A susceptibility locus rs7099208 is associated with non-obstructive azoospermia via reduction in the expression of FAM160B1
Yan Zhang,Jing Qian,Minghui Wu,Mingxi Liu,Kai Zhang,Yuan Lin,Xuejiang Guo, Zuomin Zhou,Zhibin Hu,Jiahao Sha,✉
1State Key Laboratory of Reproductive Medicine;2Departmentof Histology and Embryology;3DepartmentofEpidemiology and Biostatisticsand Key Laboratory of Modern Toxicology of Ministry of Education Nanjing MedicalUniversity,Nanjing,Jiangsu 210029,China;
4The Center for Reproductive Medicine,The Second Affiliated Hospital of Nanjing Medical University,Nanjing,Jiangsu 210011,China.
A susceptibility locus rs7099208 is associated with non-obstructive azoospermia via reduction in the expression of FAM160B1
Yan Zhang1,4,Jing Qian1,2,Minghui Wu1,2,Mingxi Liu1,2,Kai Zhang1,3,Yuan Lin1,3,Xuejiang Guo1,2, Zuomin Zhou1,2,Zhibin Hu1,3,Jiahao Sha1,2,✉
1State Key Laboratory of Reproductive Medicine;2Departmentof Histology and Embryology;3DepartmentofEpidemiology and Biostatisticsand Key Laboratory of Modern Toxicology of Ministry of Education Nanjing MedicalUniversity,Nanjing,Jiangsu 210029,China;
4The Center for Reproductive Medicine,The Second Affiliated Hospital of Nanjing Medical University,Nanjing,Jiangsu 210011,China.
Non-obstructive azoospermia(NOA)is a severe defectin male reproductive health thatoccurs in 1%of adult men.In a previous study,we identified that rs7099208 is located within the last intron of FAM160B1 at 10q25.3.In this study,we analysed expression Quantitative Trait Loci(eQTL)of FAM160B1,ABLIM1 and TRUB1,the three genes surrounding rs7099208.Only the expression level of FAM160B1 was reduced for the homozygous alternate genotype(GG)of rs7099208,butnotfor the homozygous reference or heterozygous genotypes.FAM160B1 is predominantly expressed in human testes,where itis found in spermatocytes and round spermatids.From 17 patients with NOA and five with obstructive azoospermia(OA),immunohistochemistry revealed thatexpression of FAM160B1 is reduced,orundetectable in NOA patients,butnotin OA cases ornormalmen.We conclude thatrs7099208 is associated with NOA via a reduction in the expression of FAM160B1.
non-obstructive azoospermia,obstructive azoospermia,rs7099208,FAM160B1,expression Quantitative Trait Loci,apoptosis
Introduction
Approximately 15%-20%ofcouplesare infertile,with males and females suffering equally from infertility[1]. Non-obstructive azoospermia(NOA),a severe defectin male reproductive health,occurs in 1%ofadultmen[2].It has been reported thata defectin the AZF region ofthe Y chromosome causes 10%-15%ofidiopathic NOA[3]. However,othergenetic causes of NOA remain unclear. Many geneknockoutslead to NOAin mice,butfew corresponding defectshave been observed in NOA men.
Oneexplanationoftherelationship between risk lociand diseaseisthattheselociareassociated with mRNAexpression ofrelevantgenes[4-5].Protein markersfordisease also could have been discovered by GWAS analysis[6-7].To investigate genetic variation in NOApatients,we recently conducted a genome-wide association study(GWAS)of NOA in Han Chinese based on genotyping 587,347 single nucleotide polymorphisms(SNPs)in 981 NOA casesand 1,657 controls[8].We tested promising associationsin an extended three-stage validation using 3,608 NOA cases and 5,909 controls.In thecombined analysis,meta-analysiswasused to combine the resultsof GWAS scan and three validations.We identified three risk lociassociated with NOA(rs7194 at6p21.32,rs7099208 at10q25.3,and rs13206743 at6p12.2)[9].According to ourpreviousstudy, genotype ofrs7099208 in NOA patients is 43/943/3552 (Varianthomozygote/Heterozygote/Wild type)and in controlpopulation is 35/1209/6312(Varianthomozygote/ Heterozygote/Wild type),and minor allele frequency (MAF)of rs7099208 is 0.11 in NOA patients,which is higherthan 0.08 in controlpopulation,P=6.41x10-14[9]. However,itis stillunknown whetherrs7099208 affects mRNA expression.
In this work,we tested the hypothesis that expression of risk locirelated genes is altered in testes from NOA patients.Our approach examined samples from patients with NOA and obstructive azoospermia (OA),and the germ cellline of male mice.
Materials and methods
Expression Quantitative Trait Loci(eQTL)
The publicly available RNA-seq and genotyping data of53 normaltestissamples and 167 whole blood samples from the Genotype-Tissue Expression project(GTEx, http://commonfund.nih.gov/GTEx/index)were used to assess Gene Expression Quantitative Trait Loci(eQTL) for mRNA expression of FAM160B1 and genes neighbouring SNP rs7099208[10].
Patients
This study was approved by the institutionalreview boards of each participating institution.Participants were solicited from individuals attending the First Affiliated Hospital Reproductive Centre of Nanjing Medical University for knockdown of infertility. Controls(n=7)were defined as individuals who were diagnosed with normal spermatogenesis or OA. Patients diagnosed with NOA were the experimental group(n=17).Six of the 17 formed the spermatid arrest sample,and the others were the spermatocyte arrest sample.For sample specific information see Supplementary Material.
