马铃薯遗传育种研究:现状与展望
徐建飞,金黎平
(中国农业科学院蔬菜花卉研究所/农业部薯类作物生物学与遗传育种重点实验室,北京100081)
马铃薯遗传育种研究:现状与展望
徐建飞,金黎平
(中国农业科学院蔬菜花卉研究所/农业部薯类作物生物学与遗传育种重点实验室,北京100081)
马铃薯是世界第三大粮食作物,马铃薯产业的可持续发展对保障世界和中国的粮食安全具有重要意义。优良品种是支撑马铃薯产业发展的基础。马铃薯经常遭受病虫害的侵袭和非生物胁迫,加工业的迅速发展和人们对食物营养的重视,迫切需要选育出更抗病、更耐逆、更高产、更优质和专用的马铃薯新品种。培育一个优良马铃薯品种,种质资源是基础,重要性状的遗传学是理论指导,先进的育种技术是保障,完善的推广和栽培模式是支撑。世界范围内,保存了大约65 000份马铃薯种质资源,通过对种质资源抗病、抗逆和品质方面的系统评价,并应用多种资源利用技术,将三大类约17个野生种的种质导入到普通栽培种中,应用于育种和遗传学研究。利用纯合双单倍体材料作为测序对象,马铃薯基因组序列已经被揭示,预测出了39 031个蛋白编码基因,目前更多的种质资源正在被重测序以揭示更多的等位变异。马铃薯普通栽培品种是无性繁殖四倍体作物,具有四体遗传特性,尽管如此,许多植株发育和形态、块茎品质和抗病抗逆等重要性状的遗传特性基本明确,并定位和克隆了大量重要性状相关基因。目前,马铃薯育种技术主要涵盖传统育种技术、倍性育种技术、标记辅助选择育种技术、基因工程育种技术和新兴的基因组选择育种技术。中国马铃薯遗传育种研究队伍不断壮大,品种选育取得了重大进展。荷兰马铃薯遗传育种水平居于世界前列,合作育种模式推动了商业化育种。不断完善马铃薯综合育种技术,创新育种模式和机制,充分利用现有种质资源培育突破性、专用型品种将是未来马铃薯遗传育种发展的主要方向。
马铃薯;育种;遗传学;现状;展望
0 引言
马铃薯(Solanum tuberosum)是世界第三大粮食作物和最重要的非禾本科作物。2014年全球马铃薯总产 3.82亿吨,分布在全球 158个国家和地区(http://fao.org/faostat)。马铃薯块茎营养丰富全面,含有人体必需的全部七大类营养物质,全球约10亿人将马铃薯作为主要食物食用[1]。中国是第一大马铃薯生产国,产量占世界总产量的1/4左右。马铃薯在中国各个生态区域都有广泛种植,尤其在西部贫困地区和边远山区种植面积更大,为缓解中国食物安全压力和消除地区性贫困起到了重要作用[2-3]。马铃薯起源于南美和中北美地区,经过当地人们的驯化逐渐形成可以食用的地方品种;16世纪,马铃薯经由探险家传入欧洲[4];根据古籍《长安客话》考证,马铃薯约于明朝万历年间传入中国。马铃薯不断被驯化和传播的历史,就是一部马铃薯育种史:在最初的驯化过程中,对人畜有毒的糖苷生物碱(以茄碱和卡茄碱为主)含量减少[5],匍匐茎逐渐缩短,块茎不断增大;引入欧洲后,适应长日照结薯的品种开始出现[6];随着导致爱尔兰大饥荒的晚疫病的发生,人们开始从野生种引入抗病种质培育抗病新品种;现在,为了满足马铃薯鲜食和加工市场的巨大需求,育种者选育出了大量的马铃薯新品种应用于生产。然而,马铃薯作为无性繁殖作物,经常遭受病虫害的侵袭和非生物胁迫,加之加工业的迅速发展和人们对食物营养的重视,迫切需要选育出更抗病、更耐逆、更高产、更优质和专用的马铃薯新品种。如何选育出一个优良的马铃薯品种?优异的种质资源是材料基础,重要性状的遗传学是理论指导,先进的育种方法是技术保障,完善的推广和栽培模式是应用支撑。
1 马铃薯种质资源保存和利用
种质资源是植物育种与遗传学研究的基础。马铃薯种质资源丰富,包含众多野生种和栽培种,而且种质资源的分类一直在不断变化。SPOONER等[7]在总结可以用于野生种种别界限和相互关系鉴别的形态学、分子水平、种间杂交障碍和野外观察的大量数据之后,提出马铃薯分为107个野生种和4个栽培种,这相对于HAWKES[8]提出的划分为228个野生种和 7个栽培种的分类学说发生了明显变化。
1.1 种质资源的收集和保存技术
世界范围内,目前保存了大约30大类共65 000份马铃薯种质资源[9]。世界上主要马铃薯种质资源收集和保存的机构是:国际马铃薯中心(International Potato Center,CIP)、荷兰遗传资源中心(The Centre for Genetic Resources,the Netherlands,CGN)、英国马铃薯种质资源库(Commonwealth Potato Collection,CPC)、德国马铃薯种质资源库(The IPK Potato collections at Gross Luesewitz,GLKS)、俄罗斯瓦维洛夫植物栽培科学研究所(The Vavilov Institute of Plant Industry,VIR)、美国马铃薯基因库(National Research Support Project-6,NRSP-6),除此之外,世界上其他国家如秘鲁、玻利维亚、阿根廷、智利和哥伦比亚等国都建立有马铃薯种质资源库。据估计,中国目前保存有5 000余份种质资源,以国内外育成品种和品系为主,野生种资源偏少。
马铃薯种质资源保存有多种方式,总体上来讲,野生种通常以实生种子进行保存,栽培品种(系)通常以试管苗或者田间种植的方式保存。试管苗保存技术形成于 1973年[10],由于比田间保存高效和安全,现在已经成为世界范围内种质资源保存的主要形式,其主要是利用低温(6—8℃)和山梨醇作为渗透调节剂来抑制植物生长,达到2年左右时间不用扩繁而较长时间保存资源的目的,这种技术已经成为世界上主要马铃薯种质资源库采用的通用技术。然而,试管苗保存存在耗时、高成本和因频繁更新扩繁易导致污染而造成资源丢失等诸多问题。冷冻保存技术以其低成本和长期保存的优点,逐渐开始被种质资源管理者所接受。冷冻保存是将植物以超低温(-196℃)状态长期保存在液氮中,理论上不需要定时更新保存[9]。冷冻保存最主要的问题是要避免外植体冷却过程中细胞内结冰,该项技术一直处在不断完善过程中[11-12]。目前GLKS和CIP已经采用冷冻保存技术进行马铃薯资源的保存:GLKS采用液滴冻结技术(droplet freezing technique)保存了1 341份欧洲马铃薯品种资源;CIP对197份安第斯地方品种进行了冷冻保存,但其茎段成活率和再生率都比较低。近年来,为了提高冷冻保存的可靠性和效率,研究者尝试将应用于香蕉资源的冷冻保存技术进行马铃薯资源保存,结果表明,相对于CIP和GLKS的冷冻保存技术,该方法的成活率和再生率都比较高,而且不同基因型的保存效果比较一致,可用于马铃薯资源长期保存[9]。
1.2 种质资源的评价
马铃薯种质资源遗传多样性评价主要可以分为形态学指标评价和分子水平评价。形态学指标是植物分类和品种鉴别的传统方法。在马铃薯分类学上,常涉及的茎、叶和花等形态学指标多达 53个[13]。在马铃薯品种鉴别上,世界范围内普遍采用的是国际植物新品种保护联盟(International Union for the Protection of New Varieties of Plants,UPOV)发布的DUS测试指南,其涉及的形态学相关性状指标为42个,但不同国家根据具体情况对测试指南进行了修改,例如中国测试的指标是41个,英国测试的指标是37个,而印度测试的指标是45个。随着分子生物学的发展尤其是分子标记技术的发展,种质资源的分子水平评价技术逐渐成熟,其可以快速地进行操作和分析,又可以避免由于环境变化造成的形态学指标上的变化[14]。分子水平评价主要包括应用蛋白质、分子标记和DNA序列进行遗传多样性分析,具体包括同工酶电泳、限制性酶切位点拷贝数变化(RFLP和AFLP)、基因组和质体DNA微卫星标记(SSR)、质体缺失标记、核糖体DNA非转录间隔子序列(ITS)、多倍体直系同源基因序列等[7]。近年来,随着基因组测序技术的发展,单核苷酸多态性(SNP)标记已经开始应用于种质资源评价和品种鉴别。
在马铃薯分类学研究领域,除了对种质资源进行形态学指标和分子水平评价之外,也进行生殖障碍如自交不亲和性(self-incompatibility)、单向不亲和性(unilateral incompatibility)、雄性不育性、2n配子(2n gametes)发生率和胚乳平衡数(endosperm balance numbers,EBN)的评价,并经常根据特定的研究目的,开展种质资源的生物胁迫(病害、虫害等)、非生物胁迫(干旱、霜冻和盐碱等)和品质性状(干物质、营养成分和炸片炸条等)评价。
1.3 种质资源的利用
马铃薯的起源依然存在争议,但无论马铃薯起源是源于多起源假说(multiple origin hypothesis)或是限制性起源假说(restricted origin hypothesis)[7,15],不可改变的事实是,马铃薯在自然界中主要以野生种形式存在。育种的本质是创造变异并进行选择,因此,马铃薯育种者需要不断地从野生种中寻找新的变异[16]。野生种被广泛应用于栽培种抗非生物胁迫如耐霜冻和抗低温糖化及抗生物胁迫如抗病虫(晚疫病、病毒病、青枯病、黄萎病、科罗拉多甲虫和线虫)性状改良,根据野生种与栽培种杂交从易到难变化即种质向栽培种转移从易到难又分为3个类别(表1)。此外野生种还可以提供非常丰富的等位基因多态性,拓展育种材料的遗传多样性[17]。
然而,由于野生种在进化过程中为了保持种性而与栽培种形成了诸如杂交不亲和、雄性不育和胚乳败育等生殖障碍[38],导致马铃薯野生资源利用难度增大。为了能将马铃薯野生种的优异性状转移到栽培种中,研究者开发了很多方法[17]:(1)倍性操作。倍性操作主要是通过体细胞加倍(秋水仙素处理和愈伤组织培养)和非减数配子进行染色体加倍,从而使杂交双亲或者配子体的 EBN达到相同数目后进行性状转移。有时倍性操作需要借助桥梁品种杂交来实现,S. acaule是常用的桥梁种之一[39-40]。(2)蒙导授粉(mentor pollination)与胚挽救。当花柱不亲和与胚乳败育同时发生的情况下,采用蒙导授粉和胚挽救策略有时可以获得种间杂种。蒙导授粉是指采用含有供体不易亲和花粉和介导者易亲和花粉的混合花粉进行母本授粉,借助介导者花粉与柱头识别反应而达到供体花粉成功受精目的,通常介导者的花粉具有胚斑标记,其后代容易鉴别和去除[41]。受精后,当胚不能正常发育时就需要进行胚挽救即将胚至于培养基上让其发育成熟。然而,对于马铃薯来讲,如果胚早期阶段就停止发育,胚挽救很难成功[42]。(3)激素处理。在介导者花粉缺乏胚斑等明显标记而不能进行后代选择的时候,可以利用2,4-D等生长素在授粉后24 h来处理子房从而达到获得实生种子的目的[42]。(4)正反交。在野生资源利用过程中,有些材料只有作母本或作父本才易于成功,例如当S.cardiophyllum与S.pinnatisectum杂交时,只有S. pinnatisectum作为母本才容易成功[42]。(5)亲和基因型选择。对于马铃薯来说,有时虽然2个种间可以杂交,但并不是种内的所有基因型间均可以杂交,这种种间杂交基因型的依赖性需要对杂交组合基因型进行选择以避免杂交障碍[43]。(6)体细胞融合。广义上属于倍性操作范畴,在花粉和柱头不亲和或者胚败育的情况下,体细胞杂交(somatic hybridization)或者原生质体融合(protoplast fusion)可以绕开有性杂交而进行野生资源利用。然而,体细胞杂交需要丰富的操作经验和大量的时间和物质投入,有时获得的体细胞杂种外源有利性状却不一定被导入。S.tuberosum与S.brevidens、S.bulbocastanum、S.circaeifolium、S.commersonii、S.acaule种之间的体细胞融合都见诸文献报道[17]。

表1 系统评价过或应用于马铃薯育种的野生种[16]Table 1 Wild relatives that have been evaluated and/or used in potato breeding
2 马铃薯基因组学研究
栽培马铃薯是四倍体作物,基因组高度杂合,存在严重的自交衰退现象,这给基因组学研究带来巨大挑战。揭示马铃薯基因组序列必须首先找到合适的测序材料,人们把目光投向了传统的组织培养技术。通过二倍体材料S. phureja的花药培养获得了一个单倍体材料,利用染色体加倍又获得了一个纯合的双单倍体材料DM1-3 516 R44(DM)[44]。同时,利用含有普通栽培种血缘的的二倍体杂合材料 RH89-039-16的BAC序列进行基因组序列的锚定和比较[45]。马铃薯基因组大概为844 Mb,通过DM序列的组装拼接,共获得727 Mb全基因组序列,没有完成组装的117 Mb主要是重复序列。通过 EST和已有的遗传和物理图谱上的分子标记对组装的基因组序列进行了验证。结合转录组和蛋白组学数据,从测序基因组中预测出了39 031个蛋白编码基因,其中9 875个基因存在可变剪接,这表明同一个基因即使序列不变却也存在更多的功能性变异[46]。
