Detection of Salmonella and several common Salmonella serotypes in food by loop-mediated isothermal amplification method
Zhongqiang Chen,Ke Zhang,Huan Yin,Qi Li,Lan Wang,Zhiguo Liu
College of Biology and Pharmaceutical Engineering,Wuhan Polytechnic University,Wuhan 430023,China
Abstract
Keywords:Loop-mediated isothermal amplification;Detection;Salmonella;Salmonella Choleraesuis;Salmonella Enteritidis;Salmonella Typhimurium
1.Introduction
Salmonellais a significant pathogen for humans and animals.It cannot only cause a variety of livestock and poultry diseases,but also cause food poisoning in humans[1].Currently,diseases caused bySalmonellarank first in food poisoning diseases in China[2].Among the foods that causeSalmonellapoisoning,more than 90% are meat and other animal products[3,4].Statistics show thatS.Choleraesuis,S.Enteritidis andS.Typhimurium are the major pathogenic bacteria that contaminate animal products and cause humanSalmonellapoisoning.They cause significant harm to human health and livestock development[5].
The conventional detection methods forSalmonellainclude bacterial culture,biochemical and serological identification methods.These methods are complicated,time-consuming and laborious,costly and require a variety of reagents.With the development of molecular biology and immunology methods,a number of PCR-based detection techniques have been established,such as immuno-PCR,real-time quantitative PCR and multiplex PCR[6,7],as well as immunoassay methods based on antigen.These methods are fast,sensitive and accurate.As these modern detection methods forSalmonellarequire certain equipment,technique know-how and expensive reagents,they are less economical and practical.To conduct extensive and rapid detection ofSalmonellaand effectively prevent and controlSalmonellafood poisoning at the grassroots level,the establishment of a practical,simple,rapid and sensitive detection method is necessary.
The significance of loop-mediated isothermal amplification(LAMP)method in microbial detection has been drawn increasing attentions recently[8].It was developed by Notomi et al.in 2000[9].It amplifies nucleic acids within tens of minutes by self-circulation of strand displacement DNA polymerase under isothermal conditions.With the advantages of high specificity and sensitivity,simplicity,rapidity and cheapness,it has been widely applied in many areas[10].Research shows thatinvEgene exits in allSalmonellaserotypes but not in non-Salmonellabacteria[11],whilefliC,lygDand STM4495 specifically exists inS.Choleraesuis,S.Enteritidis andS.Typhimurium,respectively[12–14].Therefore,the specific sequences ininvE,fliC,lygDand STM4495 genes were selected as the target sequences for LAMP assay.
2.Materials and methods
2.1.Bacterial strains
S.Enteritidis CICC 21513,S.Typhimurium CICC 21483,S.Choleraesuis CICC 21513,StreptococcushemolyticusCICC 10373,ShigellaflexneriCICC 21534,ShigellaboydiiCICC 21680 andShigellasonneiCICC 21594 were obtained from China Industrial Microbiology Culture Collection.Staphylococcusaureus,EscherichiacoliDH5,Bacillussubtiliswere preserved in our laboratory.
2.2.Preparation of DNA
The DNA template was prepared by boiling method[15].The singleSalmonellacolony was picked and dissolved in 100L distilled water,lysed by water bath at 100℃ for 15 min and ice bathed for 5 min.The reactants were then centrifuged at 12,000 r/min for 5 min at 4℃.The supernatant that contained DNA template was used for LAMP.
2.3.Primer design and synthesis
The LAMP reaction needs four oligonucleotide primers:forward inner primer(FIP),back inner primer(BIP),forward outer primer(F3)and back outer primer(B3).The primers targetingSalmonellainvEgene and 3 serotype-specific genesfliC,lygDand STM4495 were designed according to the sequences published in GenBank.The primer sequences are listed in Table 1.Primers were synthesized by Generay Biotech Co.,Ltd.(Shanghai,China).

