Biofilm formation in clinical isolates of nosocomial Acinetobacter baumannii and its relationship with multidrug resistance
Ebrahim Babapour,Azam Haddadi,Reza Mirnejad,Seyed-Abdolhamid Angaji,Nour Amirmozafari*Department of Microbiology,Karaj Branch,Islamic Azad University,Karaj,IranMedical Bacteriology,Molecular Biology Research Center,Baqiyatallah University of Medical Sciences,Tehran,IranDepartment of Cell and Molecular Biology,Faculty of Biological Sciences,Kharazmi University,Tehran,IranIran University of Medical Sciences,School of Medicine,Microbiology Department,Tehran,Iran
ABSTRACT
Biofilm formation in clinical isolates of nosocomial Acinetobacter baumannii and its relationship with multidrug resistance
Ebrahim Babapour1,Azam Haddadi1,Reza Mirnejad2,Seyed-Abdolhamid Angaji3,Nour Amirmozafari4*1Department of Microbiology,Karaj Branch,Islamic Azad University,Karaj,Iran
2Medical Bacteriology,Molecular Biology Research Center,Baqiyatallah University of Medical Sciences,Tehran,Iran
3Department of Cell and Molecular Biology,Faculty of Biological Sciences,Kharazmi University,Tehran,Iran
4Iran University of Medical Sciences,School of Medicine,Microbiology Department,Tehran,Iran
ABSTRACT
Keywords:
Acinetobacter baumannii Biofilm
Multidrug resistance
Nosocomial infections
ARTICLE INFO
Article history:
Received 28 Dec 2015
Received in revised form 5 Jan,2nd revised form 28 Jan 2016
Accepted 1 Feb 2016
Available online 20 Apr 2016
Original article http://dx.doi.org/10.1016/j.apjtb.2016.04.006
Tel: +98 9123877988
Fax: +98 2188058649
E-mail: amirmozafari@yahoo.com
The study was performed according to the Helsinki Declaration of 1975,as revised in 1983. All patients gave written informed consent before recruitment.
Foundation Project: Supported by an educational grant for doctoral thesis from Islamic Azad University of Karaj(grant number: 11530554922001).
Peer review under responsibility of Hainan Medical University. The journal implements double-blind peer review practiced by specially invited international editorial board members.
1. Introduction
Acinetobacter baumannii(A. baumannii)is responsible for the majority of nosocomial infections. The bacterium shows wide range antibiotics resistance towards cephalosporins,penicillins,carbapenems,fluoroquinolones and aminoglycosides. It is often isolated from respiratory tract through which it infects installed catheters. This will eventually lead to fever,pneumonia,bacteremia,and meningitis[1-3]. Among A. baumannii different virulence factors,the most important one is the ability to produce biofilms and their survival in hospital environment which is related to their high degree of antibiotic resistance[4-7]. Biofilms are complex mixtures of microbes which are predominantly attached to hard surfaces. They are often enclosed by thick polysaccharide layer which makes them resistant to antibiotics and thus very hard to eliminate [8,9]. The ability to produce biofilms causes bacteria to endure relatively hard conditions. Bacteria binding to artificial surfaces in hospitals induce long-term durability and tolerancefor dry environments and use of various metabolic resources. These features make eradication of biofilm-associated bacteria almost impossible from hospital environments[1,7,10,11]. On the other hand,sensitivity to different antibiotics as well as microbial metabolism due to biofilm formation will be reduced. This is attributable to lack of food in the biofilm depth. Slower metabolism and antibiotic resistance lead to bacterial dissemination which can create a quick critical situation[12]. This may increase the incidence of nosocomial infections caused by bacteria,especially in patients in intensive care and those in burn and surgery units[4-6]. This study focused on determination of biofilm formation in clinical isolates of A. baumannii,elucidation of their sensitivity to different antibiotics,and presenting any possible link between the ability to form biofilm and multidrug resistance(MDR)status.