通过基因组序列中的共线性同源基因区块分析发现,马铃薯基因组存在2次全基因组范围的复制事件。马铃薯基因组不同单倍型序列之间存在高度的多态性。通过RH的部分区段序列与DM对应序列比较后发现,每隔40 bp就存在一个SNP,每隔394 bp就存在一个indel;RH的2个单倍型之间的部分序列比较表明,每隔29 bp就存在一个SNP,每隔253 bp就存在一个indel[46]。
近来,抗病和抗逆尤其是耐寒特性突出的马铃薯野生种 S.commersonii的基因组序列也被揭示[47]。相对于栽培种,该种的基因组杂合程度更低,重复区段更少,抗病候选基因更少,但却包含很多栽培种不具备的耐寒相关基因。目前,只有3个材料(DM基因组序列,RH的部分单倍型序列,PI243503基因组序列)的基因组序列被揭示,这远不能揭示丰富的马铃薯原始栽培种、新型栽培种、普通栽培种以及自然界中大量存在的野生种基因组水平上的遗传多样性,进行更多单倍型测序将对马铃薯研究具有更大的作用,当前涉及同源四倍体测序的技术尝试正在进行中。
3 马铃薯重要性状遗传学和决定基因
马铃薯由于存在无性和有性结合的混合繁殖方式,造成其遗传组成高度杂合,在自然界中存在大量从二倍体到六倍体的结薯和非结薯种。现代马铃薯栽培品种是同源四倍体作物,具有四体遗传(tetrasomic inheritance)特性[48]。虽然四体遗传比较复杂,但是单基因控制的质量性状和多基因控制的数量性状依然可以应用孟德尔遗传学和数量遗传学进行分析[49]。马铃薯许多质量性状是由主效基因控制,如控制晚疫病小种专一性抗性基因 R1-R11、非小种专一性抗性基因RB/Rpi-blb1、抗PVX基因Rx、抗PVY基因Ryadg和金线虫抗性基因H1等,薯皮和花冠颜色、薯形和芽眼深浅也是由主效基因控制的[49]。对于单基因控制的质量性状来说,可以通过子代测验(progeny tests)或者分子生物学手段如高分辨率熔解曲线(high-resolution melt,HRM)来分析亲本等位基因构成,对于等位基因组成形式是单式(simplex)或者复式(duplex)亲本,后代需要对目标性状进行筛选,而对于亲本组成是三式(triplex)或者四式(quadruplex)时,在排除双交换情况下,子代全部含有目标基因。
然而,马铃薯大多数性状是由多基因控制的数量性状。数量性状的表型是由基因型和环境互作形成的,子代表型数据常呈正态分布。数量性状可以通过家系均值和方差进行分析,但不同群体的均值和方差会变化很大。在试验设计中,如果环境变异被压缩到趋近于零时,可以认为表型变异全部由遗传变异决定的[50]。数量性状有效分析例如容错性子代测验(robust progeny tests)对于群体改良、选择策略和遗传增益(genetic gain)非常重要[51],研究者已经开始将这种统计分析方法应用于马铃薯的数量性状分析[49]。马铃薯主要性状及其遗传形式见表2。
在抗病基因定位和克隆方面,晚疫病、病毒病和线虫抗性研究有较大突破。晚疫病是马铃薯第一大病害,平均导致马铃薯减产16%[77]。晚疫病抗性研究中,最先受到育种者关注的是来自于S.demissum的主效基因R1-R11[78],在2000年左右,R1、R3、R2、R4和R10都被导入到栽培品种中[79-81],但由于田间晚疫病菌小种组成的变化,这些品种在田间相继失去抗性。由于小种专一性抗性基因不断被晚疫病菌克服,人们开始寻找并分离广谱抗性的主效抗性基因,例如来自于 S.bulbocastanum的基因 Rpi-blb1/RB、Rpi-blb2和Rpi-blb3等基因接连被定位和克隆[32,81-83]。相对于主效基因,晚疫病数量抗性研究也比较深入,在全部 12条染色体上共发现至少20个QTL[48,68,84],其中不乏一些贡献率高和重复性好的抗性位点。对于晚疫病主效抗性和数量抗性关系,GEBHARDT[85]认为:失效的R基因能增加数量抗性;一些抗性QTL通常与R基因连锁存在;一些防卫信号传导基因或者防卫反应基因属于数量抗性基因的一部分。马铃薯在进化过程中,由于抗性基因和病原菌之间的互作,使得马铃薯不同种中含有丰富的抗性基因资源,有时又形成一个个抗性基因家族,例如,晚疫病抗性基因 R2基因家族,就包括晚疫抗性基因R2、R2-like、Rpi-abpt、Rpi-blb3、RD、Rpi-edn1.1、Rpi-snk1.1、Rpi-snk1.2、 Rpi-hjt1.1、Rpi-hjt1.2和Rpi-hjt1.3[86-87]。已克隆的晚疫病主效基因见表3。
自然界中大概有 40余种病毒感染马铃薯,其中PLRV和PVY危害最大,其次是PVX、PVA、PVM、PVS和PMTV,是导致马铃薯退化的主要原因[48]。目前,已经有很多病毒抗性基因被定位和克隆。花叶病毒病的抗性分为极端抗性(extreme resistance,ER)和过敏抗性(hypersensitive resistance,HR),ER抗性基因无病毒小种选择性,抗性反应通常不表现出症状,而HR抗性的抗性依赖于病毒小种组成[98-99],抗性反应表现为明显的病毒侵染部位坏死。已知的病毒抗性基因见表4。
线虫对马铃薯根茎危害极大,并可以脱离寄主在土壤中长期存留,致使防治难度极大。对栽培马铃薯危害最大的线虫是孢囊线虫(Globodera spp.),其次是根结线虫(Meloidogyne spp.)。
在S.tuberosum ssp. andigena、S.spegazzinii和S.vernei等许多种中发现了根结线虫抗性基因,其中许多基因已经被导入到了栽培马铃薯种中[48]。目前,共有 14个线虫抗性位点被定位于马铃薯的8个连锁群中[108]。已知的马铃薯线虫抗性基因或者QTL见表5。

表2 主要马铃薯性状的遗传学Table 2 Genetic control of major potato traits

表3 已经克隆的马铃薯抗晚疫病基因Table 3 Late blight resistance genes cloned from potato
马铃薯细菌性病害如青枯病(Ralstonia solanacearum)、黑胫病(Pectobacterium atrosepticum)、软腐病(Pectobacterium spp.)和疮痂病(Streptomyces spp.)的抗性一般都属于数量抗性,在病原致病机理方面研究比较深入,但在马铃薯抗性机制和抗性位点研究方面进展较小,只是通过群体进行了抗性QTL定位或者通过原生质体融合或体细胞杂交方式进行了抗性的利用。普通栽培种(S.tuberosum)还没有发现具有青枯病抗性的材料,来自于二倍体栽培种S.phureja的抗性被广泛导入到栽培种中,但在高温时,青枯病抗性并不稳定且具有小种特异性[119]。目前,研究者主要是通过原生质体融合,从S.commersonii、S.chacoense、S.stenotomum 和茄子中向马铃薯中引入了抗性种质[120-123]。研究者对马铃薯野生种软腐病的抗性进行了评价,结果表明,来源于S.paucijugum、S.brevicaule和S.commersonii的材料具有良好的软腐病抗性[124]。马铃薯疮痂病抗性机制目前还不清楚,还没有鉴定出来针对疮痂病菌的抗性基因,只筛选出一些病菌侵染后的一些防卫相关基因[125],但疮痂病耐受品种的块茎通常会有更多、更厚的木栓细胞层[126],通过体细胞无性系筛选技术(somaclonal cell selection techniques)已经育成了具有疮痂病极端抗性的马铃薯材料[127]。

表4 已知的马铃薯抗病毒病基因Table 4 Known virus resistance genes from potato

表5 已知马铃薯抗线虫基因Table 5 Known nematode resistance genes from potato
在马铃薯块茎性状研究方面,块茎颜色、薯形和芽眼深度等表观性状研究较多。块茎颜色包括薯皮颜色和薯肉颜色。研究者最初认为,四倍体马铃薯薯皮颜色由D、R、P 3个位点控制,R位点控制红色色素合成,P位点控制紫色色素合成,而D位点是薯皮特异的、控制色素合成的调控因子[128]。在二倍体群体中,也相应地存在控制薯皮颜色的I、R、P三基因系统,I相当于四倍体中的D位点,P对R显性,即I_ R_ P_呈现紫色,I_ R_pp呈现红色,ii_ _ _ _呈现白色[129]。D、R、P位点被分别定位于第10、第2和第11染色体上[130-131]。借助于番茄色素结构和调节基因的相关研究,研究者发现 R编码二氢黄酮醇还原酶(dfr),P编码类黄酮羟化酶(f 3'5' h),D编码R2R3 MYB转录因子,并通过转基因进行了功能验证,D位点与薯形主效遗传位点和芽眼深度基因 Eyd相距不远[132-136]。薯肉颜色遗传机理与薯皮颜色类似,只是组织特异性的调控因子发生了改变[132]。
块茎形状的研究历史也较长,MASSON[137]将控制圆形薯形的基因命名为Ro,并确定该基因距着丝粒12.2 cM,后来的研究表明,除了Ro影响薯形性状之外,还有其他位点的修饰效应[138],最近,研究者构建了一个包含2 157个SNP标记的遗传图谱,将薯形基因定位在第10和第2染色体上,第10染色体存在主效效应[53]。芽眼深度和薯形存在相互关联,研究发现深芽眼和圆薯形2个性状成连锁关系,深芽眼控制基因被定位在第10染色体上,与薯形主效基因大概相距4 cM[54]。
而块茎的产量、淀粉和还原糖含量、炸片颜色和损伤等复杂性状,是由多基因控制的,遗传机制比较复杂,而且易受环境影响。近年来,研究者一般通过关联分析来进行复杂性状研究。LI等[139]利用243个四倍体品种(系),对块茎产量、淀粉含量和炸片颜色进行了分子标记关联分析,发现了50个与淀粉含量相关的分子标记。马铃薯块茎成熟时,碳水化合物主要是以淀粉和少量可溶性糖贮存在块茎当中。在块茎休眠期间,由于贮藏温度较低,淀粉又会部分转化成糖类物质,即“低温糖化(cold-induced sweetening)”现象,其严重影响块茎炸条和炸片质量。块茎淀粉和糖相互转化过程中,共有约18个遗传位点参与作用,SCHREIBER等[140]利用208个四倍体材料,克隆了1个质体淀粉磷酸化酶基因PHO1a,该基因可以提升块茎淀粉含量;研究发现,马铃薯中存在多种转化酶(invertase)基因,转化酶的活性与块茎低温糖化现象显著相关,对其进行基因沉默或抑制表达可显著改善低温糖化现象[139,141-142]。酶促褐变和机械损伤严重影响块茎商品品质,URBANY等[143]通过205个四倍体品种的酶促褐变和机械损伤性状与候选基因和SSR标记关联分析,鉴定出21个贡献率的较大的分子标记或遗传位点。
马铃薯耐受非生物胁迫如抗旱、耐寒、耐热和耐盐碱的机制比较复杂。马铃薯是水分高效利用的作物,但却对干旱比较敏感[144-145]。抗旱的遗传学研究主要集中在抗旱相关QTL定位方面,结果表明,马铃薯抗旱性遗传复杂,受多个位点的影响[61,146],此外关于马铃薯抗旱的转录组学研究也较多[147-148]。马铃薯耐寒性和冷驯化能力是独立遗传控制的并且是由许多微效基因影响[149]。高温会造成块茎畸形、表皮开裂和内部坏死,而这些表型都是受独立的遗传机制控制的[59,150]。马铃薯耐盐碱也是属于多基因控制的复杂性状,应用QTL定位和转录组学进行耐盐碱机制研究见诸报道[59,151]。
4 马铃薯育种技术
4.1 传统育种技术
目前,世界范围内育成的马铃薯品种中,绝大多数是依靠传统育种技术育成的品种,传统育种技术是马铃薯育种技术的基础。马铃薯传统育种技术是在双亲杂交产生的子代基础上,进行多代无性世代性状评价和选择,进而培育出优良品系和品种的过程。在无性世代的评价过程中,由于选择压力的增大,育种群体规模逐渐缩小,但入选的每个品系植株数量却不断增加[152]。在传统育种中,亲本的选配非常关键,亲本一般为综合性状优良、个别性状需要改良的育成品种或者是经过子代测验证明能产生优良后代的育种材料,通常要求亲本性状互补[49]。由于马铃薯一些主要商品性状如块茎产量、数量、大小和比重等易受环境影响,无性世代品系通常需要进行多年多点的试验评价,所以育成一个马铃薯新品种通常需要10年左右的时间。表6是一个典型的中国北方一季作地区的传统育种流程。

表6 中国北方一季作地区传统育种流程Table 6 Conventional potato breeding program in single cropping zone of China