Table 1LAMP primer sets for amplifying invE,fliC,lygD and STM4495.
2.4.LAMP reaction and condition optimization
LAMP reaction was carried out in a 25L mixture which contained inner and outer primer sets,BstDNA polymerase,dNTPs,1×ThermoPol buffer and DNA template.The mixture withBstDNA polymerase absent was water bathed at 95℃ for 5 min,then quickly cooled on ice.After cooling,BstDNA polymerase was added.The mixture was incubated at certain temperature for tens of minutes in a thermal cycler(UNO II,Whatman Biometra,Germany),then heated to 80℃ for 10 min to terminate the reaction.The LAMP products were electrophoresed in a 1% agarose gel and visualized by ethidium bromide staining.The concentration ratio between inner and outer primer sets,reaction duration and temperature were optimized to determine the most efficient amplifying conditions of LAMP.The efficiency was analyzed by electrophoresis.
2.5.Specificity of LAMP detection
The specificity of LAMP primers was identified by LAMP detection for the 3 major strains of pathogenicSalmonellaserotypes,i.e.,S.Choleraesuis,S.Enteritidis andS.Typhimurium and seven non-Salmonellastrains.
2.6.Sensitivity of LAMP detection
The single colony ofSalmonellawas picked after culturing for 2 days,dissolved in 1 mL liquid medium and mixed.10-fold serial dilutions of the bacteria liquid was carried out with saline.Plate cultivation counting was performed with 100L bacteria liquid of each dilution.DNA was prepared by boiling.LAMP amplification was conducted by using 1L of supernatant after quantitation.
2.7.LAMP assay of Salmonella in simulated pork samples
2.5 g sterilized fresh pork homogenized in 22.5 mL sterile saline worked as the diluent forSalmonella.The bacteria liquid was 10-fold serially diluted.Samples were taken for plate cultivation counting.DNA template was prepared for LAMP.
3.Results and discussion
3.1.Optimization of LAMP reaction conditions
The LAMP products appeared as a ladder of multiple bands,while no bands was amplified from no template control.TakeS.Choleraesuis as example,the optimum concentration ratio of inner and outer primers was 3:1(Fig.1A);the typical DNA ladder appeared after amplification for 45 min,while better stability was observed after amplification for 60 min(Fig.1B);the optimal temperature was 65℃(Fig.1C).
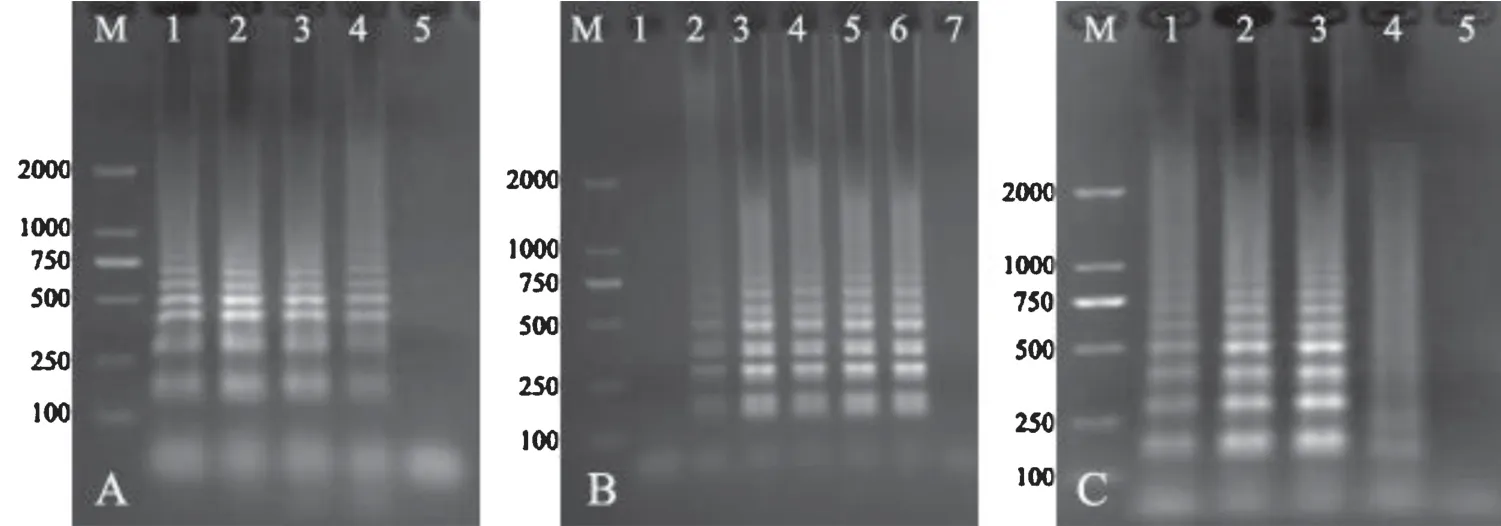
Fig.1.Optimization of LAMP reaction conditions.
