Interaction Study between DNA and Histone Proteins on Single-molecule Level using Atomic Force Microscopy
Yu-ying Liu,Peng-ye Wng,Shuo-xing Dou,Hong-feng Lv
a.College of Science,China Agricultural University,Beijing 100083,China
b.Laboratory of Soft Matter Physics,Beijing National Laboratory for Condensed Matter Physics, Institute of Physics,Chinese Academy of Sciences,Beijing 100190,China
Interaction Study between DNA and Histone Proteins on Single-molecule Level using Atomic Force Microscopy
Yu-ying Liua∗,Peng-ye Wangb,Shuo-xing Doub,Hong-feng Lva
a.College of Science,China Agricultural University,Beijing 100083,China
b.Laboratory of Soft Matter Physics,Beijing National Laboratory for Condensed Matter Physics, Institute of Physics,Chinese Academy of Sciences,Beijing 100190,China
DNA and histone protein are important in the formation of nucleosomal arrays,which are the fi rst packaging level of DNA into a more compact chromatin structure.To characterize the interactions of DNA and histone proteins,we reconstitute nucleosomes using lambda DNA and whole histone proteins by dialysis and perform direct atomic force microscopy(AFM) imaging.Compared with non-speci fi c DNA and histone binding,nucleosomes are formed within the assembled“beads-on-a-string”nucleosomal array by dialysis.These observations facilitate the establishment of the molecular mechanisms of nucleosome and demonstrate the capability of AFM for protein-DNA interaction analysis.
DNA,Histone,Atomic force microscopy,Single molecule,Dialysis
I.INTRODUCTION
DNA is packaged into the chromatin in the cell nucleus.The nucleosome,which is the repeating unit of chromatin,consists of an octamer of core histones(two each of H2A,H2B,H3,and H4)around which approximately two superhelical turns of DNA are wrapped[1]. A length of 146 double-stranded DNA base pairs(bp) wrap around this structure 1.65 times,which forms a relatively larger cylinder that is 11 nm in diameter and 5-6 nm in height.The formation of nucleosomes is the fi rst stage of DNA packing into chromatin,followed by the assembly of the“beads-on-a-string”nucleosomal array into compact chromatin fi bers[2].H1 histone is a key player in the formation of the 30 nm thick fi bers [3].In addition,adjacent core particles may interact by stacking together to form the higher-ordered chromatin structure.Assembled chromatin fi bers,approximately 30 nm in diameter,are further compacted in the cell into euchromatin and ultimately heterochromatin.The molecular mechanisms behind the formation of higherordered chromatin structures remain unclear.
DNA and histone interactions are important in the formation of nucleosomal arrays.The genetic information also has to remain accessible for DNA binding factors involved in processes such as replication,transcription,repair,and recombination.Thus,the interaction of histones and DNA dynamically mediates these two apparent functions.
Atomic force microscopy(AFM)has been applied to study the interactions between nucleosomes within the assembled beads-on-a-string nucleosomal array in the absence of H1 histone[4].The interaction between nucleosomes has been recently studied using fluorescence resonance energy transfer(FRET)microscopy[5]. Transiently formed dinucleosomes with 1 s lifetime are detected using the FRET technique.AFM has also been used to visualize the isolated mono-nucleosomes [6],chromatin f i bers,and isolated nucleosomes[7].The molecular dynamics of poly-nucleosomal arrays in solution has been analyzed using fast-scanning AFM at single-molecule level[8].DNA and protein molecules are stretched by spinning method for detection by fluorescence and AFM[9,10].The conformation of DNA molecules,ranging from hundreds of bp to thousands of bp can be observed using AFM[11].With morphological analysis[12],the length and conformation changes of DNA can be utilized to study their interactions with small molecules[13,14].
