079-1088.
[28] Rice-Evans C A,Miller N J,Paganga G.Structure-antioxidant activity relationships of flavonoids and phenolic acids[J].Free Radic Biol Med,1996,20(7):933-956.
[29] Harborne J B,Wlliams C A.Advances in flavonoid research since 1992[J].Phytochem,2000,55:481-504.
[30] Heim K E,Tagliaferro A R,Bobilya D J.Flavonoid antioxidants:chemistry,metabolism and sructure-activity relationships[J].J Nutr Biochem,2002,13(10):572-584.
[收稿2016-11-30;修回2017-01-22]
(编辑:王静)
国家自然科学基金资助项目(NO: 81402985,81560673,81660685);上海市复方中药重点实验室(上海中医药大学)开放基金资助项目(NO:2014OP01);贵州省科学技术基金资助项目(NO:黔科合J字[2015]2158号,黔科合JZ字[2015]2010号);贵州省留学人员择优资助项目(NO:黔人项目资助合同[2015]03);遵义医学院博士科研启动基金资助项目(NO:F-736,F-744)。
何芋岐,男,博士,副教授,研究方向:药物分析与系统生物学,E-mail:hyqJeff@hotmail.com;鲁艳柳,女,博士,副教授,研究方向:药物代谢与毒理学,E-mail:yanliu.lu@foxmail.com。
天然黄酮类化合物抑制CYP3A4的构效关系研究
汪 巍1,2,李 明3,艾纯芝4,杜艺玫1,2,凌 蕾1,2,马菲菲1,2,周旭美2,鲁艳柳1,何芋岐1,2
(1.遵义医学院 基础药理教育部重点实验室,贵州 遵义 563099;2.遵义医学院 药学院药学实验室,贵州 遵义 563099;3.沈阳药科大学 制药工程学院,辽宁 沈阳 110000;4.中国科学院 大连化学物理研究所生物技术部,辽宁 大连 116023)
目的 研究天然黄酮类化合物对CYP3A4的抑制活性并通过定性及三维定量分析两种方法挖掘天然黄酮结构中抑制CYP3A4活性的关键因素。方法 利用睾酮特征代谢反应表征人肝微粒体中CYP3A4的活性。在此体系中,首先选取较高浓度初筛25种黄酮类化合物的抑制活性。对于抑制活性超过50%的化合物,进行多浓度测试以获得IC50值。最后分别采用定性和三维定量两种方式分析黄酮结构与CYP3A4抑制之间的相关性。结果 初筛实验表明,黄酮苷元类化合物中的黄芩素、异鼠李素、山奈酚、白杨素、木犀草素、芹菜素、槲皮素、柚皮素以及豆蔻明9个化合物对CYP3A4表现出最强抑制作用。除了异鼠李素之外,另外8个化合物的抑制百分比高达90%。黄酮类化合物中主要是取代基的类型、羟基的数量和位置以及碳1、2位双键影响对CYP3A4的抑制活性。建立了预测性较好的反映黄酮结构及对CYP3A4抑制活性的定量计算模型。结论 本文发现了黄酮类化合物抑制CYP3A4活性的关键结构特征,同时建立了可以用于黄酮类化合物抑制CYP3A4活性的计算化学模型。
黄酮类化合物;CYP3A4;抑制;SAR;3D-QSAR
R969.2
A
1000-2715(2017)01-0006-11