The LAMP products under different reaction conditions were electrophoresed in 1% agarose gel and stained with ethidium bromide.(A)Determination of optimal concentration ratio between inner and outer primers.M,DNA marker(DL2000);Lane1–4,the concentration ratio between inner and outer primers was 4:1,3:1,2:1 and 1:1,respectively;Lane 5,negative control.(B)Optimization of reaction duration.M,DNA marker;Lane 1–6,LAMP reaction carried out for 30,45,60,75,90 and 105 min,respectively;Lane 7,negative control.(C)Optimization of reaction temperature.M,DNA marker;Lane 1–4,LAMP reaction carried out at 61,63,65 and 67℃,respectively;Lane 5,negative control.
3.2.Specificity of LAMP detection for Salmonella
3.2.1.Genus-specificityofLAMPdetectionforSalmonella
Amplification reactions were performed usinginvELAMP primers to detect the 10 strains ofSalmonellaand non-Salmonellabacteria.The LAMP results showed that the characteristic ladder bands were amplified from the DNA templates of 3 strains ofSalmonella,while not from that of the 7 non-Salmonellastrains(Fig.2).
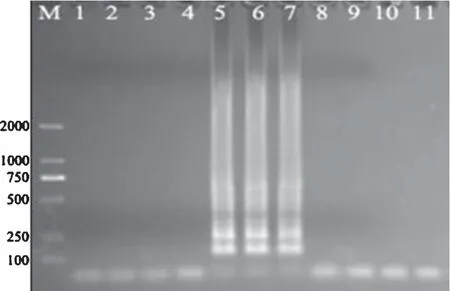
Fig.2.Genus-specificity of LAMP detection for Salmonella.
Salmonellais one of the major pathogens of food poisoning,among whichS.Choleraesuis,S.Enteritidis andS.Typhimurium are dominant.The protein encoded byinvEgene is the main virulence factor[11]and play an important role inSalmonellapathogenesis.The results indicated that theinvELAMP primers were highly specific forSalmonella.
The genus-specificity ofinvELAMP primers was determined by amplifying the 10 different bacterial strains;the products were electrophoresed in 1% agarose gel and stained with ethidium bromide.M,DNA marker(DL2000);Lane 1–10,LAMP amplifications of DNA templates fromS.aureus,E.coliDH5,S.hemolyticus,B.subtilis,S.Enteritidis,S.Typhimurium,S.Choleraesuis,S.flexneri,S.boydiiandS.sonnei,respectively;Lane 11,negative control.