Genomic DNA is a very long molecule,but the radius of gyration of the coil must fit into a very small space inside a cell or virus particle.For example,the fully extended 1.6×105bp of T4 phage DNA span 54µm. However,the T4 DNA molecule can be fitted into a virus capsid,which is approximately 100 nm in diameter,which is a 540-fold linear compression[15].Chromatin proteins and DNA are partners in controlling the genetic material within cells[16].The interaction of DNA and histone is the f i rst packaging level of DNA into a more compact chromatin structure.The interaction of DNA and histone has been studied using molecular combing method[17]and fluorescence assays[18].
In this study,we use AFM to study the interactions between DNA and histone proteins on single-moleculelevel.AFM results show the non-specif i c binding of DNA and histone.Using dialysis,the nucleosomes are formed within the assembled beads-on-a-string nucleosomal array.
II.EXPERIMENTS
Histone,Tris base,EDTA,PMSF,and NaCl were purchased from Sigma-Aldrich(USA).DNA was obtained from the Sino-America Biotechnology Company (Shanghai,China).Solutions were obtained using 18.2 MΩ deionized water,which was purif i ed using the Milli-Q water purif i cation system(Millipore Corporation,France).
For non-specif i c DNA and histone binding,histone was diluted with 18.2 MΩ deionized water.The molar ratio of DNA to histone was from 100:1 to 1:1.DNA (2 kb,4 ng/µL)and histone were incubated together in Tris-HCl buffer(10 mmol/L,pH=7.5)at total volume of 40µL for 1 h at 37◦C.
A.Nucleosomes reconstruction using dialysis
Caps from 200µL PCR tubes were cut to obtain buttons.The cut length was slightly shorter than the cap extrusions,that is,1-1.5 mm length was cut from the body of the tube.The dialysis tubing was cut into 1 cm×1 cm to 1.5 cm×1.5 cm squares for button sealing.
Then,one or two 5-cm long tubing(dialysis bag)were obtained for each dialysis.All tubings were placed into a clean cup,which was then fi lled with400-500 mL 2%NaHCO3and 1 mmol/L EDTA,and then boiled for 10 min.The solutions were then discarded,and the cups were rinsed with distilled water three times,and then fi lled with400-500 mL 1 mmol/L EDTA.The cups were then boiled again for 10 min,and then covered tightly with clean foil(to avoid air exchange)while the water was still hot.The covered cups were then stored at 4◦C until use.This procedure was based on the method used in Ref.[19].
B.Reaction set-up
The lambda DNA and histone in the start buffer (2 mol/L NaCl,5 mmol/L Tris-HCl at pH=7.5, 0.5 mmol/L EDTA,and 0.5 mmol/L PMSF)in sterile clean tube were incubated.The mass of DNA was 350 ng to 3µg in 50µL reaction buffer.The mass ratio of lambda DNA to histone was 1:0.7.The mixture was then transferred into the caps,with each cap covered with a piece of square tubing using forceps,ensuring that no bubbles were formed between the solution and tubing surface.
The caps,which were cut to make a button,were sealed.The buttons were then placed into 5 cm tubing, and~2 mL start buffer was injected into the 5 cm tubing,and then the tubing was sealed using clamps.The sealed tubing was then placed into the clean sterile cup, which was then filled with~200 mL pre-chilled dialysis buffer and 1 mL PMSF(100 mmol/L).The solution was mixed thoroughly,and the cup was then covered tightly with the foil.
The samples were stored at 4◦C for 4-6 h,and then fresh dialysis buffer with PMSF was replaced.The cup was stored in a cold room or at 4◦C overnight.
C.AFM imaging
For conventional AFM imaging using digital instruments(multi-mode AFM),the DNA nucleosome solution was diluted with binding buffer(5 mmol/L HEPES, 10 mmol/L NaCl,and 1 mmol/L MgCl2)to an appropriate concentration.To enhance the binding efficiency of DNA-histone molecules on mica surface[20],we pretreated the mica surface with spermidine.The pretreatment procedure was as follows:20µL 10 mmol/L spermidine(SpdCl3)was added on newly cleaved mica surface,which was then incubated for 5 min,washed thrice with distilled water,and blown dry in a gentle stream of nitrogen gas[20].