2. Materials and methods
2.1. Collection,separation and identification of isolates
In this cross-sectional study,a total of 650 clinical samples were taken from patients who spent at least one week of their hospitalization duration in Imam Khomeini,Milad and Motahari Hospitals(Tehran,Iran)during the years 2013-2014(second half of 2013 and first six months of 2014). The collected specimens included blood,urine,sputum,catheter,bronchial,spinal fluid[(cerebrospinal fluid)CSF],bronchoalveolar lavage,exudates and burned skin. Specimens were placed in brain-heart infusion broth medium and then transferred to microbiology lab for culture. Isolates were initially identified using standard laboratory methods including growth on blood agar(non-hemolytic),MacConkey agar medium(Merck,Germany)at 37°C showing a pink to light purple appearance,Gram stain(Gram negative coccobacilli),catalase test(positive),oxidase test (negative),oxidative-fermentative test for glucose(producing acid from glucose in oxidative state),sulfide indole motility,methyl red-Voges-Proskauer,citrate utilization,urease test,triple sugar iron(non-fermentative or alkaline/alkaline),and growth at 44°C on nutrient agar(Merck,Germany).
2.2. Molecular identification of isolates by PCR
Presence of blaOXA-51-like gene was verified using PCR according to Turton's experiment[13]. Genomic DNA was extracted by boiling method. All DNA isolates were subjected to PCR for identification of blaOXA-51 gene. Negative samples were tested for second time with the PCR. The primers were F-TAATGCTTTGATCGGCCTTG- and RTGGATTGCACTTCATCTTGG-(353 bp)[14]. A single reaction mixture contained: 1 μL DNA(0.8 mmol/L),12.5 μL ready 2×PCR Master Mix(composition of 1×solution: 0.5 mol/L Tris-HCl,1.5 mmol/L MgCl2,0.2 mmol/L dATP,0.2 mmol/L dCTP,0.2 mnol/L dGTP and 0.2 mmol/L dTTP and 0.04 IU/μL Taq)(SinaClon BioScience Co.,Iran),1 μL of 10 pmol/L of each primer and 9.5 μL of sterile ultrapure water,with final volume of 25 μL. The PCR protocol included 30 cycles of amplification under the following condition: initial denaturation at 94°C(5 min),denaturation at 94°C (45 s),annealing at 58°C(60 s),extension at 72°C(60 s),and final extension at 72°C(5 min)[14]. PCR products were electrophoresed on 1.2%agarose gel,followed by staining with safe DNA stain and illumination under UV light. Fifty bp DNA ladder was used for size determination.
2.3. Susceptibility testing of bacterial isolates to antibiotics Antibiotic susceptibilities towards eleven clinically relevant antibiotics were determined by Kirby-Bauer disc diffusion method based on Clinical and Laboratory Standards Institute guidelines using antibiotic discs supplied by Rosco Diagnostica (Denmark). The antibiotic discs were as follows: piperacilin (PIP,100 μg),ceftriaxone(CRO,30 μg),cefepime(FEP,30 μg),imipenem(IPM,10 μg),polymyxin B(PB,300 μg),
gentamicin(GM,10 μg),tetracycline(TE,30 μg),ciprofloxacin (5 μg),sulfamethoxazole-trimethoprim(SXT,75 μg),ticarcilin (TIC,75 μg),and ticarcillin/clavulanic acid(TCC,75/10 μg). Inoculums of the A. baumannii isolates(106CFU/mL)were swabbed onto Muller-Hinton agar(Merck,Germany)plates and antibiotic discs were then placed on the inoculated plates. They were incubated at 37°C for 24 h after which the growth inhibition zones around each disc were measured. Pseudomonas aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 were used as reference strains for quality control[15].
2.4. Congo red agar(CRA)method(qualitative biofilm production assay)
A simple qualitative assay for detection of biofilm,using CRA medium,was described by Freeman et al.[10]. Following inoculation,the agar plates were incubated at 37°C for 24-48 h. Appearance of black colonies with a dry crystalline consistency could be considered as strong evidence for ability to form biofilm. Each experiment was conducted in three repeats.
2.5. Tube method(quantitative biofilm production assay)
A quantitative assay for biofilm detection has been proposed by Christensen et al.[16]. A heavy inoculation of bacteria was made in a test tube containing 3 mL of tryptic soy broth (TSB). The inoculated tubes were placed in 37°C incubator for 48 h. After decanting,the bacterial cell pellets were washed with phosphate buffer saline(pH 7.3)and allowed to dry. Tubes were stained with safranin(1%)for 7 min and then washed with distilled water for 5 min. Presence of an attached film of stained material on the tube inner surface was indicative of biofilm formation. Sterile TSB tubes were used as negative control.