Processing of testicular biopsy material
This study wasapproved by the ethicscommittees of Nanjing Medical University and was conducted in accordance with national and international guidelines. A testicular biopsy was performed in all samples (n=24).A smallincision was made in the tunica albuginea to remove tissue samples,which comprised about3 mm3in total.
Indirect immunohistochemistry and immunofluorescence
Indirect immunohistochemistry and immunofluorescence were performed as previously described[11-13].To prepare sections,tissue from normal testes was fixed in Bouin fluid and dehydrated through a series of graded alcohols.The testes were then embedded in paraffin at 65°C and sections were cutfor immunohistochemistry. The testis sections were hydrated with series of graded alcohols.After endogenous peroxidase activity was blocked with 3%H2O2,non-specific binding was blocked with 10%normalgoatserum.Thesectionswereincubated overnight at 4°C with primary antibody against FAM160B1(Abgent)ata dilution of1:100.Afterextensive washes in PBS,the sections were incubated with the secondary antibody for1 hourat37°C.Next,DBA colourliquid was added to the sections and the reaction productwas generated.Afterimmunostaining,the testis sectionswerecounterstained with hematoxylin and examined undera lightmicroscope.Forimmunofluorescence, GC2 cells were fixed with 4%PFA atroom temperature and 70%Ethanolat-20°C.Cells were analysed using an epifluorescence microscope(Zeiss 710)atroom temperature and Image Pro-Plus version software(Carl Zeiss,Germany)was used for analysis and imaging. Imageswere acquired using a100×magnification oillens (Zeiss).In brief,green fluorescencewasexcitedwith a488 nm diode laserand red fluorescence with a 555 nm diode laser.Multicolour images were acquired sequentially.
PCR
Total RNA was extracted from cells with mo-FAM160B1 and mo-controltissue using an RNeasy plus Micro Kit(QIAGEN,74034).RNA was reversetranscribed into cDNA using a PrimeScript RT reagent Kit(Takara,DRR037S).PCR was performed with GoTaq Green Master Mix(Promega,M7122),and real-time PCR was performed with SYBR Premix EX Taq II(Takara,DRR081B).In PCR,the forward and reverse primers used were:MUS(F)GAGGGCTTGA TGCTCTTGGT and(R)GCTGTATGAGTCCAACC CCC;HUM(F)GGCTGCAAAGTGCCTTACAC and(R)CGAACGATGCGATGAAGCAG.
The PCR started at95°C for 30 seconds and was performed as follows:28 cycles of denaturation at 95°C for 5 seconds,annealing at 60°C for 30 seconds and elongation at 72°C for 30 seconds.
Western blotting assay
Samples containing 20μg of protein(measured by the Bradford method)were used for Western blotting.Proteins from mo-FAM160B1 cells and control samples were subjectto electrophoresis on a 10%sodium dodecylsulfate polyacrylamide gel.The proteins were then transferred from the gel onto a nitrocellulose me mbrane(GE He a lthc a re Bio-Sc ienc es AB, Uppsala,Sweden).Membranes were blocked in a solution ofphosphate-buffered saline(PBS)containing 5% non-fat milk powder for 1 hour,and then incubated overnight at 4°C with the following primary antibodies:anti-FAM160B1(1:1,000;Abgent)and anti-β-tubulin(1:2,000;Abcam).After four 10 minutes washes in PBS,the membraneswere incubated for1 hourat37°C with horseradish peroxidase-conjugated secondary antibody.Specific proteins were detected using an ECL kit and AlphaImager(FluorChem5500;Alpha Innotech,San Leandro,CA).
Cell culture and morpholino knockdown
The mouse spermatocyte cell line GC2-spd(ATCC catalogue number CRL-2196,Manassas,VA,USA) was used for in vitro studies.Growth media contained 10%foetalbovine serum(GIBCO)and 5%penicillin/ streptomycin,and was maintained in a 37°C incubator in a humidified,5%CO2atmosphere.The knockdownwas performed using morpholinos(splice blocking and translation blocking).Depending on the oligonucleotide sequence selected,morpholinos either modify pre-mRNA splicing in the nucleus,orblock translation initiation in the cytosol.After incubation for 24 hours, cells were used for RT-PCR analysis,Western blotting, cell apoptosis assays and electron microscopy analysis. Morph olino se q ue nce s we re splic e b loc king: CGTCCTAAGAAAGAACGCACACGGA;translation blocking:ACGTAAGCTTGGAGAACATCCTG TC(http://www.gene-tools.com/).
TUNEL
Apoptotic cells were examined using the terminal dideoxynucleotidyl transferase dUTP nick end labelling(TUNEL)method with the in situ celldeath detection kit,POD,according to the manufacturer’s protocol (Roche,Mannheim,Germany).
Electron microscopy
Testes were fixed in glutaraldehyde for 2 hours at room temperature.Ultra-thin sections(-90 nm)were cut parallel to the cell monolayer and stained in uranyl acetate and lead citrate.Nucleiof spermatocytes were randomly selected using an electron microscope (JEM-1010)and capture software(SIS VELETA CCD).The observer was blinded to the genotype.Statistical analysis
Forcontinuous variables,differences between two groups were tested by Studentt-testor t'-test(equalvariances notassumed).Differences between three groups were tested by ANOVA test or Kruskal-Wallis test (equalvariances notassumed).Analyses were carried outusing Statistical Analysis System software(version 9.1.3;SAS Institute,Cary,NC).A two-sided P≤0.05 wasconsidered as statistically significant.