The sample was then dropped onto the spermidinepretreated mica surface.After 5 min,the mica surface was washed thrice with 100µL Milli-Q filtered water and blown dry in a gentle stream of nitrogen gas.
The imaging was performed in air using a multimode AFM with nanoscopeIIIa controller(Digital Instruments,Santa Barbara,CA,USA)in the tappingmode.Silicon probe RTESP14 from Veeco(USA)was employed,with a resonance frequency of 315 kHz,an E scanner was used.The scan rate was 1 Hz per line,and the scan size was 2-4µm.DNA tracing and measurements were performed semi-automatically using Image J software.
III.RESULTS AND DISCUSSION
To obtain a deeper insight on the interactions of DNA and histone,we used AFM to study this interaction by employing two methods.We f i rst observed the nonspecif i c binding of DNA and histone using AFM,and then we used the dialysis method to reconstruct the nucleosomes.The DNA-nucleosome interactions were observed using AFM.
A.effect of different concentrations of histones on the non-specif i c binding of DNA
In our experiment,non-specif i c DNA and histone binding occurred when the molar ratio of DNA to histone ranged from 100:1 to 1:1.DNA(2 kb,4 ng/µL f i nal concentration in reaction solution)and histone were incubated in Tris-HCl buffer(pH=7.5)at total volume of40µL for 1 h at 37◦C.Approximately 2µL of the reaction mixture was withdrawn and incubated in 18µL binding buffer(5 mmol/L HEPES,10 mmol/L NaCl, and 1 mmol/L MgCl2)at room temperature for 5 min, and then loaded onto spermidine-pretreated mica.The DNA concentration was 4 ng/µL in the reaction solution and 0.4 ng/µL during loading onto mica surface, which is shown in Fig.1(a)-(d).
Compared with our previous studies,it was demonstrated the binding of DNA and histone at much lower molar concentration of histone in the present study. The molar concentration of DNA was much higher than that of histone.In Fig.1(a),the molar ratio of DNA to histone was 100:1;in this case,almost no histone binding to DNA molecules was observed,and single DNA molecules were distributed on the mica surface with their natural configurations(Fig.1(a)).With the increase in histone,more histones bound to DNA molecules.The binding positions on the DNA molecules were usually at random.The histones seemed to bind on some positions of DNA at f i rst,whereas in other regions,almost no histone binding was found.Most of the DNA regions also showed natural stretched configuration(Fig.1(b)).However,when the molar ratios of DNA to histone were 3:1 and 1:1,histone condensed the DNA.More histones were found to bind to single DNA molecules and induced DNA aggregation(Fig.1 (c)and(d)).The DNA-histone complexes could not be stretched because they were tightly entangled with each other.
In our previous study[21],the DNA-histone complexes were stretched and aligned using molecular combing method.At higher molar concentration ratio of histone to DNA(100:1),the DNA-histone complexes were tightly condensed into numerous spheres,as observed using fluorescence microscopy,and could not be stretched using molecular combing method.From the results of fluorescence microscopy and AFM,we deduced that the efficiency of non-specif i c binding DNA and histone was very high.In the case of fewer histones, histones bound to some positions of DNA.With the increase in histone,histones bound to other positions on the DNA.The binding of histone and DNA was sensitive to their molar concentration ratio.Histones contain a few lysine(K)residues at the N terminus.Under normal cellular conditions,the R group of lysine is positively charged,which can interact with the negatively charged phosphates in DNA.In high quantity of histone,DNA aggregation was easily induced.
In a reaction,in which purif i ed histones and DNA are simply mixed together under physiological solution conditions,at sufficiently high concentration of DNA and histone octamer so that nucleosome formation is favored at equilibrium,some nucleosomes will form as well as numerous non-nucleosomal histone-DNA complexes and aggregates.Therefore,in our second method,we used dialysis to reconstitute nucleosomes using lambda DNA and whole histone proteins.