4.2 倍性育种技术
自然界中,马铃薯大部分以不同倍性的近缘资源存在,其中74%以上是二倍体[153]。在EBN数目不同的情况下,它们难以与四倍体普通栽培马铃薯通过杂交进行优异性状转移,而倍性操作技术是进行野生资源利用的重要方法。广义上的倍性育种技术包括双单倍体(dihaploid)诱导、2n配子利用和体细胞杂交或者原生质体融合等技术。
1958年,HOUGAS等[154]通过马铃薯普通栽培种与来源S.phureja的材料杂交,成功地通过孤雌生殖诱导出双单倍体,发现S.phureja种的某些选系是诱导四倍体孤雌生殖产生单倍体的优良授粉者(pollinator)[155]。在授粉过程中,授粉者的精细胞正常进入母本子房,可以使胚乳正常发育,同时刺激未受精的卵子发育成双单倍体(2x)的胚[7]。但有时二倍体授粉者会产生2n花粉,这样就会造成母本产生孤雌生殖的双单倍体种子的同时,也会伴有四倍体种子,为解决此问题,研究者把一个产生胚斑的标记基因转入具有高诱导能力的S.phureja无性系中,这样在选择时就可以直接淘汰授粉后代中带有胚斑的四倍体杂交种子,提高了双单倍体的诱导效率[156]。HOUGAS等[157]在1960年报道了双单倍体可与24个结薯二倍体种杂交,并得到了健壮的后代。
CHASE[158]提出了利用二倍体资源的分解育种法(analytical breeding),接着研究者对利用二倍体杂种发生不减数 2n配子而产生的四倍体后代进行了评价[159],形成了分解合成育种方法并应用于育种实践。2n配子即染色体不减数的配子或者是和体细胞染色体数目一样的配子,它是由控制减数分裂的隐性基因控制的[7,160]。2n配子包括2n卵子和2n花粉,2n卵子是由于第二次减数分裂重组(second division restitution,SDR)形成的,而2n花粉主要是由于第一次减数分裂重组(first division restitution,FDR)形成的[7],并能将亲本的杂合性100%的传递给后代。2n卵子的鉴别需要借助一系列显微技术[161],这给检测带来很多不便,但实际应用过程中,二倍体母本和四倍体父本杂交,如果产生实生种子,通常就认为二倍体母本产生了2n卵子[162]。2n花粉检测比较简单,染色后通过普通光学显微镜即可检测,其大小比正常花粉几乎大了一倍并多了一个萌发孔。利用可以产生 2n配子的二倍体材料与四倍体普通栽培种杂交,可以产生四倍体实生种子。在分解合成育种中,通过种间杂交诱导四倍体栽培品种孤雌生殖产生双单倍体,其与二倍体野生种和原始栽培种杂交产生二倍体杂种,从而达到利用丰富的野生资源的目的,最终利用二倍体杂种产生的 2n配子将优良性状转育到普通四倍体品种中[163]。目前,通过分解合成育种法育成了许多具有二倍体野生种种质的栽培品种或育种材料[164-167]。
体细胞杂交是除了2n配子利用技术和双单倍体诱导技术外,绕过杂交障碍进行近缘种种质转移的重要技术,可以同时转移细胞核和细胞质基因,尤其在转移多基因控制的数量性状进行育种材料创制方面应用广泛[168]。对于体细胞杂交技术,进行原生质体分离、培养、融合和再生程序复杂,且具有种质依赖性,因此,进行原生质体融合建立一个稳定的工作体系非常重要。目前通过原生质体融合获得了很多具有抗病毒病、晚疫病和青枯病的育种材料[122,168-169]。
4.3 标记辅助选择(marker-assisted selection,MAS)
由于栽培马铃薯是高度杂合的常异花授粉四倍体作物,异交率仅为0.5%左右,其杂交后代优良基因或者染色体区段重组到一起的概率极小,这就需要育种者必须通过扩大 F1代群体数量来增加优良子代出现概率,这就极大增加了选择工作量。正常来讲,育种者要每年要评价40个左右的植株和块茎性状,从杂交到品种释放通常需要 10年左右时间[85]。随着分子遗传学和分子生物学研究的进步,标记辅助育种技术成为了加快育种进程的重要手段。由于栽培马铃薯四体遗传特性和自交造成的高度等位变异,使得马铃薯相比其他主要作物,相对缺乏可以应用于育种的分子标记[170]。马铃薯育种辅助选择标记主要集中在重要农艺性状如抗病性和块茎品质方面,在复杂性状如产量和非生物胁迫方面的分子标记开发比较缓慢。目前,已经发表的、可用于育种群体选择的分子表记如表 7
所示。
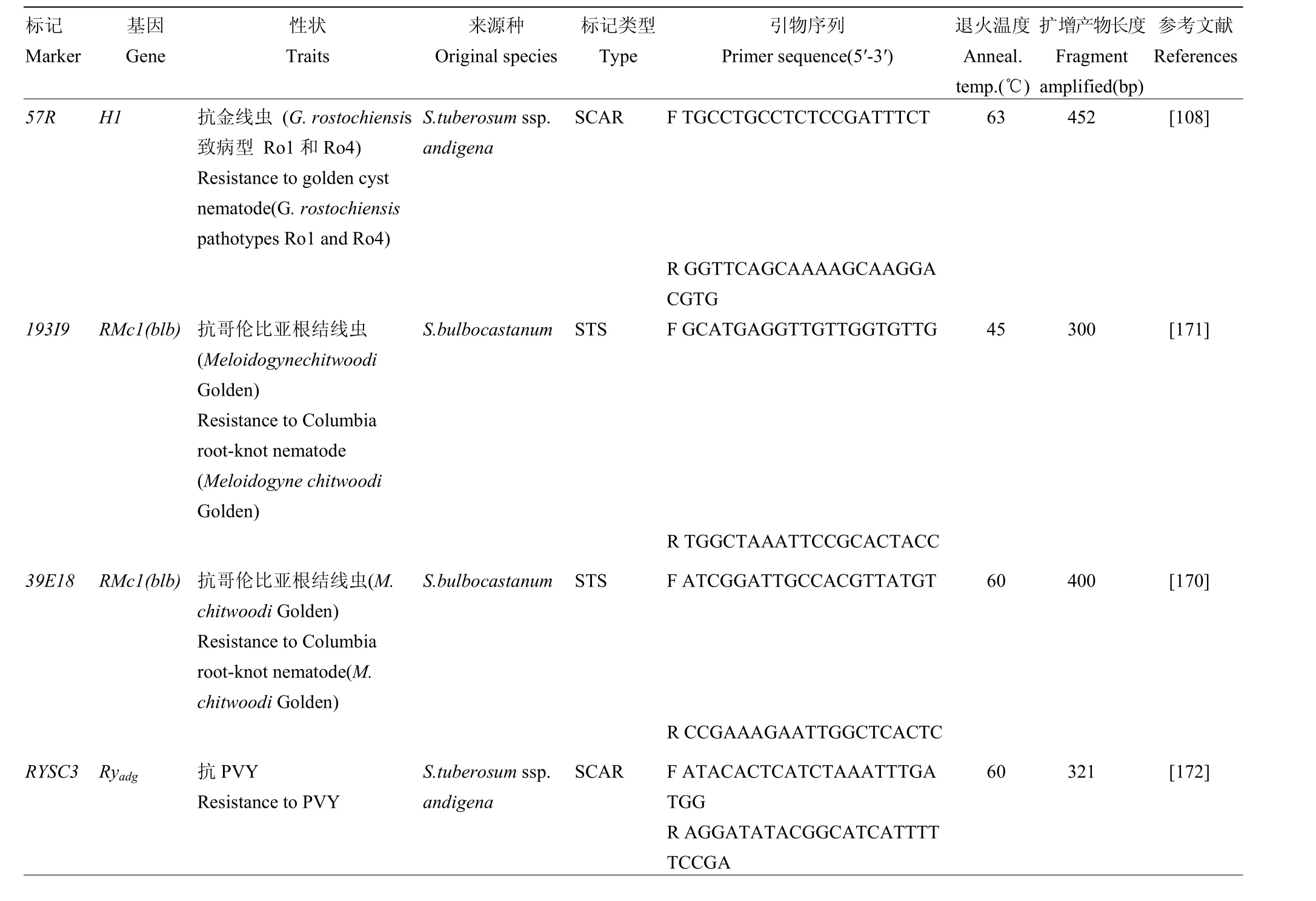
表7 可用于马铃薯标记辅助选择的分子标记Table 7 Markers used in potato breeding selection

续表7 Continued table 7

续表7 Continued table 7
在分子标记实际应用于育种群体选择时,有几点需要注意:(1)开发的分子标记的遗传背景。大多数与目标性状连锁的分子标记是在二倍体水平上开发的,而相当一部分的栽培种群体是不含有二倍体种质的,因此,四倍体栽培种群体是否含有目标性状分子标记,所属的遗传背景非常关键,成功的育种选择标记必须经过育种群体或资源的验证才能证明其有效性[186]。(2)分子标记的组合应用。一方面,由于不同遗传背景的资源的等位基因具有丰富的序列变异,对于特定性状的选择标记,需要标记组合应用才能完全追踪到目标性状,例如对于来源于S.tuberosum ssp. andigena的抗PVY的Ryadg来说,RYSC3标记具有良好的选择效果[172],但是后来的研究者利用育种群体和不同遗传背景的品种、品系进行标记验证发现,RYSC3结合RYSC4和ADG2/BbvI标记共同检测时会取得更好效果,即同时含有这3个标记,植株一定表现出对PVY抗性[187];另一方面,一部分分子标记不是依据目标性状决定基因序列开发的,而是其单侧的序列开发,因此,对此类标记,应用两侧标记同时进行选择,可提高选择准确率;此外,在标记检测过程中,应用多重PCR检测体系,即将多个标记检测融合到一个PCR反应中,可以显著提高标记检测效率[175]。
4.4 基因组选择(genomic selection,GS)
标记辅助选择自20世纪80年代开始应用以来,主要针对少数基因控制的简单性状,而对于微效多基因控制的复杂性状无能为力[188-189]。而基因组选择是检测全基因组范围的所有分子标记,而不是针对单一性状的部分标记。基因组选择主要流程是通过试验群体来估计出每个标记(通常是SNP)或者不同染色体区段的效应值,然后再利用这些效应值来计算育种群体的育种值,进而进行后代个体选择[190]。基因组选择技术自2001年开始应用于预测复杂性状的表现以来,现已应用于奶牛、玉米、棕榈和大麦等动物和植物的育种上,显著提高了育种选择效率[189,191-192]。
随着新一代测序技术(next generation sequencing,NGS)的不断改进和完善及生物信息学平台的丰富,数以万计的、以SNP为主的分子标记不断地被开发出来,这为基因组选择提供了充足的分子标记信息。以马铃薯基因组序列信息为基础,基于Illumina平台、包含20 000个SNP标记马铃薯全基因组SNP芯片已经商业化应用[193]。栽培马铃薯遗传复杂,传统育种中马铃薯存在严重的连锁累赘,优异基因的导入常常伴随着不利性状。因此在马铃薯育种中通常既要尽量保留亲本材料优异遗传背景,又要定向改良现有受体亲本的特定性状,也就是说将前景选择和背景选择结合起来创制优异新材料和选育优良新品种,而基因组选择技术是以前景选择和背景选择相结合的一项新的标记辅助选择技术。目前,全基因组SNP芯片已经应用于马铃薯种质资源遗传多样性、简单性状和复杂性状的基因定位和QTL作图,但还没有以SNP标记为基础、将基因组选择技术系统应用于马铃薯育种进程的报道,因此,开展系统的基因组选择技术研究并将之应用于材料创制和品种选育意义重大。
4.5 基因工程(genetic engineering)
传统的基因工程手段,一般通过基因转化或者基因沉默技术进行作物改良,一直是伴随着争议的研究热点。2015年,美国农业部批准了抗损伤和褐化的马铃薯品种 Innate®[194],该品种是通过 RNAi(RNA interference)技术限制了多酚氧化酶基因 PPO5和天冬酰胺酸合成酶基因Asn1的表达,从而使马铃薯加工过程中不会褐变和降低有害物质丙烯酰胺含量。最近,增加了晚疫病抗性和抗低温糖化能力的 Innate®二代改良品种也已经通过了美国食品和药品管理局的审批,正在办理环保注册手续,准备投放市场(http:// www.simplotplantsciences.com)。在马铃薯中,最早的转基因品种是针对科罗拉多甲虫(colorado potato beetles)抗性的转Bt基因Russet Burbank[195],后来又引入了病毒病抗性基因,形成商业化品种Newleaf®系列,并于1996年获得美国农业部审批投放市场[196-197],但是后来由于加工企业和消费者的排斥,不得不退出商业化种植。通过转基因技术提高马铃薯晚疫病抗性也是研究热点,晚疫病抗性基因R1、R3a、RB/Rpi-blb1都通过转基因方式引入到感病品种中并使其获得了抗性[80],甚至将3个晚疫病抗性基因Rpi-sto1、Rpi-vnt1.1和Rpi-blb3同时引入到一个马铃薯品种Désirée中[183]。
考虑到转基因产品(genetically modified organism,GMO)的争议,研究者相继提出了无标记质粒(markerfree plasmids)[198]和顺式转基因(cisgenesis)的技术策略[199]。传统的携带外源基因的转化质粒含有使细胞产生抗生素抗性的基因,以方便阳性转化材料的筛选,但人们担心这种抗生素抗性基因会漂移并整合到其他非目标生物体的基因组中,因此研究者开发了一系列无抗性选择标记的转化质粒筛选系统[200]。相对于传统的转基因策略,顺式转基因定义是:引入生物体的外源核酸片段,只是来源于本种或与本种可杂交的种具有的、自然界本来就存在的核酸片段(包括启动子和终止子),不含有任何其他不可杂交物种的外源核酸片段[201]。顺式转基因的概念实质上既包括了对无标记质粒的要求,又对质粒携带的外源基因进行了规范和限制。
近几年来,在基因工程领域,CRISPR(clustered regularly interspaced short palindromic repeats)技术炙手可热[202-203],该技术原理是依靠一种称作Cas9的蛋白酶,利用引导性RNA分子锁定目标DNA,进而对DNA进行编辑达到阻断基因表达或者插入目标基因的目的。相对于锌指核酸酶(zinc finger nucleases)和TALEN(transcription activator-like effector nucleases)技术,CRISPR技术更高效、更特异,而且成本更低,被誉为继 PCR技术之后生物科学领域的又一个革命性技术[204]。目前,CRISPR技术已经有了应用于马铃薯研究的报道[205-206]。最近,另一种称之为 NgAgo–gDNA的基因组编辑技术见诸报道[207],该技术是在一种短序列DNA引导下,NgAgo蛋白酶对特定DNA序列进行编辑,初步试验结果表明,NgAgo–gDNA相对于 CRISPR,效率更高且目标序列不受所处位置限制,但该技术结果重复性和应用效果有待进一步验证。在技术风险可控情况下,基因组编辑技术在消除马铃薯不利性状如抗低温糖化、降低丙烯酰胺和龙葵素含量等提升块茎品质方面,以及编辑马铃薯特定目标性状基因来提高抗病、抗逆能力方面将发挥重要作用。
5 中国马铃薯遗传育种现状
2014年,中国马铃薯总产量9 551.5万吨,但单产只有16.92 t·hm-2(http://fao.org/faostat),相比发达国家,单产增长的潜力依然巨大。中国马铃薯育种虽然起步较晚,但经过一代代育种者的不懈努力,从最初的品种引进到培育出一系列具有完全自主知识产权的品种,品种选育工作取得了重要突破,为中国马铃薯产业发展提供了坚实的品种支撑。
5.1 育种单位和育种历史
目前,中国主要从事马铃薯品种选育的单位超过30家,以科研院所和大学为主,从事育种的企业相对较少。中国的马铃薯育种历史始于20世纪30年代的国外品种和资源引进[3],而20世纪40年代由于战乱,育种进程基本停滞;20世纪50年代,应用了36个高产并具有晚疫病抗性的引进品种,并开始了将实生种子(true potato seed,TPS)应用于生产的尝试;20世纪60年代,中国育成了具有完全自主知识产权的马铃薯新品种,当时的育种目标以适应性、优质、抗病和高产为主;到了20世纪70年代,中国成为世界上TPS应用最广泛的国家,这对中国西南山区马铃薯发展起到了积极作用; 20世纪80年代,育种目标聚焦到抗病和高产,在此期间,共育成了克新系列、高原系列和坝薯系列等93个马铃薯新品种;20世纪90年代,育种目标开始强调早熟性、加工型新品种的选育,在此期间,共育成了中薯系列、晋薯系列、鄂薯系列、郑薯系列和青薯系列等67个马铃薯新品种;21世纪以来,中国马铃薯品种选育进程加快,共育成了 362个马铃薯新品种。