2.6. Tissue culture plate method(quantitative biofilm production assay)
This assay which was proposed by Christensen et al. is generally considered to be the gold-standard technique for biofilm detection[17]. Bacterial cells were grown overnight at 37°C in 10 mL TSB(Merck). Each well of a 96-well flat-bottomed polystyrene tissue culture plate(3 wells for each strain)was filled with 20 μL of the overnight culture(equivalent to 0.5 McFarland standard). Sterile TSB medium(180 μL)was added into each well. The biofilm producing reference strain ofA. baumannii ATCC 19606 was used as positive control. Wells inoculated with sterile broth were used as negative control. The plates were covered with a lid and incubated aerobically for 24 h at 37°C. After incubation,the content of each well was removed and the wells were carefully washed,three times,with 0.2 mL of phosphate buffer saline(pH 7.2)in order to remove free floating bacteria.
Adherent bacteria were fixed with 200 μL of 99%methanol per well for 15 min. The plates were decanted and allowed to dry. Then,plates were stained for 7 min with 0.2 mL of 2% Hucker crystal violet. Excess stain was rinsed off by washing with tap water. After the plates were air dried,the dye bound to the adherent cells was resolublized with 160 μL of 33%(v/v)glacial acetic acid per well. The optical density(OD)of each well was measured at 630 nm using ELISA reader(Stat Fax 2100).
The absorbance values were read for two times:firstly prior to addition of glacial acetic acid,and secondly after addition of glacial acetic acid. According to the absorbance values,the adherence capability of each bacterial cell was classified into the following four categories: none(−),weak(+),moderate(++),and strong(+++)adherent cells. The cut-off absorbance value (ODc)was considered as three standard deviations above the mean OD of the negative control. Tests were performed for three times and results were averaged[4-6]. The following classifications were used(Table 1):

Table 1Adherence classification based on microtiter plate method.
2.7. Statistical analysis
One way ANOVA was applied using SPSS 16 in order to compare differences among tube test method,microtiter plate under standard and modified conditions. The experiments were performed in triplicate. P values of≤0.05 were considered as significant.
3. Results
Of the 650 clinical specimens tested,195 Acinetobacter spp. were isolated. Of these,156 were A. baumannii,27 samples were Acinetobacter lwoffii(A. lwoffii)and 12 samples belonged to other species. Among the 156 A. baumannii isolates,83 were obtained from hospitalized women and the rest were from men. The patient's age groups were between 2 and 75 years old. All isolates were finally recognized by both biochemical tests as well as PCR for blaOXA-51 gene(Figure 1). A. baumannii isolates were recovered from tracheal aspirate(23%),sputum (21%),urine(17%),burned skin(17%),wound(10%),blood (5%),sterile body fluids(5%)and other samples(2%). The lowest number of isolate 2 was from bone and 3 isolates were recovered from CSF. Disc diffusion tests indicated that 95.51% of the isolates were resistant to PIP(100),94.87%to CRO(30),94.23%to FEP(30),91.03%to IPM(10),83.33%to GM(10),72.44%to TE(30),89.10%to SXT(75),93.59%to TIC(75),and 91.03%to TCC(75/10). There was a high degree of susceptibility to PB(300)(Figure 2). The results showed that 92.95%of the A. baumannii isolates examined in this study were MDR and 86.53%were extensively drug-resistant;however,none of them was pan-drug resistant. Almost 60%of the isolates were resistant to 10 of the 11 antibiotics tested and only 5 isolates were sensitive to all antibiotics.

Figure 1. PCR products of the blaOXA-51-like gene in A. baumannii.The first well: 50 bp ladder;second to eighth wells are related to the clinical isolates;the last well is for the control strain,A. baumannii ATCC 19606.

Figure 2. The antibiotic susceptibility patterns of the A. baumannii isolates.CP: Ciprofloxacin.