B.Dialysis methods to reconstitute nucleosomes using lambda DNA and whole histone proteins
At high salt concentrations,the affinity of histone octamer for DNA is signif i cantly reduced.We used gradual dialysis,which was achieved using a double-dialysis technique that relies on a micro-dialysis button inside a dialysis bag.This method ensures a reversible nucleosome assembly process.
In 2 mol/L NaCl,in which histones and DNA have negligible affinity for each other,free equilibration of the histones is achieved between different DNAs. Subsequent decreases in concentration of NaCl reversibly allow nucleosome reconstitution,which then ultimately traps and freezes-in stable nucleosomes at a sub-physiological salt concentration.As concentration of NaCl drops from 2 mol/L to 1.5-1 mol/L,tetramers develop an increasing affinity for DNA,whereas heterodimers develop signif i cant affinity only when concentration of NaCl reaches 1-0.75 mol/L[19].
We fabricated a micro-dialysis button inside a dialysis bag and performed the standard dialysis procedure according to Ref.[19].In our dialysis reaction,we used lambda DNA and purif i ed histones.In the present study,the mass ratio of lambda DNA to histone was 1:0.7(the molar ratio of histone to lambda DNA was approximately 300).The results showed that the histone binding to lambda DNA molecules was very different from their non-specif i c binding(Fig.2).DNA and histone