3.2.2.SpecificityofLAMPdetectionforthe3majorstrains ofpathogenicSalmonellaserotypes
Amplification reactions were conducted using LAMP primers forfliC,lygDand STM4495 to detect the 10 strains respectively(Fig.3).AsFliCgene encodingS.Choleraesuis flagellum protein is conservative at both ends,but multifarious in the middle area amongSalmonellaserotypes[12,16].LygDgene exists only inS.Enteritidis genome[13]and in all the genomes of 39S.Enteritidis phage types that isolated from clinical samples[17].The region including genes STM4488 to STM4497 houses a putative type II restriction enzyme that is only present inS.Typhimurium[14].Therefore,the specific sequences ininvE,fliC,lygDand STM4495 genes were selected as the target sequences for LAMP assay.The results in Fig.3 showed that the characteristic ladder bands were amplified only from the DNA templates of their correspondingSalmonellaserotypes,while not from that of the other 2 serotypes or the 7 non-Salmonellastrains.
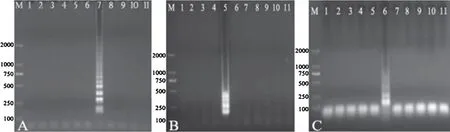
Fig.3.Specificity of LAMP detection for the 3 Salmonella serotypes.
LAMP specificity was determined by amplifying the DNA of 10 different strains using primers forfliC(A),lygD(B)and STM4495(C),respectively;the products were electrophoresed in 1% agarose gel and stained with ethidium bromide.M,DNA marker(DL2000);Lane 1–10,LAMP amplifications of DNA templates fromS.aureus,E.coliDH5,S.hemolyticus,B.subtilis,S.Enteritidis,S.Typhimurium,S.Choleraesuis,S.flexneri,S.boydiiandS.sonnei,respectively;Lane 11,negative control.
3.3.Sensitivity of LAMP detection for Salmonella
Salmonellawas 10-fold serially diluted in saline and the DNA templates of each dilution were extracted for LAMP reactions.The results exemplified byS.Choleraesuis showed that the LAMP products could be detected to 10−7dilution(Fig.4A),and the detection limit was 1.33×101CFU/mL by calculation according to the plate counting.When 0.3L SYBR Green was added into the LAMP products,fluorescence was observed under the UV light in the amplifications from the templates of 10−6dilution(Fig.4B).The detection limit of PCR forS.Choleraesuis was 1.67×102CFU/mL when assays were performed with different dilutions(Fig.4C).The detection results of the sensitivity for the other serotypes are shown in Table 2.
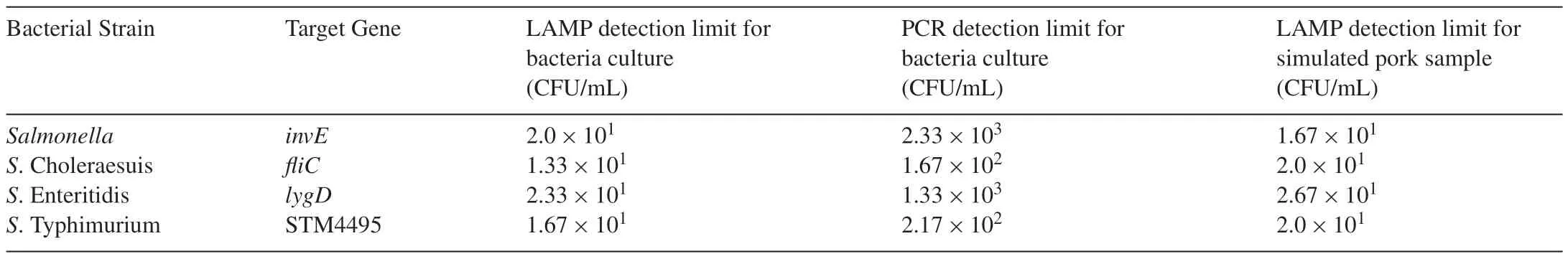
Table 2Sensitivity of LAMP and PCR for the detection of Salmonella.