Sixteen of the total 156 isolates that were cultured on the CRA medium produced black colony pigmentation which was considered to be a rough indication of the ability to produce biofilm(Figure 3). However,in the more reliable tube method,25 isolates were non-biofilm producers,55 were weak biofilm formers,51 were moderate biofilmers,and 25 isolates produced biofilms with high power(Figure 4). The results of biofilm formation in the standard microtiter plate method showed that three isolates were not able to produce biofilm,49 isolates formed biofilm weakly,91 isolates created biofilm moderately,and 13 isolates were strong biofilm producers. In the modified microtiter plate assay,one isolate was non-biofilm former,40 isolates were weak,82 isolates were moderate,and 33 isolates were strong producers of biofilms(Table 2). Of the 104 isolates in the standard microtiter plate method with moderate to strongbiofilm formation,98(94.23%)were MDR strains. Amongst the 115 isolates in modified microtiter plate method with moderate or strong biofilm forming trait,106(94.23%)were MDR. All isolates with biofilm positive trait on the CRA were MDR and amongst the 76 isolates in test tube method with moderate to high power biofilm formation,72(94.74%)were MDR.

Figure 3. Growth on CRA medium.Black pigmentation is indicative of biofilm forming phenotype.

Figure 4. Biofilm formation on the inner surfaces of glass.Tube 1: Non-adherent cell;Tube 2: Weakly adherent cell;Tube 3: Moderately adherent cell;Tube 4: Strongly adherent cell.
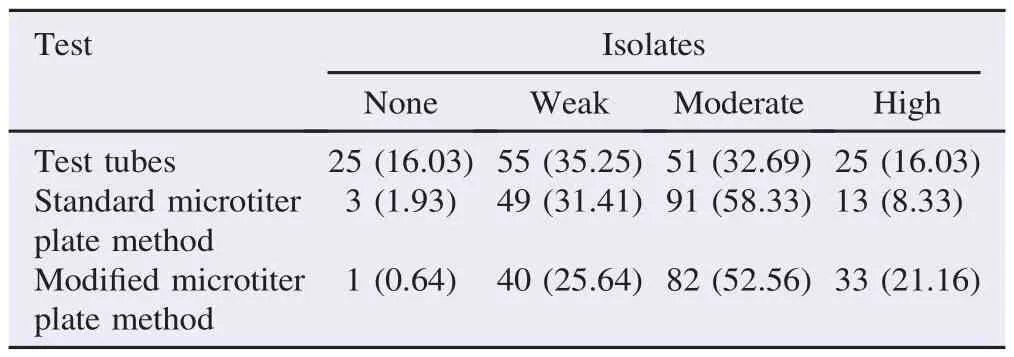
Table 2Percentage of biofilm phenotype in the A. baumannii isolates as depicted by test tubes and microtiter plate methods. n(%).
4. Discussion
A. baumannii is an opportunistic pathogen and one of the main causes of nosocomial infections during the past few decades[18]. This bacterium,especially MDR and biofilm producing strains can cause serious infectious diseases in hospitalized patients. Due to their widespread resistance to antimicrobial drugs,treatment of such infections can be very difficult. This study aimed to determine the prevalence of antibiotic resistance and biofilm-producing strains of A. baumannii isolated from clinical samples and examine the relationship between antibiotic resistance and biofilm formation using phenotypic methods.
Amongst the six hundred and fifty infected samples which were received,156 A. baumannii isolates were recovered. This constituted 80%of the total bacterial isolates. A. lwoffii comprised 13.8%of the isolates and the rest belonged to other species of Acinetobacter. This result was in agreement with the study carried out by Mostofi and her colleagues in 2010-2011 [19]. They isolated 50 bacteria,71%of which were A. baumannii,followed by A. lwoffii(17.1%)and other species(11.9%). Likewise,Constantiniu et al. reported that from a total of 24 clinical isolates,71%were A. baumannii and 29%were A. lwoffii[18]. Acinetobacter infections usually involve organ systems with high fluid content(e.g.,respiratory tract,CSF,peritoneal fluid,urinary tract). In this study,most isolates were recovered from trachea(23%)followed by pus (21%),urine(17%),burn(17%)and wound(10%). Maryam and her colleagues[20]found out the most prevalent isolates were in trachea(25%)followed by urine(19%). Mostofi et al. reported that amongst the A. baumannii cases investigated,30%were from respiratory tract infections,12%were from pus and 8%were originated from urinary tract infections[19].
In the current research,all A. baumannii isolates contained the blaOXA-51-like gene. This result was in agreement with other researchers[3,20,21]. Results of the antibiotic susceptibility tests were very close to recent similar studies in Iran including Vafaei et al. and Maryam et al.[19,20]. Mostofi et al. reported that their A. baumannii clinical strains were resistant to gentamicin(61%),cefepime(95%),ciprofloxacin(88%),and imipenem(76%). However,A. baumannii has retained susceptibility to polymyxin B despite resistance to other antibacterial agents. Maryam and her colleagues indicated that their A. baumannii isolates showed resistance to sulfamethoxazole(90%),imipenem(92%),gentamicin(84%),and tetracycline(89%). Mak et al.[22]claimed that polymyxin B is the most effective drug in controlling this bacterium.