5.2 种质资源和育成品种
目前,中国各单位共保存了包括国内审定品种、国外引进品种、育种品系、原始栽培种和野生种等在内的5 000余份种质资源,这些资源大多数以试管苗形式保存,少部分以实生种子和块茎形式保存,其中2 000份左右的资源被系统评价过。资源保存数量以中国农业科学院蔬菜花卉研究所和国家马铃薯改良中心(依托于黑龙江省农业科学院克山分院)为最多。
截止到2016年底,根据品种审定部门公告统计,中国共审定马铃薯品种611个(含国外引进品种),其中绝大多数为鲜食品种,而加工品种以国外引进品种为主。20世纪60年代育成的克新1号依然是中国种植面积最大的品种。
5.3 育种目标和品种轮换
高产、稳产、抗病、耐贮和优质是中国马铃薯最重要的育种目标,专用品质好、薯形好、芽眼浅、早熟、高产、抗病、抗逆是重点选择方向。不同的栽培区育种目标各不相同,北方一作区以中熟和晚熟品种选育为主,东北地区尤其注重抗晚疫病和黑胫病,华北和西北地区注重耐旱、抗土传病害、晚疫病和病毒病;中原二季作区以早熟或块茎膨大快、对日照长度不敏感的品种选育为主,早熟、高产、休眠期短、抗病毒病和疮痂病是主要的育种目标;对于西南一二季混作区的高海拔地区,主要是培育高抗晚疫病、癌肿病和粉痂病的中晚熟和晚熟品种,而对于中低海拔地区,则为以抗晚疫病、病毒病的中熟和早熟品种选育为主;在南方冬作区,品种选育聚焦日照长度反应不敏感、抗晚疫病和耐湿、耐寒和耐弱光的中、早熟品种。
在育种进程中随着育种目标的不断调整和新品种推广应用,中国主栽品种先后经历了四批次的品种轮换:(1)1950—1970年,17世纪以来从欧美国家引入中国的部分品种成为了适应当地条件的地方品种如河坝洋芋、深眼窝、广灵里外黄和乌洋芋。20世纪30和 40年代引进筛选出胜利(Triumph)、卡它丁(Katahdin)和巫峡等品种成为了20世纪50年代的主栽品种。到了20世纪60年代,主栽品种逐渐被晚熟品种米拉(Mira)、疫不加(Epoka)和阿奎拉(Aquila)及早熟品种白头翁(Anemone)所替代;20世纪 60年代后期,自主育成的虎头、跃进和晋薯2号等几十个抗晚疫病高产品种与上述引进品种一起成为主栽品种。(2)第二次品种轮换发生在20世纪80年代,自主育成的晚熟品种克新1号、坝薯8号和高原7号,早熟品种郑薯2号、郑薯4号和坝薯9号,以及国外引进品种费乌瑞它(Favorita)、台湾红皮(Cardinal)、底西芮(Désirée)和中心 24(CIP24)逐渐成为主栽品种。(3)第三次品种轮换发生在 1980年至 2000年期间,20世纪80和90年代育成的早熟新品种东农303、中薯2号、郑薯5号、郑薯6号和川芋早等品种种植面积增长迅速,引进品种大西洋(Atlantic)、夏波蒂(Shepody)、阿格瑞亚(Agria)和斯诺登(Snowden)等加工专用品种和冀张薯5号(Kondor)、抗疫白(Kennebec)等鲜食品种应用于生产,到2000年左右实现了第三次品种轮换。(4)2000年后,加大了专用和早熟新品种选育,育成了新品种300多个,其中中薯3号和中薯5号等早熟品种种植面积稳定增长,2006年首次国家级审定了炸片加工专用品种中薯10号和中薯11号,2010年前后实现了品种的第四次轮换。
在品种轮换过程中,品种的遗传背景被不断拓宽。1983年前,利用6个常用亲本多籽白(292-20)、卡它丁、疫不加、米拉、白头翁和小叶子育成品种 74个,占这一时期审定品种总数的68.8%,而2005年以前,利用上述6个亲本共育成了156个品种,占同时期审定品种总数的45%。另外,国外品种作为亲本资源在中国马铃薯品种选育中占有重要地位,2012年前育成的 379个审定品种中,含北美亲本血缘的占13.7%、欧洲亲本血缘的占35.9%,含国际马铃薯中心亲本血缘的占17.9%。中国马铃薯品种类型不断丰富,在产量、品质、抗病和外观性状上有较大改良。
6 荷兰马铃薯育种水平位居世界前列的关键因素分析
2014年,荷兰马铃薯总产710.03万吨,不及中国总产量8%,但其单产达到45.66 t·hm-2,近中国单产的3倍(http://fao.org/faostat)。荷兰的马铃薯育种和栽培水平位居世界前列,下面简要分析一下其育种特点。
6.1 参与育种模式(participatory plant breeding)
荷兰马铃薯育种体系是大学或研究机构、育种公司和农民育种者(farmer breeder/hobby breeder)共同组建而成的,形成了别具荷兰特色马铃薯参与育种模式。荷兰马铃薯育种主要由商业育种公司作为主体完成,而农民育种者在育种体系中扮演重要角色。荷兰有近一半的农民种植马铃薯,农民是荷兰马铃薯育种的重要参与者。2009年,荷兰409个品种用于种薯生产,其中293个品种是荷兰本国育成的,而这293个品种中,有一半左右的品种是由农民育种者选育出来的,占荷兰种薯生产面积的44%[208]。荷兰有19家较大的马铃薯育种公司,其中13家公司拥有自己的育种体系,14家公司与农民育种者进行品种选育合作。年繁殖实生苗数量超过5万的、大的育种公司,一般都是既有自己的育种体系,又和农民育种者具有合作关系,而年繁殖实生苗数量小于1.5万的、小的育种公司仅仅依靠自由育种者(与公司没有合作关系)以及外国育种者进行品种选育。
大多数优秀的农民育种者都拥有可进行种薯生产的、50—80 hm2规模的农场,他们具有丰富的优质种薯生产和优良单株选择经验。通常每年冬天,与公司合作的农民育种者会收到来自育种公司的、含有系谱信息的实生种子和实生苗家系,然后他们根据各自偏好的育种目标,在与育种公司充分讨论的基础上进行材料选择,然后农民育种者对这些材料进行田间种植、性状评价和选择,3年以后,大约有1%的株系保留下来并返回育种公司,进行后续的多地多年的田间评价及抗病性、品质等性状的室内评价,有时育种公司会根据品种市场需求情况,直接将相关品系送往目标市场国家进行评价。经过这样的程序,育成1个马铃薯品种需要大概12年时间。育种公司和农民育种者的合作育种流程见表8[209]。

表8 荷兰育种公司和农民育种者的合作育种流程Table 8 Potato breeding program in a collaborative model of company and farmer breeder
6.2 公立科研机构在育种体系中的角色
荷兰设有很多涉及马铃薯遗传育种和种薯质量检测的公立科研机构和管理部门,下面以瓦赫宁根大学及研究中心(Wageningen University and Research,WUR)为例说明一下公立科研机构在育种体系中的角色。在马铃薯遗传育种研究方面,WUR主要从事以种质扩增、改良和创新为核心的前育种(pre-breeding)研究,利用最新的遗传学研究成果,进行种质资源评价、亲本选配和杂交及育种材料创制和低代品系培育,具体工作分为传统育种和有机育种 2个体系同时进行。传统育种体系中,无性一代至无性三代品系均会保留一部分块茎网棚内种植,以免退化;有机育种体系各世代材料均在符合有机食品种植要求的地块评价,一般不在网棚内备份繁种。对于传统和有机育种体系的无性世代评价,WUR只负责早代品系的分子标记(晚疫病、病毒病和线虫抗性为主)、田间农艺性状和块茎品质性状的评价,一般在第四个无性世代评价时就将品系转交给商业育种公司进行后续工作。WUR每年也会将部分杂交实生种子直接提供给公司和农民育种者进行筛选和评价[210],同时面向育种公司和农民育种者进行育种技术和田间选择技术的培训。
6.3 有机育种体系
荷兰的传统育种体系和中国差别较小,而有机育种体系相对传统育种体系有较大差别。有机育种体系是指其培育出来的马铃薯品种符合有机食品生产标准要求,因此,其育种目标就更具挑战性。对于有机栽培的马铃薯品种总的要求是在短的生育期内快速结薯,在病害侵袭和单纯有机肥施用情况下,尽可能多的获得较高经济产量[211],因此,荷兰有机马铃薯育种目标分为几个层次:(1)必须具备性状:叶片和块茎具有良好的晚疫病抗性,良好的氮肥利用效率;(2)应该具备性状:立枯病抗性,早疫病抗性,PVY抗性,早熟性,长休眠期;(3)最好具备性状:疮痂病和粉痂病抗性,植株快速封垄以压制杂草,银腐病抗性,食味好。为了达到以上育种目标,又在不借助转基因技术的情况下,在早代育种群体即开始应用分子标记进行以抗病虫为主的目标性状辅助选择,以加快育种进度。
7 展望
随着生物科学的迅猛发展,基础研究领域取得的科研成果越来越快地直接在应用研究领域应用并取得成效,这直接促使了作物综合育种技术的不断成熟和完善,同时创新育种模式和机制,利用现有种质资源培育突破性、专用型品种将是未来马铃薯遗传育种发展的主要方向。
7.1 基因组学数据在遗传育种上的应用
马铃薯基因组序列已经被揭示,虽然获得的全基因组序列只是源于2个种的测序材料,这相对于自然界中存在广泛变异的马铃薯种质资源来说只是冰山一角,但基因组序列对马铃薯重要性状遗传学的促进作用已经显现。DM参考基因组序列促进了很多重要性状基因的定位和克隆,基于其开发的SNP芯片已经商业化应用。目前,研究者正在着手进行马铃薯种质资源重测序和攻克四倍体栽培种测序技术。在可预见的未来,随着包含更多等位变异信息的基因组序列信息的揭示,马铃薯主要性状尤其是多基因控制的、复杂性状的遗传机制将逐渐被揭示,这会大大促进基于全基因组水平上的标记选择技术在育种群体评价和选择上的应用,从而为育种者在亲本组合选配上提供分子水平上的依据,并在早代群体中即可快速锁定目标品系,加快育种进度。
7.2 种质资源收集和改良重要性凸显
利用基因工程技术进行品种改良,在技术风险、食物安全、环境保护甚至社会伦理上一直存在争议,即使在技术风险得到控制的情况下,转基因食品的释放是依然一个慎重和缓慢的过程。因此,为了培育突破性新品种,收集和挖掘现有马铃薯种质资源优异性状,将是一个长期的基础性工作。中国不是马铃薯起源国,而且涉及马铃薯起源地区的国家已经限制一些重要资源的释放,这增加了资源引进的难度。作为应对措施,应当一方面通过合法途径从世界上主要马铃薯种质资源库进行资源引进;另一方面,要对中国现有种质资源进行系统评价,挖掘其应用潜力。
7.3 综合育种技术逐渐形成
目前,中国乃至世界范围内种植的马铃薯品种主要是依靠传统育种技术培育而成的,随着生物学研究尤其是分子生物学领域的快速发展,马铃薯育种技术必将逐渐整合现有技术方法并吸纳最新基因组学研究成果形成综合育种技术。具体来说,马铃薯综合育种技术,将以杂交为基础的传统育种技术为基础,以2n配子利用技术和体细胞杂交为主的倍性操作技术为资源改良和创制的途径,以简单性状和复杂性状追踪及利用的标记辅助选择技术和基因组选择技术为提升亲本组配和后代选择效率的工具,以培育抗病、抗逆、高产、优质、专用马铃薯优良品种为目标,全面促进马铃薯优良品种选育进度。
7.4 专用型品种选育需求持续增大
随着马铃薯加工业的迅速发展、环境友好的生产方式的转变及人们对食品安全和营养的关注度增加,选育特定市场和可持续农业发展需求的专用型品种尤显必要。选育耐旱、耐瘠薄、低成本种植品种,可以提高华北、西北和西南等占中国马铃薯总种植面积70%左右的、土壤瘠薄、经济不发达主产区的马铃薯旱作生产能力,促进生态脆弱区域水、光、温、土等自然资源的综合利用;选育优质丰产早熟品种,可以在中原二季作区和南方冬作区进行设施和保护地种植,提高种植效益;选育抗病、耐贮藏、优质品种,可以减少东北、西南、西北、华北等地区气传和土传病害重发区农药施用、产量和贮藏损失,提高马铃薯商品性;选育优质加工专用品种,改良品种的块茎干物质、淀粉、还原糖等加工品质特性,可以提高加工原料生产能力,促进马铃薯加工业稳定、持续发展。
7.5 商业化育种模式的尝试
具有中国自主知识产权的马铃薯品种主要是公益性科研院所和大学育成的,马铃薯企业鲜有完善的育种体系,多是以种薯繁殖、销售和加工为主。在市场经济中,企业会最先觉察到市场需求并开发相关产品,利用其商业推广体系,可以快速将产品推向市场,荷兰马铃薯育种模式的成功证明了这一点。随着马铃薯品种应用的市场导向机制的不断完善,以及科研院所和企业人才交流及技术合作的不断加强,企业会在中国马铃薯品种选育中扮演越来越重要的角色,但公益性育种机构与企业的合作机制有待于进一步探索。
[1] MULLINS E, MILBOURNE D, PETTI C, DOYLE-PRESTWICH B M, MEADE C. Potato in the age of biotechnology. Trends in Plant Science, 2006, 11(5): 254-260.