Results
FAM160B1correlates negatively with the homozygous alternate(GG)of rs7099208.
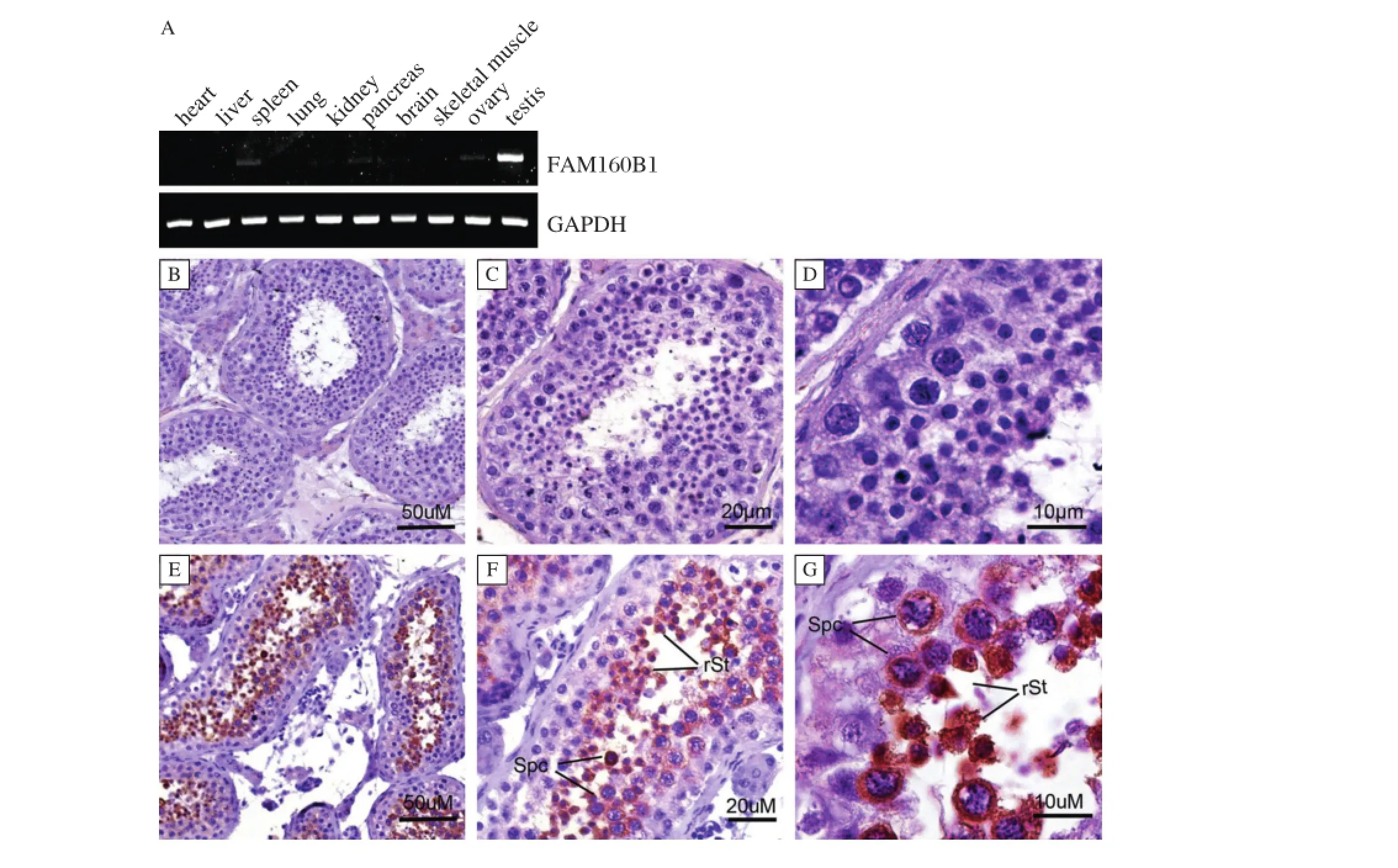
Fig.2 FAM160B1gene expression in Human.A:The expression of FAM160B1 in multi-organization successively:heart,liver,spleen,lung,kidney pancreas,brain,skeletalmuscle,ovarian and testes.GAPDH was used as an internalcontrol.B-G:Immunohistochemistry of FAM160B1 in the normal Human testes.B,C,D:the negative control;E,F,G:Human testis.FAM160B1 located in post-meiotic germ cells containing spermatocytes and round spermatids.
To investigate whetherrs7099208 is associated with expression ofmRNA,we analysed expression eQTL of genes surrounding rs7099208.Three genes at10q25.3,FAM160B1,ABLIM1 and TRUB1,are candidates whose mRNA expression may be affected by rs7099208.We assessed the various genotypes of rs7099208 and the expression of FAM160B1,ABLIM1 and TRUB1 in normal human testes,using data from the Genotype-Tissue Expression project(GTEx).The eQTL data revealthatthe mean mRNAexpression level of FAM160B1 in testes with the homozygous alternate (GG)genotype ofrs7099208 is less than that of the homozygous reference(AA)or heterozygous(AG) genotypes(Fig.1A).The mean expression levels of ABLIM1 and TRUB1 were not affected by the genotype of rs7099208(Fig.1B and C).In addition,the expression level of FAM160B1 in peripheral blood was not associated with the rs7099208 genotype(P=0.8)(Fig.1D).These results suggestthat rs7099208 is associated with mRNA expression ofgene FAM160B1 in testes.