octamer(nucleosomes)were formed using dialysis method.Numerous single histones octamer were bound on single lambda DNA molecule(Fig.2).We also measured the size of these nucleosomes,which were between 12 and 21 nm.DNA and histones formed the“beadson-a-string”nucleosomal array using dialysis.Compared with other studies[22,23],nucleosomes were often formed with short DNA fragments,which typically ranged from 146 bp to 207 bp,with a high-affinity nucleosome positioning sequence[23].In the present study, although we used the longer lambda DNA without highaffinity nucleosome positioning sequence,nucleosomes were also formed.We directly observed using AFM that nucleosome could be formed on lambda DNA.Nucleosomes were well resolved along with the linear lambda DNA.When the stretched force on DNA molecules was small,the strands of DNA molecule tended to bend and aggregated on the mica surface.In the aggregation regions(Fig.2),histone and DNA showed much larger spots(black arrow)than single nucleosomes(red arrow).The aggregations suggested that attractive interactions between the nucleosomes existed,thereby leading to array compaction.

FIG.1 Non-specif i c binding DNA and histone.The molar ratio of DNA to histone ranged from(a)100:1,(b)10:1,(c)3:1, to(d)1:1.The scale bar is 400 nm in(a)-(d).(e)Montage of AFM images of typical DNA-histone complex of different modes are shown in the black-line box.Scale bar is 200 nm.