[2] 屈冬玉, 谢开云, 金黎平, 庞万福, 卞春松, 段绍光. 中国马铃薯产业发展与食物安全. 中国农业科学, 2005, 38(2): 358-362.
QU D Y, XIE K Y, JIN L P, PANG W F, BIAN C S, DUANG S G. Development of potato industry and food security in China. Scientia Agricultura Sinica, 2005, 38(2): 358-362. (in Chinese)
[3] 金黎平, 屈冬玉, 谢开云, 卞春松, 段绍光. 我国马铃薯种质资源和育种技术研究进展. 种子, 2003, 5: 98-100.
JIN L P, QU D Y, XIE K Y, BIAN C S, DUAN S G. Advances of potato germplasm and breeding technology in China. Seed, 2003, 5: 98-100. (in Chinese)
[4] HAWKES J G, FRANCISCO-ORTEGA J. The early history of the potato in Europe. Euphytica, 1993, 70(1): 1-7.
[5] JOHNS T, ALONSO J G. Glycoalkaloid change during the domestication of the potato, Solanum Section Petota. Euphytica, 1990, 50(3): 203-210.
[6] GHISLAIN M N, ÚÑEZ J, HERRERA M R, SPOONER D M. The single Andigenum origin of Neo-Tuberosum potato materials is not supported by microsatellite and plastid marker analyses. Theoretical and Applied Genetics, 2009, 118(5): 963-969.
[7] SPOONER D M, GHISLAIN M, SIMON R, JANSKY S H, GAVRILENKO T. Systematics, diversity, genetics, and evolution of wild and cultivated potatoes. The Botanical Review, 2014, 80(4): 283-383.
[8] HAWKES J G. The Potato: Evolution, Biodiversity, and Genetic Resources.Washington D. C.: Smithsonian Institution Press, 1990.
[9] PANTA A, PANIS B, YNOUYE C, SWENNEW R, ROCA W M. Development of a PVS2 droplet vitrification method for potato cryopreservation. CryoLetters, 2014, 35(3): 255-266.
[10] ROCA W M, ESPINOZA N O, ROCA M R, BRYAN J E. A tissue culture method for the rapid propagation of potatoes. American Potato Journal, 1978, 55(12): 691-701.
[11] GONZALEZ-ARNAO M T, PANTA A, ROCA W M, ESCOBAR R H, ENGELMANN F. Development and large scale application of cryopreservation techniques for shoot and somatic embryo cultures of tropical crops. Plant Cell, Tissue and Organ Culture, 2007, 92(1): 1-13.
[12] KACZMARCZYK A, ROKKA V M, KELLER E R J. Potato shoot tip cryopreservation, a review. Potato Research, 2010, 54(1): 45-79.
[13] VAN DEN BERG R G, MILLER J T, UGARTE M L, KARDOLUS J P, VILLAND J, SPOONER D. Collapse of morphological species in the wild potato Solanum brevicaule complex (Solanaceae: sect. Petota). American Journal of Botany, 1998, 85(1): 92-109.
[14] COOKE R J. New approaches to potato variety identification. Potato Research, 1999, 42(3): 529-539.
[15] SALAMAN R N. The early European potato: Its character and place of origin. Journal of the Linnean Society(Botany), 1946, 53: 1-27.
[16] CASTAÑEDA-ÁLVAREZ N P, DE HAAN S, JUÁREZ H, KHOURY C K, ACHICANOY H A, SOSA C C, BERNAU V, SALAS A, HEIDER B, SIMON R, MAXTED N, SPOONER D M. Ex situ conservation priorities for the wild relatives of potato (Solanum L. Section Petota). PLoS ONE, 2015, 10: e0122599.
[17] JANSKY S. Overcoming hybridization barriers in potato. Plant Breeding, 2006, 125(1): 1-12.
[18] ESTRADA R N. Frost resistant potato hybrids via Solanum acaule, Bitt. Diploid-Tetraploid crosses. American Potato Journal, 1980, 57(12): 609-619.
[19] SUÁREZ S, CHAVES E, CLAUSEN A, FRANCO J. Solanum tuber-bearing species resistance behavior against Nacobbus aberrans. Journal of Nematology, 2009, 41: 5-10.
[20] WATANABE K N, ORRILLO M, VEGA S, MASUELLI R, ISHIKI K. Potato germplasm enhancement with disomic tetraploid Solanum acaule. II. Assessment of breeding value of tetraploid F1hybrids between tetrasomic tetraploid S. tuberosum and S. acaule. Theoretical and Applied Genetics, 1994, 88(2): 135-140.
[21] CARPUTO D, CARDI T, SPEGGIORIN M, ZOINA A, FRUSCIANTE L. Resistance to blackleg and tuber soft rot in sexual and somatic interspecific hybrids with different genetic background. American Potato Journal, 1997, 74(3): 161-172.
[22] FROST K E, JANSKY S H, ROUSE D I. Transmission of Verticillium wilt resistance to tetraploid potato via unilateral sexual polyploidization. Euphytica, 2006, 149(3): 281-287.
[23] JANSKY S H, HAMERNIK A, BETHKE P C. Germplasm release: Tetraploid clones with resistance to cold-induced sweetening. American Journal of Potato Research, 2011, 88(3): 218-225.
[24] SANTINI M, CAMADRO E L, MARCELLÁN O N, ERAZZÚ L E. Agronomic characterization of diploid hybrid families derived from crosses between haploids of the common potato and three wild Argentinian tuber-bearing species. American Journal of Potato Research, 2000, 77(4): 211-218.
[25] BRADSHAW J E, RAMSAY G. Utilisation of the commonwealth potato collection in potato breeding. Euphytica, 2005, 146(1): 9-19.
[26] TUCCI M, CARPUTO D, BILE G, FRUSCIANTE L. Male fertility and freezing tolerance of hybrids involving Solanum tuberosum haploids and diploid Solanum species. Potato Research, 1996, 39(3): 345-353.
[27] LINDQVIST-KREUZE H, CARBAJULCA D, GONZALEZESCOBEDO G, PÉREZ W, BONIERBALE M. Comparison of transcript profiles in late blight-challenged Solanum cajamarquense and B3C1potato clones. Molecular Plant Pathology, 2010, 11(4): 513-530.
[28] BRADSHAW J E, BRYAN G J, RAMSAY G. Genetic resources (including wild and cultivated Solanum species) and progress in their utilisation in potato breeding. Potato Research, 2006, 49(1): 49-65.
[29] NARANCIO R, ZORRILLA P, ROBELLO C, GONZALEZ M, VILARÓ F, PRITSCH C, RIZZA M D. Insights on gene expression response of a characterized resistant genotype of Solanum commersonii Dun. against Ralstonia solanacearum. European Journal of Plant Pathology, 2013, 136(4): 823-835.
[30] JO K R, ARENS M, KIM T Y, JONGSMA M A, VISSER R G F, JACOBSEN E, VOSSEN J H. Mapping of the S. demissum late blight resistance gene R8 to a new locus on chromosome IX. Theoretical and Applied Genetics, 2011, 123(8): 1331-1340.
[31] VILLAMON F G, SPOONER D M, ORRILLO M, MIHOVILOVICH E, PÉREZ W, BONIERBALE W. Late blight resistance linkages in a novel cross of the wild potato species Solanum paucissectum (series Piurana). Theoretical and Applied Genetics, 2005, 111(6): 1201-1214.
[32] VAN DER VOSSEN E, SIKKEMA A, HEKKERT B, GROS J, STEVENS P, MUSKENS M, WOUTERS D, PEREIRA A, STIEKEMA W, ALLEFS S. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. The Plant Journal, 2003, 36: 867-882.
[33] NAESS K S, BRADEEN M J, WIELGUS M S, HABERLACH T G, MCGRATH M J, HELGESON J P. Resistance to late blight in Solanum bulbocastanum is mapped to chromosome 8. Theoretical and Applied Genetics, 2000, 101(5): 697-704.
[34] LAFERRIERE T L, HELGESON P J, ALLEN C. Fertile Solanum tuberosum+S. commersonii somatic hybrids as sources of resistance to bacterial wilt caused by Ralstonia solanacearum. Theoretical and Applied Genetics, 1999, 98(8): 1272-1278.
[35] CARDI T, D'AMBROSIO E, CONSOLI D, PUITE K J, RAMULU K S. Production of somatic hybrids between frost-tolerant Solanum commersonii and S. tuberosum: characterization of hybrid plants. Theoretical and Applied Genetics, 1993, 87(1): 193-200.
[36] ESTRADA N. Utilization of Solanum brevidens to Transfer PLRV Resistance into the Cultivated Potato, Solanum tuberosum. London: Royal Botanical Gardens, 1991.
[37] THIEME R, RAKOSY-TICAN E, GAVRILENKO T, ANTONOVA O, SCHUBERT J, NACHTIGALL M, HEIMBACH U, THIEME T. Novel somatic hybrids (Solanum tuberosum L. + Solanum tarnii) and their fertile BC1 progenies express extreme resistance to potato virus Y and late blight. Theoretical and Applied Genetics, 2008, 116(5): 691-700.
[38] CAMADRO E L, CARPUTO D, PELOQUIN S J. Substitutes for genome differentiation in tuber-bearing Solanum: Interspecific pollenpistil incompatibility, nuclear-cytoplasmic male sterility, and endosperm. Theoretical and Applied Genetics, 2004, 109(7): 1369-1376.
[39] DIONNE L A. Studies on the use of Solanum acaule as a bridge between Solanum tuberosum and species in the series Bulbocastana,Cardiophylla and Pinnatisecta. Euphytica, 1963, 12(3): 263-269.
[40] HERMSEN J G T. Crossability, fertility and cytogenetic studies in Solanum acaule × Solanum bulbocastanum. Euphytica, 1966, 15(2): 149-155.
[41] SINGSIT C, HANNEMAN R E. Rescuing abortive inter-EBN potato hybrids through double pollination and embryo culture. Plant Cell Reports, 1991, 9(9): 475-478.
[42] CHEN Q, LYNCH D, PLATT H W, LI H Y, SHI Y, LI H J, BEASLEY J, RAKOSY-TICAN L, THEME R. Interspecific crossability and cytogenetic analysis of sexual progenies of Mexican wild diploid 1EBN species Solanum pinnatisectum and S. cardiophyllum. American Journal of Potato Research, 2004, 81(2): 159-169.
[43] WATANABE K N, ORRILLO M, VEGA S, VALKONEN J P T, PEHU E, HURTADO A, TANKSLEY S D. Overcoming crossing barriers between nontuber-bearing and tuber-bearing Solanum species: Towards potato germplasm enhancement with a broad spectrum of solanaceous genetic resources. Genome, 1995, 38: 27-35.
[44] PAZ M M, VEILLEUX R E. Influence of culture medium and in vitro conditions on shoot regeneration in Solanum phureja monoploids and fertility of regenerated doubled monoploids. Plant Breeding, 1999, 118(1): 53-57.
[45] VAN OS H, ANDRZEJEWSKI S, BAKKER E, BARRENA I, BRYAN G J. Construction of a 10,000-marker ultradense genetic recombination map of potato: Providing a framework for accelerated gene isolation and a genomewide physical map. Genetics, 2006, 173(2):1075-1087.
[46] POTATO GENOME SEQUENCING CONSORTIUM. Genome sequence and analysis of the tuber crop potato. Nature, 2011, 475(7355): 189-195.
[47] AVERSANO R, CONTALDI F, ERCOLANO M R, GROSSO V, IORIZZO M, TATINO F, XUMERLE L, AVANZATO C, FERRARINI A, DELLEDONNE M, SANSEVERINO W, CIGLIANO R A, CAPELLA-GUTIERREZ G T, FRUSCIANTE L, BRADEEN J M, CARPUTO D. The Solanum commersonii Genome Sequence provides insights into adaptation to stress conditions and genome evolution of wild potato relatives. The Plant Cell, 2015, 27(4): 954-968.
[48] GEBHARDT C, VALKONEN J P T. Organization of genes controlling disease resistance in the potato genome. Annual Review of Phytopathology, 2001, 39: 79-102.
[49] SLATER A T, COGAN N O I, HAYES B J, SCHULTZ L, DALE M F B, BRYAN G J, FORSTER J W. Improving breeding efficiency in potato using molecular and quantitative genetics. Theoretical and Applied Genetics, 2014, 127(11): 2279-2292.
[50] KEARSEY M J, POONI H S. The Genetical Analysis of Quantitative Traits. Cheltenham: Stanley Thornes Ltd, 1998.
[51] MOOSE S P, MUMM R H. Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiology, 2008, 147(3): 969-977.
[52] VAN ECK H J. Genetics of Morphological and Tuber Traits. Amsterdam: Elsevier, 2007.
[53] PRASHAR A, HORNYIK C, YOUNG V, MCLEAN K, SHARMA S K, DALE M F B, BRYAN G J. Construction of a dense SNP map of a highly heterozygous diploid potato population and QTL analysis of tuber shape and eye depth. Theoretical and Applied Genetics, 2014, 127(10): 2159-2171.
[54] LI X Q, DE JONG H, DE JONG D M, DE JONG W S. Inheritance and genetic mapping of tuber eye depth in cultivated diploid potatoes. Theoretical and Applied Genetics, 2005, 110(6): 1068-1073.
[55] DE JONG H. Inheritance of russeting in cultivated diploid potatoes. Potato Research, 1981, 24(3): 309-313.
[56] KLOOSTERMAN B, ABELENDA J A, GOMEZ M M, OORTWIJN M, DE BOER J M, KOWITWANICH K, HORVATH B M, VAN ECK H J, SMACZNIAK C, PRAT S, VISSER R G, BACHEM C W. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature, 2013, 495(7440): 246-250.
[57] CELIS-GAMBOA C, STRUIK P C, JACOBSEN E, VISSER R G F. Temporal dynamics of tuber formation and related processes in a crossing population of potato (Solanum tuberosum). Annals of Applied Biology, 2003, 143(2): 175-186.