FAM160B1was predominantly expressed in human spermatocytes and round spermatids
Using RT-PCR,expression of FAM160B1 mRNA was assessed in multiple tissue,including heart,liver, spleen,lung,kidney pancreas,brain,skeletalmuscle, ovarian and testes.Expression of FAM160B1 was predominantly observed in testes(Fig.2A).Using immunohistochemistry,we detected FAM160B1 in male germ cells including spermatocytes and round spermatids in normal human testes(Fig.2E-G;negative controls Fig.2B-D).
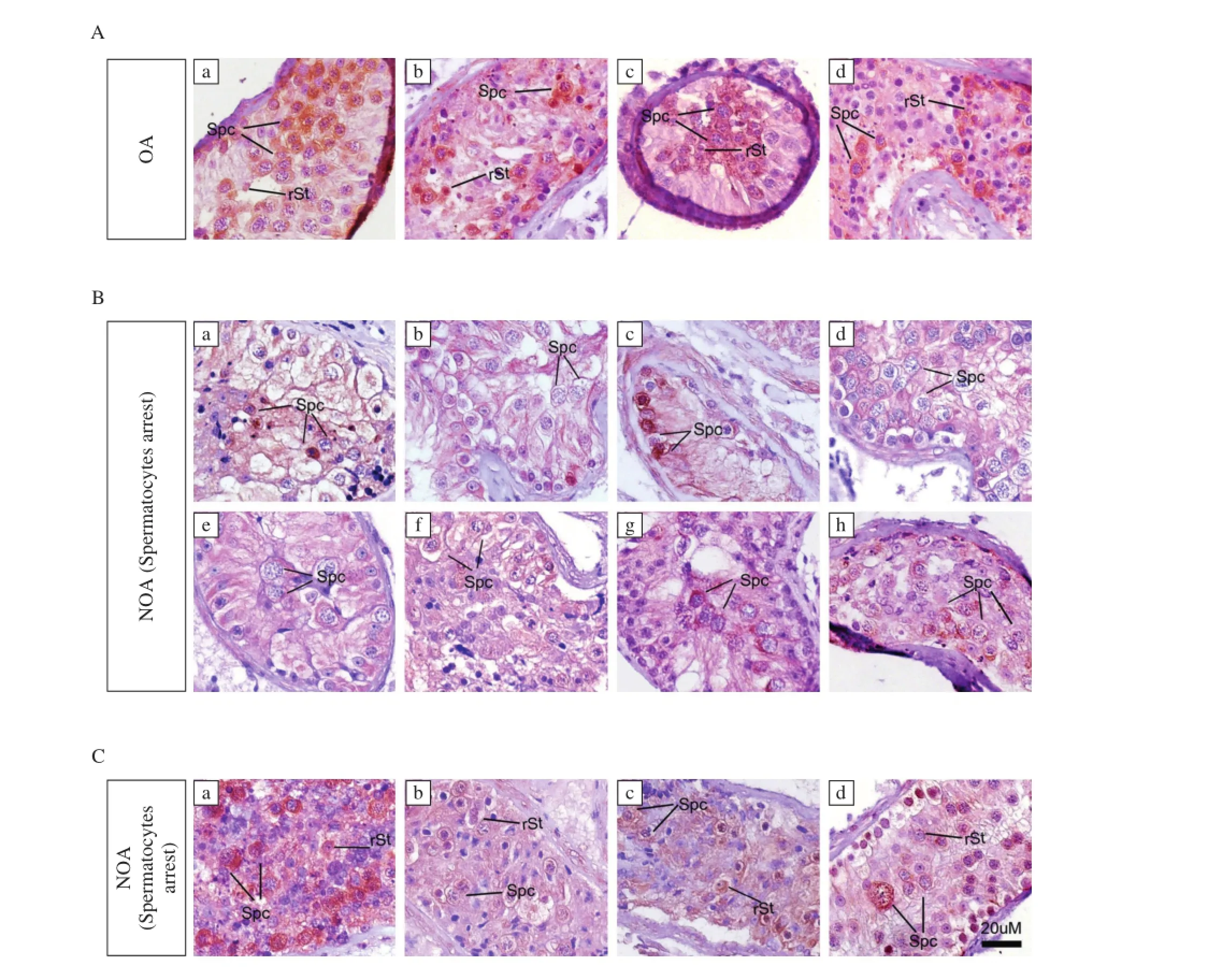
Fig.3Immunohistochemistry of FAM160B1in the OA testes and NOAtestes.A:a-d:4 cases ofthe OA.B:in spermatocyte arrestgroup of NOA testes(withoutspermatid and mature spermatozoa butcontained spermatocyte).a-h:8 cases of the spermatocyte arrest.C:in spermatid arrestgroup of NOA testes(withoutmature spermatozoa but contained round spermatid).a-d:4 cases of the spermatid arrest.Spc:spermatocyte,rSt:round spermatid.
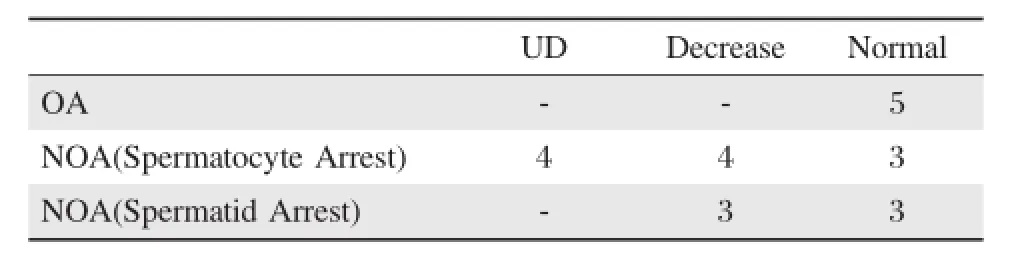
Table1 Immunohistochemical analysis of FAM160B1 in testis section of NOA patients and control.