Recent studies showed an overall increasing trend of resistance to antibiotics. Houang and his colleagues reported that their isolates showed 43%resistance to imipenem which was in accordance with our result[11]. Soroush et al. indicated that 40.6%(59/145)of their A. baumannii isolates were identified as MDR[23]. This finding indicates that antimicrobial resistance of A. baumannii in Iran has increased,which may very well affect the antimicrobial resistance of this organism worldwide.
Biofilm production was initially detected by CRA method. The result showed that 10%of the bacteria were biofilm producer which was in agreement with other studies[10]. As stated by Hassan and his colleagues,49%of bacteria were able to form biofilm moderately or with high power and the rest lacked power of biofilm formation or produced weakly[12]. The results of standard microtiter plate using a qualitative approach is also almost in agreement with Kaleem's study. In his study,63%of the bacteria produced biofilm and the rest were not able to do so[12]. Dheepa et al. studied biofilm production and their results showed that 60%of their 50 isolates produced biofilm [6]. Likewise,another study demonstrated 62%of the 51 isolates formed biofilm[24]. As a conclusion,all of them are in agreement with results of this current study. Dheepa et al. in their study that was conducted in 2011 and on 50 isolates of A. baumannii,stated that biofilms assays were performed by three methods: tube method,standard microtiter plate and modified microtiter plate[6]. The percentages of biofilm formation obtained in our study were very close to those reported by Dheepa et al.[6]. Abdi-Ali et al. reported on biofilm production using test tube and modified microtiter plate methods[4]. They worked on 100 isolates of A. baumannii and obtained the following results in the test tube method: 18%no power,42%as weakly,18%as moderate and 22%as high power. The results of modified microtiter plate method were as follows: 25%without biofilm formation,41%as weak,10%as moderate and 18%as high power values. These results were somewhat different from the values obtained in this study. This may be due to the difference in the number of clinical isolates from different sources[4]. What is common in all these studies is the observed intrinsic ability of this bacterium to form biofilms[4,5,10]. Although biofilm assays are not difficult to perform,accurate readings of the test tube results are somewhat subjective. However,it should be noted that certain changes in the tube test procedure may improve the interpretation of the results. After reading the OD of the standard microtiter plates by ELISA reader,if we add 160 μL of glacial acetic(33%v/v)to each well and again read the absorption,in this case,a qualitative study will be converted to a quantitative one. In fact,the shift from qualitative to a quantitative microtiter plate method can prove to be a more reliable assay. The modified microtiter plate assay is a very sensitive,accurate,and reproducible method and can be used as a reliable quantitative assay to determine biofilm formation. The addition of acetic acid permits measurement of the attached bacteria both on the bottom as well as on the inner walls of the wells. Only 160 μL of 33%(v/v)glacial acetic acid was added per well. This will help to evade interference with stained matter at the liquid-air interface,which is not considered to be indicative of biofilm formation in the tube tests[4].
Analyses of the results of this study also indicate that there is a significant correlation between power of biofilm formation and antibiotic resistance;considering that,more than 90%of the bacteria with the ability to form biofilms were MDR. This result is in accordance with Hassan et al. study[12]. However,we noted that those with weak or no power of biofilm formation showed higher resistance to other antibiotic. This is also in agreement with other studies[4,5,10]. This can be due to different mechanisms that may cause resistance to antibiotics [4,5]. Resistance patterns amongst hospital-acquired bacterial pathogens are often different from country to country and also within a single country over time[4].
In conclusion,the results showed that most clinical isolates of A. baumannii have the ability to produce biofilms. This could potentially increase colonization of antibiotic-resistant bacteria in hospital environments. Increased hospital acquired infections can result in increased morbidity and mortality. Thus,a continuous monitoring of antibiotic susceptibility in A. baumannii isolated from different clinical sources in every region seems necessary. These studies can provide useful information for selection of appropriate antimicrobial chemotherapy as well as adopting a suitable method for prevention control. The modified microtiter plate assay seems to be the most reliable and appropriate technique for detection of biofilm formation.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgments
This research was supported by an educational grant for doctoral thesis from Islamic Azad University of Karaj(grant number: 11530554922001). Authors wish to thank Mrs. Maryam Rahmanii for her assistance in the collection and identification of the bacteria. Special thanks also go to the laboratory technicians in Imam Khomeini,Milad and Motahari hospitals (Tehran,Iran)for their sincere help in providing the human specimens.