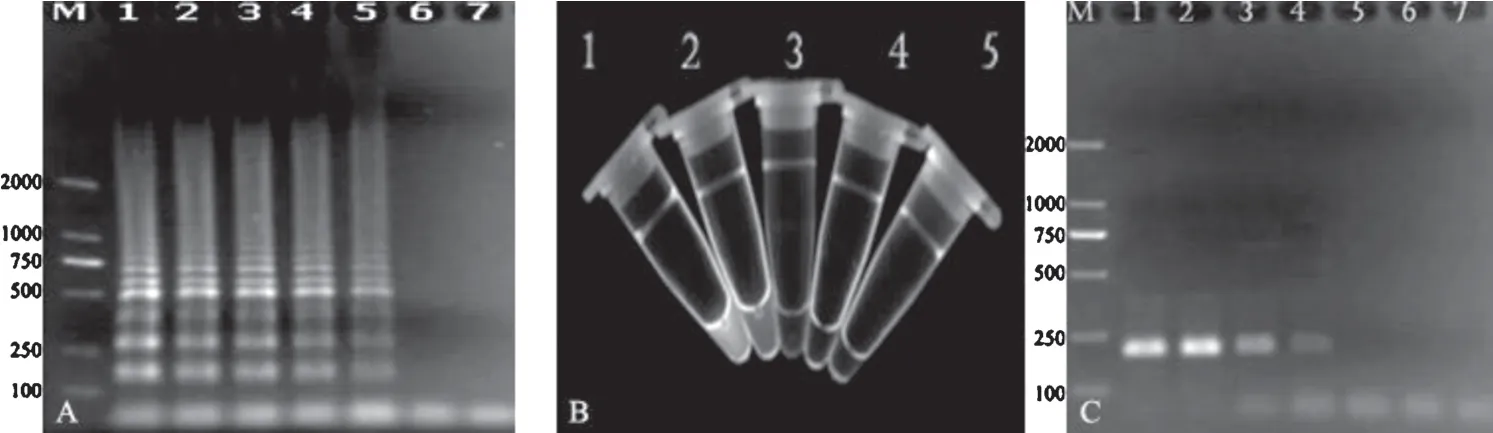
Fig.4.Sensitivity of LAMP detection.
(A)The electrophoresis analysis of LAMP products ofS.Choleraesuis culture with 10-fold serial dilutions.M,DNA marker(DL2000);Lane 1–6,S.Choleraesuis culture with 10−3,10−4,10−5,10−6,10−7and 10−8dilutions,respectively;Lane 7,negative control.(B)SYBR GREEN fluorescence analysis of LAMP products ofS.Choleraesuis.Tube 1–4,S.Choleraesuis culture with 10−5,10−6,10−7and 10−8dilutions,respectively;Tube 5,no template control.(C)The electrophoresis analysis of PCR products ofS.Choleraesuis with serial dilutions,with the positive band of 221 bp.M,DNA marker(DL2000);Lane 1–6,S.Choleraesuis culture with 10−3,10−4,10−5,10−6,10−7and 10−8dilutions,respectively;Lane 7,negative control.
3.4.LAMP detection for Salmonella in simulated pork samples
Salmonellawas 10-fold serially diluted with sterilized pork homogenate and DNA was extracted from each dilution for LAMP assay.The results exemplified byS.Choleraesuis showed that the LAMP products could be detected to 10−6dilution(Fig.5),and the detection limit was 2.0×101CFU/mL by calculation according to the plate counting.