Expression of FAM160B1 was reduced or undetectable in testes from NOA patients
Then,we examined FAM160B1 expression in patients within NOA and OA testes.Arrestofspermatocytes and spermatids are common phenotypes of NOA. Our NOA samples were sorted to the spermatocyte arrestgroup(withoutspermatid and mature spermatozoa but contained spermatocyte)and spermatid arrest group(without mature spermatozoa but contained round spermatid).For all five patients with OA, FAM160B1 was detected in spermatocytes and round spermatids(Fig.3A).Immunohistochemical analysis of 11 cases of spermatocyte arrest testes(Fig.3B) revealed thatonly three of 11 showed normalpositive FAM160B1 signals in spermatocyte(Fig.3B g,h and j),and the FAM160B1 signal in spermatocyte was undetectable in four cases(Fig.3B b,d,e and k). On the other hand,in spermatocyte and round spermatid(Fig.3C),three in six cases of spermatid arrest testes showed normal positive FAM160B1 signals (Fig.3C a,d and f).In the remaining seven NOA samples,including spermatocytes and round spermatids arrestpatients(Fig.3B c,f and i;Fig.3C b,c and e),FAM160B1 signals were decreased relative to controls.These results suggestthat FAM160B1 expression is associated with NOA defects(Table 1).
Analysis of FAM160B1in mouse germ cells
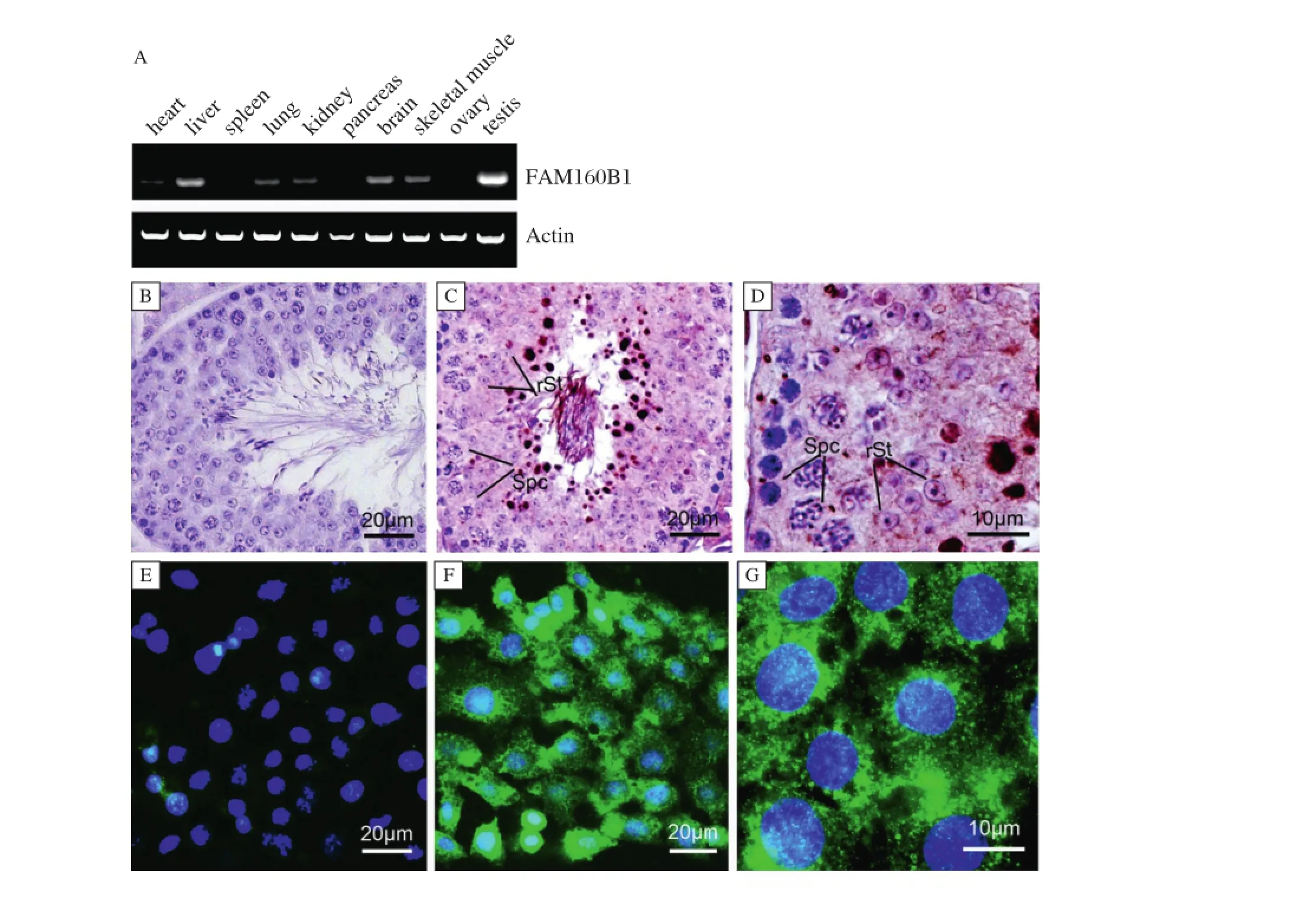
Fig.4 FAM160B1gene expression in Mice.A:The expression of FAM160B1 in multi-organization successively:heart,liver,spleen,lung,kidney pancreas,brain,skeletalmuscle,ovarian and testes.Actin was used as an internalcontrol.B-D:immunohistochemistry of FAM160B1 in the mice testes.B:the negative control.C,D:FAM160B1 showed punctate signals in germ cells including spermatocytes(Spc)and round sperm(rSt),and highly expressed in elongated sperm.E-G:Expression of FAM160B1 in GC2 cells.E:the negative control.F,G:FAM160B1 located in the cytoplasm and partof the nucleus.