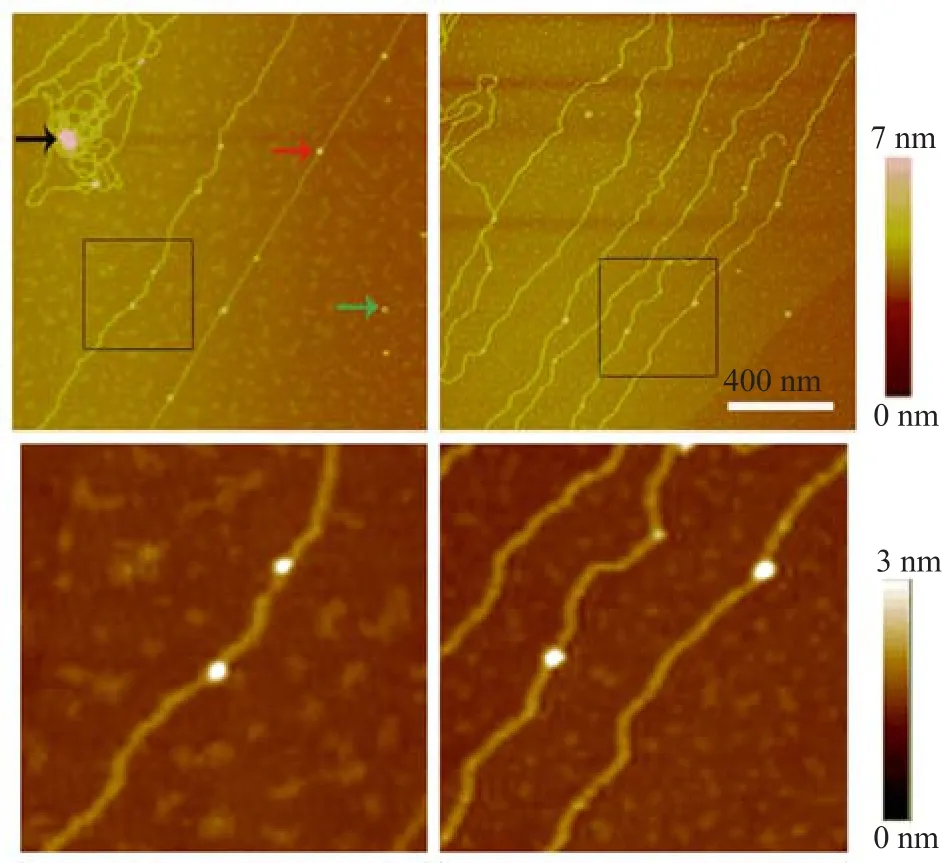
FIG.2 Nucleosomal array reconstituted from lambda DNA and core histones using salt dialysis on the mica surface in air.The sizes of nucleosomes(red arrow)in these images are from 12 nm to 21 nm.The single histone on the mica surface is indicated by the green arrow.Scale bar is 400 nm. Enlarged images of the region enclosed in black rectangles are shown in the lower panel.
After we optimized the condition for DNA-histone binding on the mica surface,nucleosomes became well resolved along with the stretched DNA.Nucleosomal arrays were reconstituted from lambda DNA and core histones by salt dialysis and observed using AFM on mica in air(Fig.3).In addition,nucleosomes with relatively large sizes were also observed in the same sample.Some nucleosomes are approximately 28 nm to 47 nm(an example is indicated by an arrow in Fig.3). Given that H1 fraction of histone was present in the sample,higher-order chromatin was probably formed. AFM analysis of the sizes of nucleosomes observed in our experiments is shown in Fig.4.
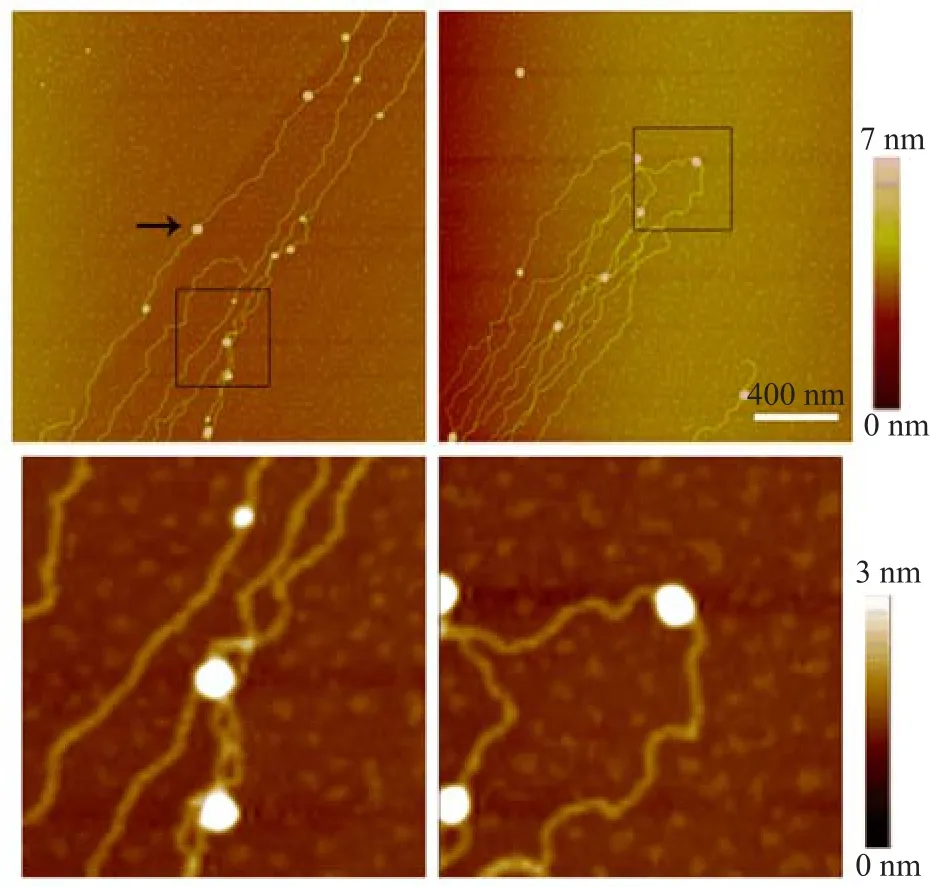
FIG.3 AFM images of nucleosome reconstitution using dialysis method.The size of some nucleosomes in this image is~28 nm to 47 nm.Scale bar is 400 nm.Enlarged images of the region enclosed in black rectangles are shown in the lower panel.