[58] BERG J H, EWING E E, PLAISTED R L, MCMURRY S, BONIERBALE M W. QTL analysis of potato tuber dormancy. Theoretical and Applied Genetics, 1996, 93(3): 317-324.
[59] LEVY D, VEILLEUX R E. Adaptation of potato to high temperatures and salinity-a review. American Journal of Potato Research, 2007, 84(6): 487-506.
[60] ORTIZ R, HUAMAN Z. Inheritance of Morphological and Tuber Characteristics. Wallingford, UK: CAB International, 1994.
[61] ANITHAKUMARI A M, NATARAJA K N, VISSER R G F, LINDEN C G. Genetic dissection of drought tolerance and recovery potential by quantitative trait locus mapping of a diploid potato population. Molecular Breeding, 2012, 30(3): 1413-1429.
[62] GANGADHAR B H, YU J W, SAJEESH K, PARK S W. A systematic exploration of high-temperature stress-responsive genes in potato using large-scale yeast functional screening. Molecular Genetics and Genomics, 2013, 289(2): 185-201.
[63] ZHU X, RICHAEL C, CHAMBERLAIN P, BUSSE J S. BUSSAN AJ, JIANG J, BETHKE P C. Vacuolar invertase gene silencing in potato (Solanum tuberosum L.) improves processing quality by decreasing the frequency of sugar-end defects. PLoS ONE, 2014, 9(4): e93381.
[64] Marczewski W, Hennig J, Gebhardt C. The potato virus S resistance gene Ns maps to potato chromosome VIII. Theoretical and Applied Genetics, 2002, 105(4): 564-567.
[65] RITTER E, DEBENER T, BARONE A, SALAMINI F, GEBHARDT C. RFLP mapping on potato chromosomes of two genes controlling extreme resistance to potato virus X (PVX). Molecular Genetics and Genomics, 1991, 227(1): 81-85.
[66] HÄMÄLÄINEN H J, WATANABE N K, VALKONEN T J P, ARIHARA A, PLAISTED L R, PEHU E, MILLER L, SLACK A S. Mapping and marker-assisted selection for a gene for extreme resistance to potato virus Y. Theoretical and Applied Genetics, 1997, 94(2): 192-197.
[67] MARCZEWSKI W, FLIS B, SYLLER J, SCHÄFER-PREGL R, GEBHARDT C. A major quantitative trait locus for resistance to Potato leafroll virus is located in a resistance hotspot on potato chromosome XI and is tightly linked to N-gene-like markers. Molecular Plant-Microbe Interactions, 2001, 14(12): 1420-1425.
[68] SIMKO I, JANSKY S, STEPHENSON S, SPOONER D M. Genetics of Resistance to Pests and Diseases. Amsterdam: Elsevier, 2007.
[69] BROWN C R, YANG C P, MOJTAHEDI H, SANTO G S, MASUELLI R. RFLP analysis of resistance to Columbia root-knot nematode derived from Solanum bulbocastanum in a BC2population. Theoretical and Applied Genetics, 1996, 92(5): 572-576.
[70] PHILLIPS M S. Inheritance of Resistance to Nematodes. Wallingford: CAB International, 1994.
[71] RODEWALD J, TROGNITZ B. Solanum resistance genes against Phytophthora infestans and their corresponding avirulence genes. Molecular Plant Pathology, 2013, 14(7): 740-757.
[72] SANTA CRUZ J H, HAYNES K G, CHRIST B J. Effects of one cycle of recurrent selection for early blight resistance in a diploid hybrid Solanum phureja-S. stenotomum population. American Journal of Potato Research, 2009, 86(6): 490-498.
[73] BURKHART C R, CHRIST B J, HAYNES K G. Non-additive genetic variance governs resistance to fusarium dry rot in a diploid hybrid potato population. American Journal of Potato Research, 2007, 84(3): 199-204.
[74] DEES M W, LYSØE E, ALSHEIKH M, DAVIK J, BRURBERG M B. Resistance to Streptomyces turgidiscabies in potato involves an early and sustained transcriptional reprogramming at initial stages of tuber formation. Molecular Plant Pathology, 2015, 17(5): 703-713.
[75] PAGET M F, ALSPACH P A, GENET R A, APIOLAZA L A. Genetic variance models for the evaluation of resistance to powdery scab (Spongospora subterranea f. sp. subterranea) from long-term potato breeding trials. Euphytica, 2014, 197(3): 369-385.
[76] WASTIE R L. Inheritance of Fungal Diseases of Tubers. Wallingford: CAB International, 1994.
[77] VLEESHOUWERS V G, RAFFAELE S, VOSSEN J H, CHAMPOURET N, OLIVA R, SEGRETIN M E, RIETMAN H, CANO L M, LOKOSSOU A, KESSEL G, PEL M A, KAMOUN S. Understanding and exploiting late blight resistance in the age of effectors. Annual Review of Phytopathology, 2011, 49: 507-531.
[78] Black W, Mastenbroek C, Mills W R, Peterson L C. A proposal for an international nomenclature of races of Phytophthora infestans and of genes controlling immunity in Solanum demissum derivatives. Euphytica, 1953, 2(3):173-179.
[79] VAN DER LEE T, TESTA A, VAN'T KLOOSTER J, VAN DEN BERG-VELTHUIS G, GOVERS F. Chromosomal deletion in isolates of Phytophthora infestans correlates with virulence on R3, R10, and R11 potato lines. Molecular Plant-Microbe Interactions, 2001, 14(12): 1444-1452.
[80] PARK T H, VLEESHOUWERS V G A A, JACOBSEN E, VAN DER VOSSEN E, VISSER R G F. Molecular breeding for resistance to Phytophthora infestans (Mont.) de Bary in potato (Solanum tuberosum L.): A perspective of cisgenesis. Plant Breeding, 2009, 128(2): 109-117.
[81] VAN DER VOSSEN E A, GROS J, SIKKEMA A, MUSKENS M, WOUTERS D, WOLTERS P, PEREIRA A, ALLEFS S. The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. The Plant Journal, 2005, 44(2): 208-222.
[82] PARK T H, GROS J, SIKKEMA A, VLEESHOUWERS V G, MUSKENS M, ALLEFS S, JACOBSEN E, VISSER R G, VAN DER VOSSEN E A. The late blight resistance locus Rpi-bib3 from Solanum bulbocastanum belongs to a major late blight R gene cluster on chromosome 4 of potato. Molecular Plant-Microbe Interactions, 2005, 18(7): 722-729.
[83] SONG J, BRADEEN J M, NAESS S K, RAASCH J A, WIELGUS S M, HABERLACH G T, LIU J, KUANG H, AUSTIN-PHILLIPS S, BUELL C R, HELGESON J P, JIANG J. Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(16): 9128-9133.
[84] DANAN S, VEYRIERAS J B, LEFEBVRE V. Construction of apotato consensus map and QTL meta-analysis offer new insights into the genetic architecture of late blight resistance and plant maturity traits. BMC Plant Biology, 2011, 11: 16.
[85] GEBHARDT C. Bridging the gap between genome analysis and precision breeding in potato. Trends in Genetics, 2013, 29(4): 248-256.
[86] CHAMPOURET N. Functional genomics of Phytophthora infestans effectors and Solanum resistance genes[D]. Wageningen: Wageningen University, 2010.
[87] ZHANG K, XU J, DUAN S G, PANG W F, BIAN C S, LIU J, JIN L. NBS profiling identifies potential novel locus from Solanum demissum that confers broad-spectrum resistance to Phytophthora infestans. Journal of Integrative Agriculture, 2014, 13(8): 1662-1671. [88] BALLVORA A, ERCOLANO M R, WEISS J, MEKSEM K, BORMANN C A, OBERHAGEMANN P, SALAMINI F, GEBHARDT C. The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. The Plant Journal, 2002, 30(3): 361-371.
[89] LOKOSSOU A A, PARK T H, VAN ARKEL G, ARENS M, RUYTER-SPIRA C, MORALES J, WHISSON S C, BIRCH P R, VISSER R G, JACOBSEN E, VAN DER VOSSEN E A. Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Molecular Plant-Microbe Interactions, 2009, 22(6): 630-641.
[90] HUANG S, VAN DER VOSSEN E A, KUANG H, VLEESHOUWERS V G, ZHANG N, BORM T J, VAN ECK H J, BAKER B, JACOBSEN E, VISSER R G. Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. The Plant Journal, 2005, 42(2): 251-261.
[91] LI G, HUANG S, GUO X, LI Y, YANG Y, GUO Z, KUANG H, RIETMAN H, BERGERVOET M, VLEESHOUWERS V G, VAN DER VOSSEN E A, QU D, VISSER R G, JACOBSEN E, VOSSEN J H. Cloning and characterization of R3b: Members of the R3 superfamily of late blight resistance genes show sequence and functional divergence. Molecular Plant-Microbe Interactions, 2011, 24(10): 1132-1142.
[92] OOSUMI T, ROCKHOLD D R, MACCREE M M, DEAHL K L, MCCUE K F, BELKNAP W R. Gene Rpi-bt1 from Solanum bulbocastanum confers resistance to late blight in transgenic potatoes. American Journal of Potato Research, 2009, 86(6): 456-465.
[93] LOKOSSOU A A, RIETMAN H, WANG M, KRENEK P, VAN DER SCHOOT H, HENKEN B, HOEKSTRA R, VLEESHOUWERS V G, VAN DER VOSSEN E A, VISSER R G, JACOBSEN E, VOSMAN B. Diversity, distribution, and evolution of Solanum bulbocastanum late blight resistance genes. Molecular Plant-Microbe Interactions, 2010, 23(9): 1206-1216.
[94] VLEESHOUWERS V G, RIETMAN H, KRENEK P, CHAMPOURET N, YOUNG C, OH S K, WANG M, BOUWMEESTER K, VOSMAN B, VISSER R G, JACOBSEN E, GOVERS F, KAMOUN S, VAN DER VOSSEN E A. Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE, 2008, 3(8): e2875.
[95] WANG M, ALLEFS S, BERG R G, VLEESHOUWERS V G A A, VOSSEN E A G, VOSMAN B. Allele mining in Solanum: Conserved homologues of Rpi-blb1 are identified in Solanum stoloniferum. Theoretical and Applied Genetics, 2008, 116(7): 933-943.
[96] FOSTER S J, PARK T H, PEL M, BRIGNETI G, SLIWKA J, JAGGER L, VAN DER VOSSEN E, JONES J D. Rpi-vnt1.1, a Tm-2(2) homolog from Solanum venturii, confers resistance to potato late blight. Molecular Plant-Microbe Interactions, 2009, 22(5): 589-600.
[97] ŚLIWKA J, ŚWIĄTEK M, TOMCZYŃSKA I, STEFAŃCZYK E, CHMIELARZ M, ZIMNOCH-GUZOWSKA E. Influence of genetic background and plant age on expression of the potato late blight resistance gene Rpi-phu1 during incompatible interactions with Phytophthora infestans. Plant Pathology, 2013, 62(5): 1072-1080.
[98] BARKER H. Extreme resistance to potato virus V in clones of Solanum tuberosum that are also resistant to potato viruses Y and A: Evidence for a locus conferring broad-spectrum potyvirus resistance. Theoretical and Applied Genetics, 1997, 95(8): 1258-1262.
[99] JONES R A C. Strain group specific and virus specific hypersensitive reactions to infection with potyviruses in potato cultivars. Annals of Applied Biology, 1990, 117(1): 93-105.
[100] HAMALAINEN J H, KEKARAINEN T, GEBHARDT C, WATANABE K N, VALKONEN J P. Recessive and dominant genes interfere with the vascular transport of potato virus A in diploid potatoes. Molecular Plant-Microbe Interactions, 2000, 13(4): 402-412.
[101] DE JONG W, FORSYTH A, LEISTER D, GEBHARDT C, BAULCOMBE D C. A potato hypersensitive resistance gene against potato virus X maps to a resistance gene cluster on chromosome 5. Theoretical and Applied Genetics, 1997, 95(1): 246-252.
[102] MARCZEWSKI W, HENNIG J, GEBHARDT C. The potato virus S resistance gene Ns maps to potato chromosome VIII. Theoretical and Applied Genetics, 2002, 105(4): 564-567.
[103] TOMMISKA J T, HÄMÄLÄINEN H J, WATANABE N K, VALKONEN T J P. Mapping of the gene Nxphuthat controlshypersensitive resistance to potato virus X in Solanum phureja IvP35. Theoretical and Applied Genetics, 1998, 96(6): 840-843.
[104] MARCZEWSKI W, FLIS B, SYLLER J, STRZELCZYK-ŻYTA D, HENNIG J, GEBHARDT C. Two allelic or tightly linked genetic factors at the PLRV.4 locus on potato chromosome XI control resistance to potato leafroll virus accumulation. Theoretical and Applied Genetics, 2004, 109(8): 1604-1609.
[105] VELÁSQUEZ A C, MIHOVILOVICH E, BONIERBALE M. Genetic characterization and mapping of major gene resistance to potato leafroll virus in Solanum tuberosum ssp. andigena. Theoretical and Applied Genetics, 2007, 114(6): 1051-1058.
[106] HÄMÄLÄINEN H J, WATANABE N K, VALKONEN T J P, ARIHARA A, PLAISTED L R, PEHU E, MILLER L, SLACK A S. Mapping and marker-assisted selection for a gene for extreme resistance to potato virus Y. Theoretical and Applied Genetics, 1997, 94(2): 192-197.
[107] BRIGNETI G, GARCIA-MAS J, BAULCOMBE C D. Molecular mapping of the potato virus Y resistance gene Rystoin potato. Theoretical and Applied Genetics, 1997, 94(2): 198-203.
[108] FINKERS-TOMCZAK A, BAKKER E, DE BOER J, VAN DER VOSSEN E, ACHENBACH U, GOLAS T, SURYANINGRAT S, SMANT G, BAKKER J, GOVERSE A. Comparative sequence analysis of the potato cyst nematode resistance locus H1 reveals a major lack of co-linearity between three haplotypes in potato (Solanum tuberosum ssp.). Theoretical and Applied Genetics, 2011, 122(3): 595-608.
[109] GEBHARDT C, MUGNIERY D, RITTER E, SALAMINI F, BONNEL E. Identification of RFLP markers closely linked to the H1 gene conferring resistance to Globodera rostochiensis in potato. Theoretical and Applied Genetics, 1993, 85(5): 541-544.