In mice,the expression of FAM160B1 mRNA was assessed in multiple tissue typesand found predominantlyin testes as the same within human(Fig.4A).Using immunohistochemistry,FAM160B1 wasdetected in spermatocytes and the laterstage(Fig.4C,D;Supplementary Figure,which is available online;negative controlin Fig.4B).To investigate the biologicalrole of FAM160B1 in germ cells in vitro,murine germ cellline GC2 was used.Immunofluorescence of FAM160B1 located to thecytoplasm and partofthenucleusin cellline GC2(Fig.4F,G;negative control Fig.4E).
Two morpholinos(splice blocking and translation blocking)were used to knockdown expression of FAM160B1 in GC2 cells.Following incubation with 10μM splice blocking morpholinos for 24 hours,the number of GC2 cells decreased(Fig.5A,B and C). Knockdown efficiency of splice blocking reached approximately 90%at the mRNA level(Fig.5D). Similar results were observed after knockdown with translation blocking morpholinos(Fig.5E,F andG).Translation blocking of FAM160B1 also showed high efficiency(Fig.5H).
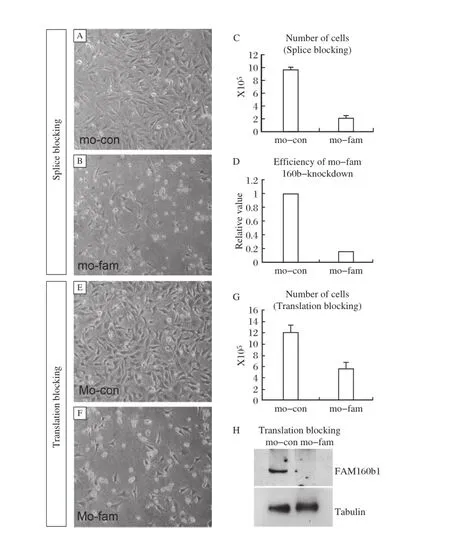
Fig.5 FAM160B1 was knocked down in murine germ cell lines GC2 with morpholinos.A,B:knockdown with splice blocking morpholinos;C:cell counting experiments between control(mo-con)and FAM160B1(mo-fam).**P<0.01.D:Verification of the efficiency of knockdown after morpholinos blocking.Actin was used as an internalcontrol.**P<0.01.E,F:knockdown with translation blocking morpholinos.G: cell counting experiments between control(Mo-con)and FAM160B1(Mo-fam).**P<0.01.H:Verification of the efficiency of knockdown after morpholinos blocking.Tubulin was used as an internal control.

Fig.6TUNEL analysis and electron microscopy after transfection with morpholinos specific to FAM160B1.A,C:Positive signals mo-controland mo-FAM160B1 with TUNEL analysis.B,D:Larger image of A,B.E-L:Electron microscopy under morpholinos inhibiting of the two groups.E,I:the control,F,J:Larger image of E,I.G,K:in perinuclear of cells there were more lipofuscin(black arrow),lipid droplets(red arrow), vacuoles(white arrow),etc.H,L:Larger image of G,K.
TUNEL analysis was performed 24 hours after knockdown with morpholinos.Positive TUNEL signals increased significantly(Fig.6A-D).Using electron microscopy,we found an accumulation of lipofuscin,lipid droplets and vacuoles in the perinuclear space of GC2 cells after inhibiting FAM160B1 (Fig.6E-L).These results suggest that FAM160B1 knockdown induced murine germ cell apoptosis.
Discussion
Only a few cases of NOA are caused by gene mutations,for example,seipin mutation[14].Due to the high incidence of both NOA and SNPs,itis reasonable to hypothesise that the main genetic cause of NOA is the collaborative effectof SNPs.In recentyears,susceptibility loci associated with multiple diseases have been identified in GWAS,including breast cancer[15-16], colorectalcancer[17-18],endometrialcancer[19],follicular lymphoma[20],gastric cancer[21],hepatocellular carcinoma[22-23],Hodgkin's lymphoma[24],lung cancer[25], melanoma[26],nasopharyngealcarcinoma[27],neuroblastoma[28],ovarian cancer[5,29],pancreatic cancer[30-31], prostate cancer[32-37],renal cell carcinoma[38],thyroid cancer[39],urinary bladdercancer[4]and congenitalheart malformations[40].In our previous study,NOA susceptibility lociwere identified based on GWAS data from thousands of patients[8-9].However,the mechanisms of disease remained unclear.Here,we analysed eQTL of genes in the region ofan NOA susceptibility locus and used the data arising to identify the disease mechanism. The success of our approach suggests that GWAS could be a key to efficientcharacterisation ofgene variations involved in NOA.Recently,we discovered several potential protein markers for cancers using GWAS analysis[6-7].