Compared with dialysis,we also performed an experiment in which purif i ed histones and DNA were simply mixed together under physiological solution conditions.However,the molar ratio of histone to DNA was 200.Histones condensed DNAs into short linear f i bers, which self-assemble into numerous mesh networks on the mica surface.Many beads were also found on the DNA f i bers(Fig.5).DNA and histone combined with each other very easily because of their opposite charge polarities.After many repetitions of the experiment, the results showed that lambda DNA-histone complexes self-assembled into network structures on mica surface.The morphologies of DNA-histone complexes were related to various factors such as the force of nitrogen f l ow on the surface.
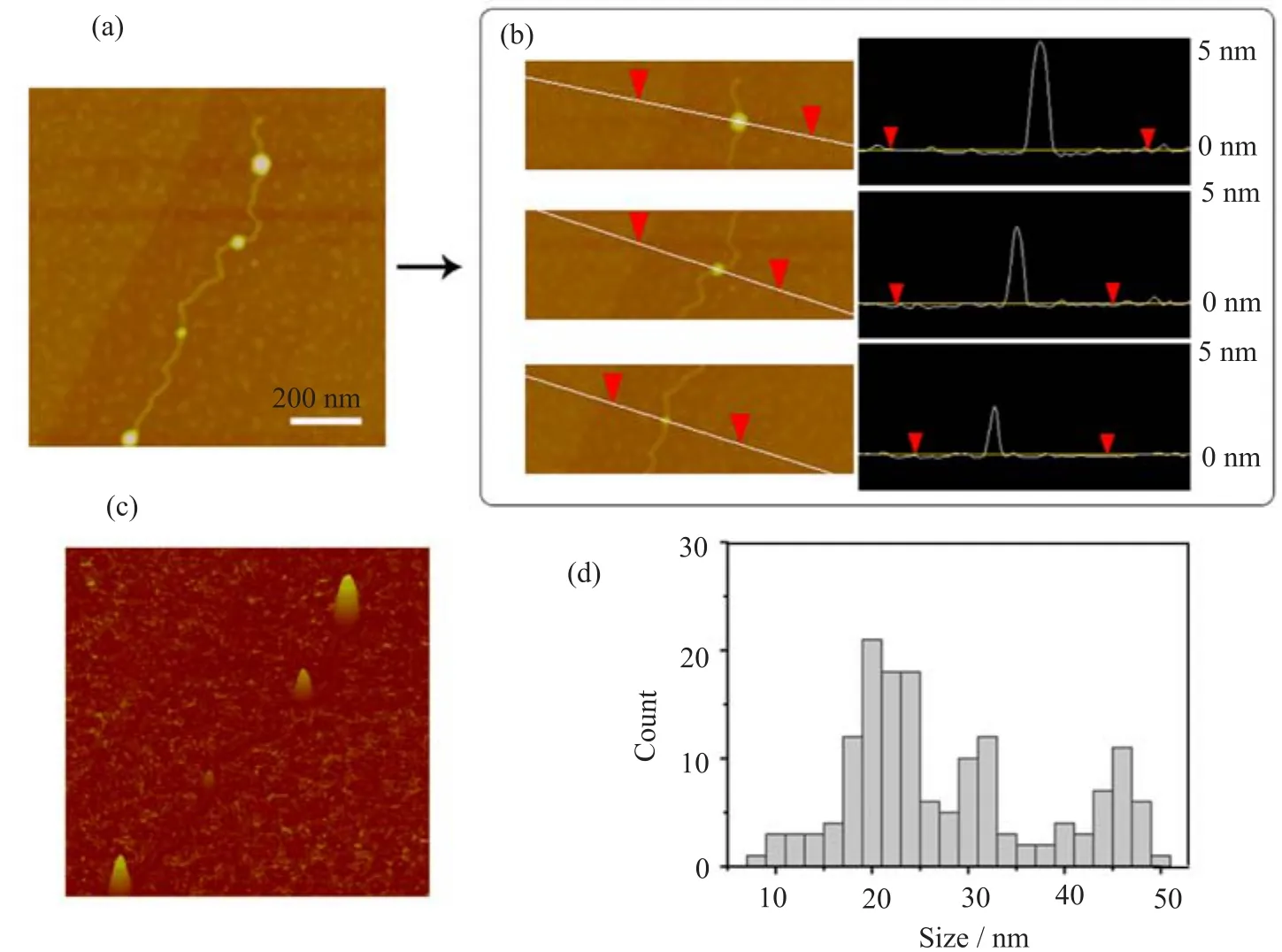
FIG.4 AFM analysis of nucleosomes reconstituted by dialysis method.(a)Nucleosomes with typical sizes in our study were formed randomly on lambda DNA due to H1 fraction in histone samples.(b)Line profile across the nucleosomes showing the vertical sizes measured by AFM.(c)AFM image of(a)with three dimensional views.(d)Histogram representing distribution of horizontal sizes of nucleosomes(n=155).
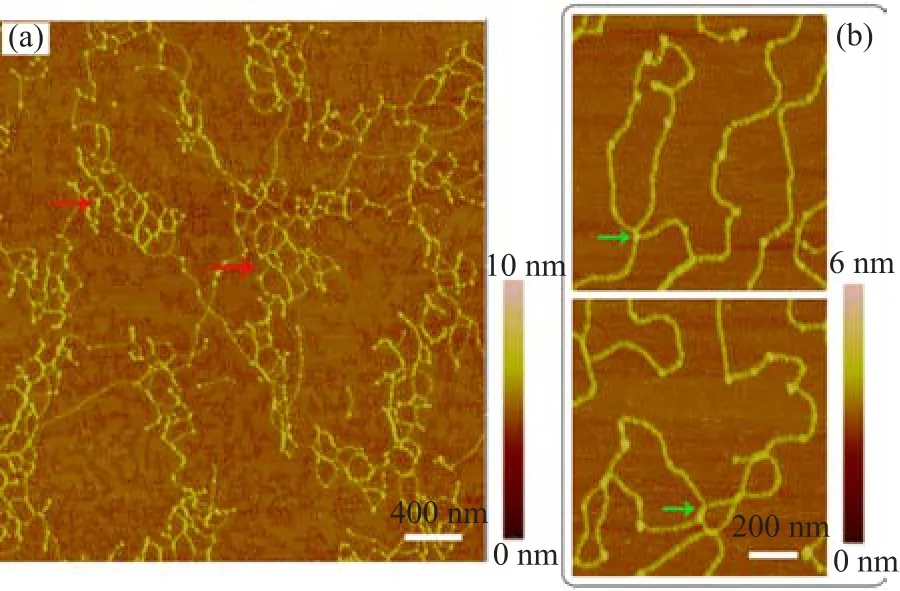
FIG.5 AFM images of lambda DNA-histone complexes. The molar ratio of histone to DNA is 200.(a)Unique mesh networks,which were formed by lambda DNA and histone, are shown(red arrow).(b)The enlarged images show junctions(green arrow)of DNA fragment linked by histone complexes in the black-line box.