[110] KREIKE C M, KONING J R A, VINKE J H, OOIJEN J W, STIEKEMA W J. Quantitatively-inherited resistance to Globodera pallida is dominated by one major locus in Solanum spegazzinii. Theoretical and Applied Genetics, 1994, 88(6): 764-769.
[111] VAN DER VOORT R J, WOLTERS P, FOLKERTSMA R, HUTTEN R, VAN ZANDVOORT P, VINKE H, KANYUKA K, BENDAHMANE A, JACOBSEN E, JANSSEN R, BAKKER J. Mapping of the cyst nematode resistance locus Gpa2 in potato using a strategy based on comigrating AFLP markers. Theoretical and Applied Genetics, 1997, 95(5): 874-880.
[112] BRADSHAW E J, HACKETT A C, MEYER C R, MILBOURNE D, MCNICOL W J, PHILLIPS S M, WAUGH R. Identification of AFLP and SSR markers associated with quantitative resistance to Globodera pallida (Stone) in tetraploid potato (Solanum tuberosum subsp. tuberosum) with a view to marker-assisted selection. Theoretical and Applied Genetics, 1998, 97(1): 202-210.
[113] ROUPPE VAN DER VOORT J, VAN DER VOSSEN E, BAKKER E, OVERMARS H, VAN ZANDVOORT P, HUTTEN R, KLEIN LANKHORST R, BAKKER J. Two additive QTLs conferring broad-spectrum resistance in potato to Globodera pallida are localized on resistance gene clusters. Theoretical and Applied Genetics, 2000, 101(7): 1122-1130.
[114] JACOBS J M E, ECK H J, HORSMAN K, ARENS P F P, VERKERK-BAKKER B, JACOBSEN E, PEREIRA A, STIEKEMA W J. Mapping of resistance to the potato cyst nematode Globodera rostochiensis from the wild potato species Solanum vernei. Molecular Breeding, 1996, 2(1): 51-60.
[115] LEISTER D, BALLVORA A, SALAMINI F, GEBHARDT C. A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nature Genetics, 1996, 14(4): 421-429.
[116] KREIKE C M, KONING J R A, VINKE J H, OOIJEN J W, GEBHARDT C, STIEKEMA W J. Mapping of loci involved in quantitatively inherited resistance to the potato cyst-nematode Globodera rostochiensis pathotype Ro1. Theoretical and Applied Genetics, 1993, 87(4): 464-470.
[117] KREIKE C M, KOK-WESTENENG A A, VINKE J H, STIEKEMA W J. Mapping of QTLs involved in nematode resistance, tuber yield and root development in Solanum sp. Theoretical and Applied Genetics, 1996, 92(3): 463-470.
[118] VAN DER VOORT R J, LINDEMAN W, FOLKERTSMA R, HUTTEN R, OVERMARS H, VAN DER VOSSEN E, JACOBSEN E, BAKKER J. A QTL for broad-spectrum resistance to cyst nematode species (Globodera spp.) maps to a resistance gene cluster in potato. Theoretical and Applied Genetics, 1998, 96(5): 654-661.
[119] TUNG P X. Genetic variation for bacterial wilt resistance in a population of tetraploid potato. Euphytica, 1992, 61(1): 73-80.
[120] YU Y, YE W, HE L, CAI X, LIU T, LIU J. Introgression of bacterial wilt resistance from eggplant to potato via protoplast fusion and genome components of the hybrids. Plant Cell Reports, 2013, 32(11): 1687-1701.
[121] KIM-LEE H, MOON J S, HONG Y J, KIM M S, CHO H M. Bacterial wilt resistance in the progenies of the fusion hybrids between haploid of potato and Solanum commersonii. American Journal of Potato Research, 2005, 82(2): 129-137.
[122] CHEN L, GUO X, XIE C, HE L, CAI X, TIAN L, SONG B, LIU J.Nuclear and cytoplasmic genome components of Solanum tuberosum + S. chacoense somatic hybrids and three SSR alleles related to bacterial wilt resistance. Theoretical and Applied Genetics, 2013, 126(7): 1861-1872.
[123] FOCK I, COLLONNIER C, LAVERGNE D, VANIET S, AMBROISE A, LUISETTI J, KODJA H, SIHACHAKR D. Evaluation of somatic hybrids of potato with Solanum stenotomum after a long-term in vitro conservation. Plant Physiology & Biochemistry, 2007, 45(3/4): 209-215.
[124] CHUNG Y S, HOLMQUIST K, SPOONER D M, JANSKY S H. A test of taxonomic and biogeographic predictivity: Resistance to soft rot in wild relatives of cultivated potato. Phytopathology, 2011, 101(2): 205-212.
[125] WANNER L A, KIRK W W. Streptomyces – from basic microbiology to role as a plant pathogen. American Journal of Potato Research, 2015, 92(2): 236-242.
[126] KHATRI B B, TEGG R S, BROWN P H, WILSON C R. Temporal association of potato tuber development with susceptibility to common scab and Streptomyces scabiei-induced responses in the potato periderm. Plant Pathology, 2011, 60(4): 776-786.
[127] WILSON C R, TEGG R S, WILSON A J, LUCKMAN G A, EYLES A, YUAN Z Q, HINGSTON L H, CONNER A J. Stable and extreme resistance to common scab of potato obtained through somatic cell selection. Phytopathology, 2010, 100(5): 460-467.
[128] SALAMAN R N. The inheritance of colour and other characters in the potato. Journal of Genetics, 1910, 1(1): 7-46.
[129] JONG H D. Inheritance of anthocyanin pigmentation in the cultivated potato: A critical review. American Potato Journal, 1991, 68(9): 585-593.
[130] GEBHARDT C, RITTER E, DEBENER T, SCHACHTSCHABEL U, WALKEMEIER B, UHRIG H, SALAMINI F. RFLP analysis and linkage mapping in Solanum tuberosum. Theoretical and Applied Genetics, 1989, 78(1): 65-75.
[131] VAN ECK H J, JACOBS J M E, DIJK J, STIEKEMA W J, JACOBSEN E. Identification and mapping of three flower colour loci of potato (S. tuberosum L.) by RFLP analysis. Theoretical and Applied Genetics, 1993, 86(2): 295-300.
[132] DE JONG W S, EANNETTA N T, DEJONG D M, BODIS M. Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theoretical and Applied Genetics, 2004, 108(3): 423-432.
[133] DE JONG W S, DE JONG D M, DE JONG H, KALAZICH J, BODIS M. An allele of dihydroflavonol 4-reductase associated with the ability to produce red anthocyanin pigments in potato (Solanum tuberosum L.). Theoretical and Applied Genetics, 2003, 107(8): 1375-1383.
[134]JUNG C S, GRIFFITHS H M, DE JONG D M, CHENG S, BODIS M, DE JONG W S. The potato P locus codes for flavonoid 3′,5′-hydroxylase. Theoretical and Applied Genetics, 2004, 110(2): 269-275.
[135] ZHANG Y, CHENG S, JONG D M, GRIFFITHS H, HALITSCHKE R, DE JONG W S. The potato R locus codes for dihydroflavonol 4-reductase. Theoretical and Applied Genetics, 2009, 119(5): 931-937.
[136] JUNG C S, GRIFFITHS H M, JONG D M, CHENG S, BODIS M, KIM T S, DE JONG W S. The potato developer (D) locus encodes an R2R3 MYB transcription factor that regulates expression of multiple anthocyanin structural genes in tuber skin. Theoretical and Applied Genetics, 2009, 120(1): 45-57.
[137] MASSON M F. Mapping, combining abilites, heritabilities and heterosis with 4x × 2x crosses in potato. Madison: University of Wisconsin-Madison, 1985.
[138] DE JONG H, BURNS V J. Inheritance of tuber shape in cultivated diploid potatoes. American Journal of Potato Research, 1993, 70: 267-283.
[139] LI L, PAULO M J, STRAHWALD J, LÜBECK J, HOFFERBERT H R, TACKE E, JUNGHANS H, WUNDER J, DRAFFEHN A, EEUWIJK F, GEBHARDT C. Natural DNA variation at candidate loci is associated with potato chip color, tuber starch content, yield and starch yield. Theoretical and Applied Genetics, 2008, 116(8): 1167-1181.
[140] SCHREIBER L, NADER-NIETO A C, SCHONHALS E M, WALKEMEIER B, GEBHARDT C. SNPs in genes functional in starch-sugar interconversion associate with natural variation of tuber starch and sugar content of potato (Solanum tuberosum L.). Genes Genomes Genetics, 2014, 4(10): 1797-1811.
[141] WIBERLEY-BRADFORD A E, BUSSE J S, JIANG J, BETHKE P C. Sugar metabolism, chip color, invertase activity, and gene expression during long-term cold storage of potato (Solanum tuberosum) tubers from wild-type and vacuolar invertase silencing lines of Katahdin. BMC Research Notes, 2014, 7: 801.
[142] LIN Y, LIU T, LIU J, LIU X, OU Y, ZHANG H, LI M, SONNEWALD U, SONG B, XIE C. Subtle regulation of potato acid invertase activity by a protein complex of invertase, invertase inhibitor, and sucrose nonfermenting1-related protein kinase. Plant Physiology, 2015, 168(4): 1807-1819.
[143] URBANY C, STICH B, SCHMIDT L, SIMON, BERDING H, JUNGHANS H, NIEHOFF K, BRAUN A, TACKE E, HOFFERBERT H, LÜBECK J, STRAHWALD J, GEBHARDT C. Associationgenetics in Solanum tuberosum provides new insights into potato tuber bruising and enzymatic tissue discoloration. BMC Genomics, 2011, 12(1): 1-14.
[144] MONNEVEUX P, RAMÍREZ D A, PINO M. Drought tolerance in potato (S.tuberosum L.): Can we learn from drought tolerance research in cereals? Plant Science, 2013, 205-206: 76-86.
[145] WEISZ R, KAMINSKI J, SMILOWITZ Z. Water deficit effects on potato leaf growth and transpiration: Utilizing fraction extractable soil water for comparison with other crops. American Potato Journal, 1994, 71(12): 829-840.
[146]ANITHAKUMARI A M, DOLSTRA O, VOSMAN B, VISSER R G F, LINDEN C G. In vitro screening and QTL analysis for drought tolerance in diploid potato. Euphytica, 2011, 181(3): 357-369.
[147] KONDRAK M, MARINCS F, ANTAL F, JUHASZ Z, BANFALVI Z. Effects of yeast trehalose-6-phosphate synthase 1 on gene expression and carbohydrate contents of potato leaves under drought stress conditions. BMC Plant Biology, 2012, 12: 74.
[148] ZHANG N, YANG J, WANG Z, WEN Y, WANG J, HE W, LIU B, SI H, WANG D. Identification of novel and conserved microRNAs related to drought stress in potato by deep sequencing. PLoS ONE, 2014, 9(4): e95489.
[149] STONE J M, PALTA J P, BAMBERG J B, WEISS L S, HARBAGE J F. Inheritance of freezing resistance in tuber-bearing Solanum species: Evidence for independent genetic control of nonacclimated freezing tolerance and cold acclimation capacity. Proceedings of the National Academy of Sciences of the United States of America, 1993, 90(16): 7869-7873.
[150] AHN Y, ZIMMERMAN J L. Introduction of the carrot HSP17.7 into potato (Solanum tuberosum L.) enhances cellular membrane stability and tuberization in vitro. Plant, Cell & Environment, 2006, 29(1): 95-104.
[151] SIMKO I, COSTANZO S, HAYNES K G, CHRIST B J, JONES R W. Linkage disequilibrium mapping of a Verticillium dahliae resistance quantitative trait locus in tetraploid potato (Solanum tuberosum) through a candidate gene approach. Theoretical and Applied Genetics, 2004, 108(2): 217-224.
[152] BRADSHAW J E, MACKAY G R. Breeding Strategies for Clonally Propagated Potatoes. Wallingford: Cab International, 1994.
[153] 金黎平, 杨宏福. 马铃薯遗传育种中的染色体倍性操作. 农业生物技术学报, 1996, 1: 70-75.
JIN L P, YANG H F. Chromosomal manipulation in potato genetics and breeding. Journal of Agricultutal Biotechnology, 1996, 1: 70-75. (in Chinese)
[154] HOUGAS R W, PELOQUIN S J. The potential of potato haploids in breeding and genetic research. American Journal of Potato Research, 1958, 35: 701-707.
[155] HOUGAS R W, PELOQUIN S J, GABERT A C. Effect of seed-partent and pollinator on frequency of haploids in Solannum tuberosum. Crop Science, 1964, 4: 593-595.
[156] HERMSEN J G T, VERDENIUS J. Selection from Solanum tuberosum group phureja of genotypes combining high-frequency haploid induction with homozygosity for embryo-spot. Euphytica, 1973, 22(2): 244-259.
[157] HOUGAS R W, PELOQUIN S J. Crossability of Solannum tuberosum haploids with diploid Solanum species. European Potato Journal, 1960, 3: 325-330.
[158] CHASE S C. Analytical breeding of Solanum tuberosum. Canadian Journal of Genetics and Cytology, 1963, 5: 359-363.
[159] YEH B P, PELOQUIN S J, HOUGAS R W. Meiosis in Solanum tuberosum haploids and haploid-haploid F1hybrids. Canadian Journal of Genetics and Cytology, 1964, 6: 393-402.
[160] 屈冬玉, 朱德蔚, 王登社, 高占旺, Ramanna M S, Jacobsen E. 马铃薯2n配子发生的遗传分析. 园艺学报, 1995, 22(1): 61-66. QU D Y, ZHU D W, WANG D S, GAO Z W, RAMANNA M S, JACOBSEN E. Genetic analysis of 2n pollen formation in potato. Acta Horticulturae Sinica, 1995, 22(1): 61-66. (in Chinese)
[161] STELLY D M, PELOQUIN S J, PALMER R G, CRANE C F. Mayer’s hemalum-methy salicylate: A stain-clearing technique for observations within whole ovules. Stain Technology, 1984, 59: 155-161.