Our aim of this study is to investigate the relationship between a NOA susceptibility locus rs7099208 and genes near this region,like FAM160B1.Based on eQTL data from GTEx,we found thatrs7099208 has a mild effect on expression of FAM160B1. Moreover,we demonstrated that FAM160B1 is essential for germ cell survival and reduce expression in germ cells of some NOA patients.Our study makes a connection between a mild effect of SNP(from GWAS,like rs7099208)and spermatogenic gene (like FAM160B1).It should be noticed that common SNPs(MAF>1%)widely exist in both patients and normal population.Mild effect may not cause a hugechange of single gene expression,but combination of mild effects from several SNPs could contribute a severe consequence and lead to a disease(like NOA).
Previous studies have shown that in NOA patients minor allele frequency(MAF)of rs7099208 is higher than the control population.Mild effect of rs7099208 may not cause a dramatic change of FAM160B1 because variant homozygote of rs7099208 exists in normal population.Although the mechanism of regulation of FAM160B1 is unclear,lack of FAM160B1 leads to germ cell apoptosis.We found the expression of FAM160B1 is changed in NOA patients.We concluded that varianthomozygote of rs7099208 caused mild effect on spermatogenic gene FAM160B1.
Unlike severe effectcaused by gene mutation,common SNPs affectgene expression may notbe contributed by singe SNP variant butcombinations of mild effect of SNPs.The functional combinations of SNPs are still under discovery.Our data provide a new way to investigate spermatogenesis associated SNPs. The fullunderstanding of FAM160B1 in spermatogenesis would be worthy to investigate in further studies.
Acknowledgment
This work was supported by the grants from the 973 program(2011CB944304 and 2015CB943003).
[1]Sharpe RM.Sperm counts and fertility in men:a rocky road ahead.Science&Society Series o n Sex and Science[J].EMBO Reports,2012,13(5):398-403.
[2]Maduro MR,Lamb DJ.Understanding new genetics of male infertility[J].J Urol,2002,168(5):2197-2205.
[3]Ferlin A,Moro E,Garolla A,etal.Human male infertility and Y chromosome deletions:role of the AZF-candidate genes DAZ,RBM and DFFRY[J].Hum Reprod,1999, 14(7):1710-1716.
[4]Wu X,Ye Y,Kiemeney LA,etal.Genetic variation in the prostate stem cellantigen gene PSCA conferssusceptibility to urinary bladder cancer[J].Nat Genet,2009,41(9):991-995.
[5]Goode EL,Chenevix-Trench G,Song H,etal.A genomewide association study identifies susceptibility loci for ovarian cancerat2q31 and 8q24[J].NatGenet,2010,42(10): 874-879.
[6]Liu M,Chen J,Hu L,etal.HORMAD2/CT46.2,a novel cancer/testis gene,is ectopically expressed in lung cancer tissues[J].Mol Hum Reprod,2012,18(12):599-604.
[7]Liu M,Hu Z,Qi L,et al.Scanning of novel cancer/testis proteins by human testis proteomic analysis[J].Proteomics, 2013,13(7):1200-1210.
[8]Hu Z,Xia Y,Guo X,et al.A genome-wide association study in Chinese men identifies three risk loci for nonobstructive azoospermia[J].Nat Genet,2012,44(2): 183-186.
[9]Hu Z,LiZ,Yu J,etal.Association analysis identifies new risk locifornon-obstructive azoospermia in Chinese men[J]. NatCommun,2014,5:3857.
[10]The Genotype-Tissue Expression(GTEx)project[J].Nat Genet.2013,45(6):580-585.
[11]Liu M,Hales BF,Robaire B.Effects of four chemotherapeutic agents,bleomycin,etoposide,cisplatin,and cyclophosphamide,on DNA damage and telomeres in a mouse spermatogonial cell line[J].Biol Reprod.2014,90(4):72.
[12]Liu M,Shi X,Bi Y,etal.SHCBP1L,a conserved protein in mammals,is predominantly expressed in male germ cells and maintains spindle stability during meiosis in testis[J].Mol Hum Reprod.2014,20(6):463-475.
[13]Liu M,QiL,Zeng Y,etal.Transientscrotalhyperthermia induces lipid droplet accumulation and reveals a different ADFP expression pattern between the testes and liver in mice[J].PLoS One.2012,7(10):e45694.
[14]Jiang M,Gao M,Wu C,et al.Lack of testicular seipin causes teratozoospermia syndrome in men[J].Proc Natl Acad Sci U S A.2014,111(19):7054-7059.
[15]Thomas G,Jacobs KB,KraftP,etal.A multistage genome--wide association study in breastcanceridentifiestwo new risk alleles at 1p11.2 and 14q24.1(RAD51L1)[J].Nat Genet, 2009,41(5):579-584.
[16]Turnbull C,Ahmed S,Morrison J,et al.Genome-wide association study identifies five new breast cancer susceptibility loci[J].Nat Genet.2010,42(6):504-507.
[17]Tomlinson IP,Webb E,Carvajal-Carmona L,etal.A genome-wide association study identifies colorectalcancersusceptibility locion chromosomes 10p14 and 8q23.3[J].Nat Genet,2008,40(5):623-630.
[18]Houlston RS,Cheadle J,Dobbins SE,etal.Meta-analysisof three genome-wide association studiesidentifies susceptibility lociforcolorectalcancer at1q41,3q26.2,12q13.13 and 20q13.33[J].Nat Genet,2010,42(11):973-977.