References
[1]Akers K,Chaney C,Barsoumian A,Beckius M,Zera W,Yu X,et al. Aminoglycoside resistance and susceptibility testing errors in Acinetobacter baumannii-calcoaceticus complex. J Clin Microbiol 2010;48(4): 1132-8.
[2]Aliakbarzade K,Farajnia S,Karimi Nik A,Zarei F,Tanomand A. Prevalence of aminoglycoside resistance genes in Acinetobacter baumannii isolates. Jundishapur J Microbiol 2014;7(10): e11924.
[3]Ardebili A,Lari AR,Talebi M. Correlation of ciprofloxacin resistance with the AdeABC efflux system in Acinetobacter baumannii clinical isolates. Ann Lab Med 2014;34(6): 433-8.
[4]Abdi-Ali A,Hendiani S,Mohammadi P,Gharavi S. Assessment of biofilm formation and resistance to imipenem and ciprofloxacin among clinical isolates of Acinetobacter baumannii in Tehran. Jundishapur J Microbiol 2014;7(1): e8606.
[5]de Breij A,Gaddy J,van der Meer J,Koning R,Koster A,van den Broek P,et al. CsuA/BABCDE-dependent pili are not involved in the adherence of Acinetobacter baumannii ATCC 19606T to human airway epithelial cells and their inflammatory response. Res Microbiol 2009;160: 213-8.
[6]Dheepa M,Rashme VL,Appalaraju B. Comparison of biofilm production and multiple drug resistance in clinical isolates of Acinetobacter baumanii from a tertiary care hospital in South India. Int J Pharm Biomed Sci 2011;2(4): 103-7.
[7]Espinal P,Martí S,Vila J. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfacees. J Hosp Infect 2012;80(1): 56-60.
[8]Magiorakos AP,Srinivasan A,Carey RB,Carmeli Y,Falagas ME,Giske CG,et al. Multidrug-resistant,extensively drug-resistant and pan drug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012;18(3): 268-81.
[9]M'hamedi I,Hassaine H,Bellifa S,Lachachi M,Kara Terk I,Djeribi R. Biofilm formation by Acinetobacter baumannii isolated from medical devices at the intensive care unit from medicaldevices at the intensive care unit of the University Hospital of Tlemcen(Algeria). Afr J Microbiol Res 2014;8(3): 270-6.
[10]Freeman DJ,Falkiner FR,Keane CT. New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol 1989;42: 872-4.
[11]Houang ET,Sormunen RT,Lai L,Chan CY,Leong AS. Effect of desiccation on the ultrastructural appearances of Acinetobacter baumannii and Acinetobacter lwoffii. J Clin Pathol 1998;51(10): 786-8.
[12]Hassan A,Usman J,Kaleem F,Omair M,Khalid A,Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz J Infect Dis 2011;15(4): 305-11.
[13]Turton JF,Woodford N,Glover J,Yarde S,Kaufmann ME,Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol 2006;44: 2974-6.
[14]Woodford N,Ellington MJ,Coelho JM,Turton JF,Ward ME,Brown S,et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents 2006;27(4): 351-3.
[15]Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing;Twenty-Fourth Informational Supplement. Wayne: Clinical and Laboratory Standards Institute;2014.[Online]Available from: http://ncipd.org/ control/images/NCIPD_docs/CLSI_M100-S24.pdf[Accessed on 20th December,2015]
[16]Christensen GD,Simpson WA,Bisno AL,Beachey EH. Adherence of slime producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun 1982;37: 318-26.
[17]Christensen GD,Simpson WA,Younger JA,Baddour LM,Barrett FF,Melton DM,et al. Adherence of coagulase negative staphylococci to plastic tissue cultures: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol 1995;22: 996-1006.
[18]Constantiniu S,Romaniuc A,Iancu LS,Filimon R,Taras¸i I. Cultural and biochemical characteristics of Acinetobacter spp. strains isolated from hospital units. J Prev Med 2004;12: 35-42.