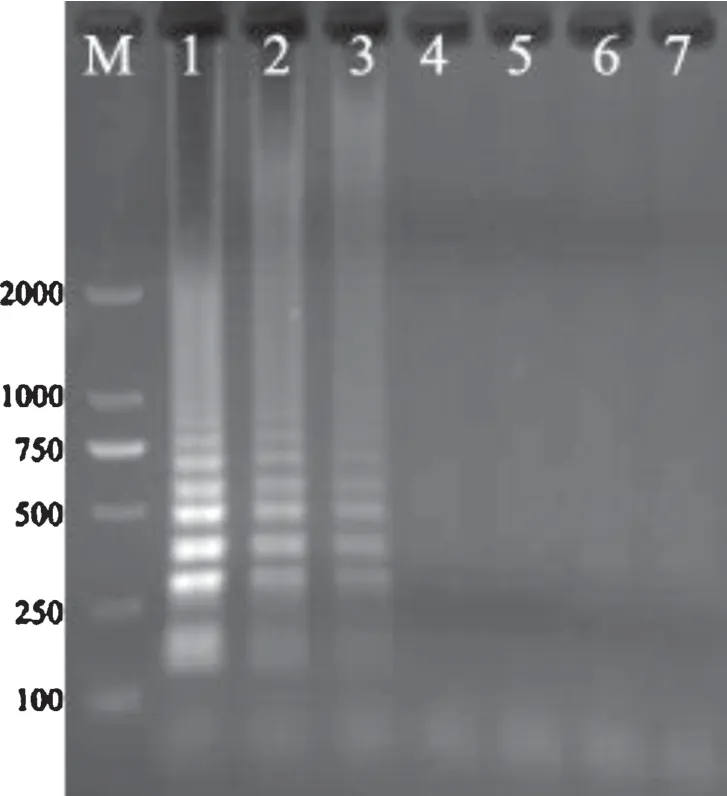
Fig.5.LAMP detection of S. Choleraesuis in simulated pork samples.
The products were electrophoresed in 1% agarose gel and stained with ethidium bromide.M,DNA marker(DL2000);Lane 1–4,LAMP detection ofS.Choleraesuis diluted to 10−4,10−5,10−6and 10−7,respectively,with sterilized pork homogenate;Lane 5,negative control;Lane 6–7,LAMP with sterilized pork homogenate as template.
4.Conclusion
In this study,we demonstrated that LAMP amplification forinvEgene could successfully detectedS.Enteritidis,S.Choleraesuis andS.Typhimurium withinvEin their genome,while could not detect non-Salmonellastrains.And LAMP amplifications forfliC,lygDand STM4495 genes could detect their correspondingSalmonellaserotypes respectively,while did not cross-reacted with the irrelevantSalmonellaserotypes or non-Salmonellastrains.This indicated that theinvELAMP primers and the sequences ininvE,fliC,lygDand STM4495 genes were highly specific forSalmonella.
The detection limit ofSalmonellawas 2.0×101CFU/mL when LAMP assay was exploited forinvEgene,which was 2.33×103CFU/mL by PCR forinvE.The results indicated that LAMP assay was more sensitive than PCR.
The LAMP reactions can be accomplished at a constant temperature of 63–65℃ within 45–70 min and terminated by inactivate the enzyme at℃for10 min.Just a thermostat water bath will work.Sample consumption is less and DNA template preparation is simple.Furthermore,identification methods of LAMP products are simple and various,such as observing precipitation by naked eyes,adding SYBR Green and agarose gel electrophoresis.
In conclusion,the LAMP detection method for pathogenicSalmonellais specific,sensitive,simple,rapid and costeffective.It has wide application prospect in the rapid detection ofS.Choleraesuis,S.Enteritidis,S.Typhimurium and other pathogenicSalmonellain food.
Acknowledgments
This work was supported by Wuhan Science and Technology Planning Project(201070934341),Wuhan Agricultural Technology Innovation Project(201120637175),and Wuhan Polytechnic University Major Incubation Planning(2011z01).
- 食品科学与人类健康(英文)的其它文章
- Phenolics from grapefruit peels inhibit HMG-CoA reductase and angiotensin-I converting enzyme and show antioxidative properties in endothelial EA.Hy 926 cells
- Carvacrol attenuates N-nitrosodiethylamine induced liver injury in experimental Wistar rats
- New perspectives on probiotics in health and disease
- A review:Health promoting lactic acid bacteria in traditional Indonesian fermented foods
- GUIDE FOR AUTHORS