[162] ERAZZÚ L E, CAMADRO E L. Direct and indirect detection of 2n eggs in hybrid diploid families derived from haploid tbr × wild species crosses. Euphytica, 2006, 155(1): 57-62.
[163] WEBER B, JANSKY S. Resistance to Alternaria solani in hybrids between a Solanum tuberosum haploid and S. raphanifolium. Phytopathology, 2012, 102: 214-221.
[164] QU D, ZHU D, RAMANNA M S, JACOBSEN E. A comparison of progeny from diallel crosses of diploid potato with regard to the frequencies of 2n-pollen grains. Euphytica, 1995, 92(3): 313-320.
[165] MURPHY A M, JONG H, TAI G C C. Transmission of resistance to common scab from the diploid to the tetraploid level via 4x-2x crosses in potatoes. Euphytica, 1995, 82(3): 227-233.
[166] PARK T H, KIM J B, HUTTEN R C B, VAN ECK H J, JACOBSEN E, VISSER R G F. Genetic positioning of centromeres using half-tetrad analysis in a 4x-2x cross population of potato. Genetics, 2007, 176(1): 85-94.
[167] MENDIBURU A O, PELOQUIN S J. The significance of 2n gametesin potato breeding. Theoretical and Applied Genetics, 1977, 49(2): 53-61.
[168] WEISZ R, KAMINSKI J, SMILOWITZ Z. Interspecific somatic hybrids Solanum villosum (+) S. tuberosum, resistant to Phytophthora infestans. Journal of Plant Physiology, 2013, 170(17): 1541-1548.
[169] THIEME R, RAKOSY-TICAN E, NACHTIGALL M, SCHUBERT J, HAMMANN T, ANTONOVA O, GAVRILENKO T, HEIMBACH U, THIEME T. Characterization of the multiple resistance traits of somatic hybrids between Solanum cardiophyllum Lindl. and two commercial potato cultivars. Plant Cell Reports, 2010, 29(10): 1187-1201.
[170] LUO Z W, HACKETT C A, BRADSHAW J E, MCNICOL J W, MILBOURNE D. Construction of a genetic linkage map in tetraploid species using molecular markers. Genetics, 2001, 157(3): 1369-1385.
[171] ZHANG L H, MOJTAHEDI H, KUANG H, BAKER B, BROWN C R. Marker-assisted selection of columbia root-knot nematode resistance introgressed from Solanum bulbocastanum. Crop Science, 2007, 47(5): 2021-2026.
[172] KASAI K, MORIKAWA Y, SORRI V A, VALKONEN J P, GEBHARDT C, WATANABE K N. Development of SCAR markers to the PVY resistance gene Ryadgbased on a common feature of plant disease resistance genes. Genome, 2000, 43(1): 1-8.
[173] SORRI A V, WATANABE N K, VALKONEN T J P. Predicted kinase-3a motif of a resistance gene analogue as a unique marker for virus resistance. Theoretical and Applied Genetics, 1999, 99(1): 164-170.
[174] FULLADOLSA A C, NAVARRO F M, KOTA R, SEVERSON K, PALTA J P, CHARKOWSKI A O. Application of marker assisted selection for potato virus Y resistance in the university of wisconsin potato breeding program. American Journal of Potato Research, 2015, 92(3): 444-450.
[175] MORI K, SAKAMOTO Y, MUKOJIMA N, TAMIYA S, NAKAO T, ISHII T, HOSAKA K. Development of a multiplex PCR method for simultaneous detection of diagnostic DNA markers of five disease and pest resistance genes in potato. Euphytica, 2011, 180(3): 347-355.
[176] SZAJKO K, STRZELCZYK-ŻYTA D, MARCZEWSKI W. Ny-1 and Ny-2 genes conferring hypersensitive response to potato virus Y (PVY) in cultivated potatoes: Mapping and marker-assisted selection validation for PVY resistance in potato breeding. Molecular Breeding, 2014, 34(1): 267-271.
[177]WITEK K, STRZELCZYK-ŻYTA D, HENNIG J, MARCZEWSKI W. A multiplex PCR approach to simultaneously genotype potato towards the resistance alleles Ry-f sto and Ns. Molecular Breeding, 2006, 18(3): 273-275.
[178] MARCZEWSKI W, STRZELCZYK-ŻYTA D, HENNIG J, WITEK K, GEBHARDT C. Potato chromosomes IX and XI carry genes for resistance to potato virus M. Theoretical and Applied Genetics, 2006, 112(7): 1232-1238.
[179] KIM H, LEE H, JO K, MORTAZAVIAN S M M, HUIGEN D J, EVENHUIS B, KESSEL G, VISSER R G F, JACOBSEN E, VOSSEN J H. Broad spectrum late blight resistance in potato differential set plants MaR8 and MaR9 is conferred by multiple stacked R genes. Theoretical and Applied Genetics, 2012, 124(5): 923-935.
[180] XU J, WANG J, PANG W F, BIAN C S, DUAN S G, LIU J, HUANG S, JIN L, QU D. The potato R10 resistance specificity to late blight is conferred by both a single dominant R gene and quantitative trait loci. Plant Breeding, 2013, 132(4): 407-412.
[181] COLTON L M, GROZA H I, WIELGUS S M, JIANG J. Marker-assisted selection for the broad-spectrum potato late blight resistance conferred by gene RB derived from a wild potato species. Crop Science, 2006, 46(2): 589-594.
[182] WANG M, ALLEFS S, BERG R G, VLEESHOUWERS V G A A, VOSSEN E A G, VOSMAN B. Allele mining in Solanum: Conserved homologues of Rpi-blb1 are identified in Solanum stoloniferum. Theoretical and Applied Genetics, 2008, 116(7): 933-943.
[183] ZHU S, LI Y, VOSSEN J H, VISSER R G F, JACOBSEN E. Functional stacking of three resistance genes against Phytophthora infestans in potato. Transgenic Research, 2012, 21(1): 89-99.
[184] SANETOMO R, HOSAKA K. A maternally inherited DNA marker, descended from Solanum demissum (2n = 6x = 72) to S. tuberosum (2n = 4x = 48). Breeding Science, 2011, 61(4): 426-434.
[185] 朱文文, 徐建飞, 李广存, 段绍光, 刘杰, 卞春松, 庞万福, De Jong W, 金黎平. 马铃薯块茎形状基因CAPS标记的开发与验证. 作物学报, 2015, 41(10): 1529-1536.
ZHU W W, XU J F, LI G C, DUAN S G, LIU J, BIAN C S, PANG W F, DE JONG W, JIN L P. Development and verification of a CAPS marker linked to tuber shape gene in potato. Acta Agronomica Sinica, 2015, 41(10): 1529-1536. (in Chinese)
[186] MILCZAREK D, FLIS B, PRZETAKIEWICZ A. Suitability of molecular markers for selection of potatoes resistant to Globodera spp. American Journal of Potato Research, 2011, 88(3): 245-255.
[187] WHITWORTH J L, NOVY R G, HALL D G, CROSSLIN J M, BROWN C R. Characterization of broad spectrum potato virus Y resistance in a Solanum tuberosum ssp. andigena-derived population and select breeding clones using molecular markers, grafting, and field inoculations. American Journal of Potato Research, 2009, 86(4):286-296.
[188] BERNARDO R. Molecular markers and selection for complex traits in plants: Learning from the last 20 years. Crop Science, 2008, 48(5): 1649-1664.
[189] HEFFNER E L, LORENZ A J, JANNINK J L, SORRELLS M E. Plant breeding with genomic selection: Gain per unit time and cost. Crop Science, 2010, 50(5): 1681-1690
[190] MEUWISSEN T H, HAYES B J, GODDARD M E. Prediction of total genetic value using genome-wide dense marker maps. Genetics, 2001, 157(4): 1819-1829.
[191] WONG C K, BERNARDO R. Genomewide selection in oil palm: Increasing selection gain per unit time and cost with small populations. Theoretical and Applied Genetics, 2008, 116(6): 815-824.
[192] ZHONG S, DEKKERS J C, FERNANDO R L, JANNINK J L. Factors affecting accuracy from genomic selection in populations derived from multiple inbred lines: A Barley case study. Genetics, 2009, 182(1): 355-364.
[193] VOS P, UITDEWILLIGEN J, VOORRIPS R, VISSER, R, VAN ECK H. Development and analysis of a 20K SNP array for potato (Solanum tuberosum ): An insight into the breeding history. Theoretical and Applied Genetics, 2015, 128(12): 2387-2401.
[194] WALTZ E. USDA approves next-generation GM potato. Nature Biotechnology, 2015, 33(1): 12-13.
[195] PERLAK F J, STONE T B, MUSKOPF Y M, PETERSEN L J, PARKER G B, MCPHERSON S A, WYMAN J, LOVE S, REED G, BIEVER D, FISCHHOFF D A. Genetically improved potatoes: Protection from damage by Colorado potato beetles. Plant Molecular Biology, 1993, 22(2): 313-321.
[196] REED G L, JENSEN A S, RIEBE J, HEAD G, DUAN J J. Transgenic Bt potato and conventional insecticides for Colorado potato beetle management: Comparative efficacy and non-target impacts. Entomologia Experimentalis et Applicata, 2001, 100(1): 89-100.
[197] LAWSON E C, WEISS J D, THOMAS P E, KANIEWSKI W K. NewLeaf Plus® Russet Burbank potatoes: replicase-mediated resistance to potato leafroll virus. Molecular Breeding, 2001, 7(1): 1-12.
[198] SOUBRIER F, CAMERON B, MANSE B, SOMARRIBA S, DUBERTRET C, JASLIN G, JUNG G, CAER C L, DANG D, MOUVAULT J M, SCHERMAN D, MAYAUX J F, CROUZET J. pCOR: A new design of plasmid vectors for nonviral gene therapy. Gene Therapy, 1999, 6(8): 1482-1488.
[199] SCHOUTEN H J, KRENS F A, JACOBSEN E. Cisgenic plants are similar to traditionally bred plants: international regulations for genetically modified organisms should be altered to exempt cisgenesis. EMBO Reports, 2006, 7(8): 750-753.
[200] OLIVEIRA P H, MAIRHOFER J. Marker-free plasmids for biotechnological applications - implications and perspectives. Trends Biotechnology, 2013, 31(9): 539-547.
[201] JACOBSEN E, SCHOUTEN H J. Cisgenesis, a new tool for traditional plant breeding, should be exempted from the regulation on genetically modified organisms in a step by step approach. Potato Research, 2008, 51(1): 75-88.
[202] JANSEN R, EMBDEN J D A V, GAASTRA W, SCHOULS L M. Identification of genes that are associated with DNA repeats in prokaryotes. Molecular Microbiology, 2002, 43(6): 1565-1575.
[203] HUANG S, WEIGEL D, BEACHY R N, LI J. A proposed regulatory framework for genome-edited crops. Nature Genetics, 2016, 48(2): 109-111.
[204] LEDFORD H. CRISPR, the disruptor. Nature, 2015, 522(7554): 20-24.
[205] BUTLER N M, ATKINS P A, VOYTAS D F, DOUCHES D S. Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS ONE, 2015, 10(12): e0144591.
[206] WANG S, ZHANG S, WANG W, XIONG X, MENG F, CUI X. Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Reports, 2015, 34(9): 1473-1476.
[207] GAO F, SHEN X Z, JIANG F, WU Y, HAN C. DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nature Biotechnology, 2016, 34(7): 768-773.
[208] ALMEKINDERS C J M, MERTENS L, LOON J P, LAMMERTS VAN BUEREN E T. Potato breeding in the Netherlands: A successful participatory model with collaboration between farmers and commercial breeders. Food Security, 2014, 6(4): 515-524.
[209] VAN BUEREN E T L. A collaborative breeding strategy for organic potatoes in the Netherlands. Ecology and Farming: International IFOAM-Magazine, 2010, 2: 50-53.
[210] VAN BUEREN L E T, ENGELEN C, HUTTEN R. Participatory potato breeding model involving organic farmers and commercial breeding companies in the Netherlands. Corvallis: 7 t h Organic Seed Growers Conference, 2014.
[211] TIEMENS-HULSCHER M, DELLEMAN J, EISING J, VAN BUEREN E T L. Potato Breeding: A practical Manual for the Potato Chain. Den Haag: Ardappelwereld BV, 2013.
(责任编辑 李莉)
Advances and Perspectives in Research of Potato Genetics and Breeding
XU JianFei, JIN LiPing
(Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences/Key Laboratory of Biology and Genetic Improvement of Tuber and Root Crops, Ministry of Agriculture, Beijing 100081)
Potato, the third most important food crop, plays a key role in global and China’s food security. Improvement of varieties is a base for sustainable development of potato industry. Potatoes frequently suffer from diverse biotic and abiotic stress, so it is urgent to breed new varieties with better disease resistance, stress tolerance, tuber yield and quality as well as specific usage to meet the needs of potato processing and people nutrition. Potato breeding is a system combining germplasm evaluation and utilization, major traits genetics analysis, breeding technology application and variety extension and crop management together. Within a global conservation strategy there are about 65,000 accessions. Using a homozygous doubled-monoploid potato clone, 86% of the 844-megabase genome sequence are revealed and assembled, and 39,031 protein-coding genes are predicted. At present, re-sequencing of potato accessions is in process. Common cultivated potato is an asexual propagation tetraploid with tetrasomic inheritance and high heterozygosity. Nevertheless, inheritance of many major traits involving plant development and morphology, tuber quality, disease resistance and stress tolerance are revealed. A lot of genes determining potato major traits aremapped and cloned. Potato breeding technology involves conventional breeding, ploidy manipulation, marker-assisted selection, genetic engineering and promising genomic selection for complex traits. Since 1949, China potato breeding has achieved great progress that is reflected on growth of number of registered varieties. Dutch potato breeding ranks among the best in world and participatory potato breeding model is a successful practice for commercial breeding. In the future, it is a trend to breed superior and specific purpose varieties based on improvement of integrated breeding technology, innovation of breeding model and germplasm utilization.
potato; breeding; genetics; perspectives; advances
2016-08-10;接受日期:2017-01-17
国家现代农业产业技术体系建设专项(CARS-10)
联系方式:徐建飞,E-mail:xujianfei@caas.cn。通信作者金黎平,E-mail:jinliping@caas.cn