[19]Mostofi S,Mirnejad R,Masjedian F. Multi-drug resistance in Acinetobacter baumannii strains isolated from the clinical specimens of three hospitals in Tehran-Iran. Afr J Microbiol Res 2011;5: 3579-982.
[20]Maryam R,Golnaz YZ,Mojgan O,Malihe T,Nour A. Identification of five phylogenic groups of carbapenemase(blaOXA-23,24,51,58,143)in Acinetobacter baumannii strains isolated from clinical samples in Iran by multiplex PCR. Der Pharma Chem 2015;7(7): 11-6.
[21]Asadollahi P,Akbari M,Soroush S,Taherikalani M,Asadollahi K,Sayehmiri K,et al. Antimicrobial resistace patterns and their encoding genes among Acinetobacter baumannii strains isolated from burned patients. Burns 2012;38: 1198-203.
[22]Mak JK,Kim MJ,Pharm J,Tapsall J,White PA. Antibiotic resistance determinants in nosocomial strains of multidrugresistant Acinetobacter baumannii. J Antimicrob Chemother 2009;63: 47-54.
[23]Soroush S,Haghi-Ashtiani MT,Taheri-Kalani M,Emaneini M,Aligholi M,Sadeghifard N,et al. Antimicrobial resistance of nosocomial strain of Acinetobacter baumannii in Children's Medical Center of Tehran: a 6- year prospective study. Acta Med Iran 2010;48(3): 178-84.
[24]Rao RS,Karthika RU,Singh SP,Shashikala P,Kanungo R,Jayachandran S,et al. Correlation between biofilm production and multiple drug resistance in imipenem resistant clinical isolates of Acinetobacter baumannii. Indian J Med Microbiol 2008;26: 333-7.
Objective:To check biofilm formation by Acinetobacter baumannii(A. baumannii)clinical isolates and show their susceptibility to different antibiotics and investigate a possible link between establishment of biofilm and multidrug resistance.
Methods:This study was performed on clinical samples collected from patients with nosocomial infections in three hospitals of Tehran. Samples were initially screened by culture and biochemical tests for the presence of different species of Acinetobacter. Identifications were further confirmed by PCR assays. Their susceptibilities to 11 antibiotics of different classes were determined by disc diffusion method according to Clinical and Laboratory Standards Institute guidelines. The ability to produce biofilm was investigated using methods: culture on Congo red agar,microtiter plate,and test tube method.
Results:From the overall clinical samples,156 specimens were confirmed to contain A. baumannii. The bacteria were highly resistant to most antibiotics except polymyxin B. Of these isolates,10.26%were able to produce biofilms as shown on Congo red agar. However,the percentage of bacteria with positive biofilm in test tube,standard microtiter plate,and modified microtiter plate assays were 48.72%,66.66%,and 73.72%,respectively. At least 92%of the biofilm forming isolates were multidrug resistant.
Conclusions:Since most of the multidrug resistant strains produce biofilm,it seems necessary to provide continuous monitoring and determination of antibiotic susceptibility of clinical A. baumannii. This would help to select the most appropriate antibiotic for treatment.
*Corresponding author:Nour Amirmozafari,Professor of Microbiology,Iran University of Medical Sciences,School of Medicine,Microbiology Department,Tehran,Iran.
 Asian Pacific Journal of Tropical Biomedicine2016年6期
Asian Pacific Journal of Tropical Biomedicine2016年6期
- Asian Pacific Journal of Tropical Biomedicine的其它文章
- Knowledge,attitude and practice of healthcare workers concerning Crimean-Congo hemorrhagic fever in Western Iran
- Epidemiological situation and molecular identification of cercarial stage in freshwater snails in Chao-Phraya Basin,Central Thailand
- Potential drug-drug interactions in pediatric wards of Gondar University Hospital,Ethiopia∶A cross sectional study
- Prevalence of latent eosinophilia among occupational gardeners at Babcock University,Nigeria
- Preliminary studies of acute and sub-chronic toxicity of the aqueous extract of Guibourtia tessmannii(Harms)J. Leonard stem barks(Caesalpiniaceae)in mice and rats
- Evaluation of the anticonvulsant activity of the essential oil of Myrothamnus moschatus in convulsion induced by pentylenetetrazole and picrotoxin