[19]Spurdle AB,Thompson DJ,Ahmed S,et al.Genomewide association study identifies a common variant associated with risk of endometrial cancer[J].Nat Genet, 2011,43(5):451-454.
[20]Conde L,Halperin E,Akers NK,etal.Genome-wide association study offollicularlymphoma identifies a risk locus at 6p21.32[J].Nat Genet,2010,42(8):661-664.
[21]Shi Y,Hu Z,Wu C,et al.A genome-wide association study identifies new susceptibility locifornon-cardia gastric cancer at3q13.31 and 5p13.1[J].Nat Genet,2011,43(12): 1215-1218.
[22]Zhang H,Zhai Y,Hu Z,et al.Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers[J].Nat Genet,2010,42(9):755-758.
[23]Kumar V,Kato N,Urabe Y,etal.Genome-wide association study identifies a susceptibility locus forHCV-induced hepatocellular carcinoma[J].Nat Genet,2011,43(5):455-458.
[24]Enciso-Mora V,Broderick P,Ma Y,etal.Agenome-wide association study of Hodgkin's lymphoma identifies new susceptibility loci at 2p16.1(REL),8q24.21 and 10p14 (GATA3)[J].Nat Genet,2010,42(12):1126-1130.
[25]Hu Z,Wu C,Shi Y,et al.A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese[J].Nat Genet, 2011,43(8):792-796.
[26]Bishop DT,Demenais F,Iles MM,et al.Genome-wide association study identifies three loci associated with melanoma risk[J].Nat Genet,2009,41(8):920-925.
[27]Bei JX,Li Y,Jia WH,et al.A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci[J].Nat Genet,2010,42(7):599-603.
[28]Capasso M,Devoto M,Hou C,et al.Common variations in BARD1 influ en ce su scep tibility to high-risk neuroblastoma[J].Nat Genet,2009,41(6):718-723.
[29]Bolton KL,Tyrer J,Song H,et al.Common variants at 19p13 are associated with susceptibility to ovarian cancer[J].Nat Genet,2010,42(10):880-884.
[30]Petersen GM,Amundadottir L,Fuchs CS,et al.A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1,1q32.1 and 5p15.33[J].Nat Genet,2010,42(3):224-228.
[31]Amundadottir L,Kraft P,Stolzenberg-Solomon RZ,et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer[J].Nat Genet,2009,41(9):986-990.
[32]Gudmundsson J,Sulem P,Gudbjartsson DF,etal.Genomewide association and replication studies identify four variants associated with prostate cancer susceptibility[J].Nat Genet,2009,41(10):1122-1126.
[33]Haiman CA,Chen GK,Blot WJ,etal.Genome-wide association study ofprostate cancerin men of African ancestry identifies a susceptibility locus at 17q21[J].Nat Genet, 2011,43(6):570-573.
[34]Eeles RA,Kote-JaraiZ,Al Olama AA,etal.Identification of seven new prostate cancer susceptibility locithrough a g e n o m e-wid e asso c iatio n stu d y[J].Na t Ge n et, 2009,41(10):1116-1121.
[35]Thomas G,Jacobs KB,Yeager M,et al.Multiple loci identified in a genome-wide association study of prostate cancer[J].Nat Genet,2008,40(3):310-315.
[36]Yeager M,Chatterjee N,Ciampa J,etal.Identification of a new prostate cancer susceptibility locus on chromosome 8q24[J].Nat Genet,2009,41(10):1055-1057.
[37]Eeles RA,Kote-JaraiZ,Giles GG,etal.Multiple newly identified lociassociated with prostate cancer susceptibility[J]. NatGenet,2008,40(3):316-321.
[38]Purdue MP,Johansson M,Zelenika D,et al.Genomewide association study of renal cell carcinoma identifies two susceptibility locion 2p21 and 11q13.3[J].Nat Genet, 2011,43(1):60-65.
[39]Gudmundsson J,Sulem P,Gudbjartsson DF,etal.Common variantson 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations[J].NatGenet,2009,41(4):460-464.
[40]Hu Z,Shi Y,Mo X,et al.A genome-wide association study identifies two risk loci for congenital heart malformations in Han Chinese populations[J].Nat Genet, 2013,45(7):818-821.
✉Corresponding authors:Dr.Jiahao Sha,State Key Laboratory of Reproductive Medicine,Department of Histology and Embryology, Nanjing Medical University,Nanjing,Jiangsu 210029,China.Tel/fax: +86-25-86862908/+86-25-86862908,E-mail:shajh@njmu.edu.cn.
Received 28 February 2015,Revised 08 March 2015,Accepted 06 May 2015,Epub 08 August 2015
R698+.2,Documentcode:A
The authors reported no conflict of interests.
©2015 by the Journal of Biomedical Research.All rights reserved.
10.7555/JBR.29.20150035
 THE JOURNAL OF BIOMEDICAL RESEARCH2015年6期
THE JOURNAL OF BIOMEDICAL RESEARCH2015年6期
- THE JOURNAL OF BIOMEDICAL RESEARCH的其它文章
- The evolving concept of physiologicalischemia training vs. ischemia preconditioning
- PCSK9 and triglyceride-rich lipoprotein metabolism
- Generation of monoclonalantibodies against n-3 fatty acid desaturase
- Dominant-negative inhibition of glucose-dependent insulinotropic polypeptide impairs function ofβcells in transgenic pigs
- Waldenstrom′s macroglobulinemia associated with Hodgkin′s lymphoma: a case report
- Neuroanatomy and clinicalanalysis of the cervicalsympathetic trunk and longus colli