IV.CONCLUSION
To obtain more detailed information on the interaction of DNA and histone proteins,we used lambda DNA and whole histone proteins to reconstitute nucleosomes using dialysis.The nucleosomes were observed using AFM.We also compared the non-specif i c binding of DNA and histone to the reconstituted nucleosomes. AFM can detect single biomacromolecules with a high signal-to-noise ratio on atomically f l at biocompatible support surfaces,such as mica.AFM can also provide valuable information on the interaction of certain proteins with DNA and signif i cant changes in the topological form of DNA.Formation of nucleosomes in the lambda DNA was easily detected.The following conclusions can be drawn:(i)Compared with other studies, in which short DNA molecules with high-affinity nucleosome positioning sequence were used,we used long lambda DNA molecules without high-affinity nucleosome positioning sequences to form nucleosomes.Nucleosomes were formed within the assembled beads-ona-string nucleosomal array using dialysis.(ii)The nonspecif i c binding of DNA and histone was also studied using AFM.The binding of histone and DNA was sensitive to their molar concentration ratio.DNA-histone complexes aggregated together and self-assembled into network structures on mica surface at high molar concentration of histone.
V.ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China(No.11274374),the NationalBasic Research Program of China(No.2009CB930704), and the Basic Scientif i c Research Foundation of China Agricultural University(No.2012QJ026).
[1]K.E.VanHolde,Chromatin,Berlin:Springer-Verlag KG,219(1988).
[2]J.C.Hansen,Annu.Rev.Biophys.Biomol.Struct.31, 361(2002).
[3]F.Thoma,T.Koller,and A.Klug,J.Chem.Biol.83, 403(1979).
[4]F.Montel,H.Menoni,M.Castelnovo,J.Bednar,S. Dimitrov,D.Angelov,and C.Faivre-Moskalenko,Biophys.J.97,544(2009).
[5]J.Y.Lee,S.J.Wei,and T.H.Lee,J.Biol.Chem.286, 11099(2011).
[6]G.Binnig,C.F.Quate,and C.Gerber,Phys.Rev.Lett. 56,930(1986).
[7]J.Zlatanova and S.H.Leuba,J.Mol.Biol.331,1 (2003).
[8]Y.Suzuki,Y.Higuchi,K.Hizume,M.Yokokawa,S. Yoshimura,K.Yoshikawa,and K.Takeyasu,Ultramicroscopy 110,682(2010).
[9]H.Yokota,J.Sunwoo,M.Sarikaya,G.Engh,and R. Aebersold,Anal.Chem.71,4418(1999).
[10]M.L.Bennink,D.N.Nikova,K.O.Werf,and J.Greve, Analytica.Chimica.Acta 479,3(2003).
[11]L.Sui,K.Zhao,M.N.Ni,J.Y.Guo,F.Q.Kong,C. M.Hui,X.Q.Lu,and P.Zhou,Chin.Phys.Lett.22, 1010(2005).
[12]H.B.Wang,X.F.Zhou,H.J.An,Y.C.Guo,J.L. Sun,Y.Zhang,and J.Hu,Chin.Phys.Lett.24,644 (2007).
[13]X.M.Hou,X.H.Zhang,K.J.Wei,J.Chao,S.X.Dou, W.C.Wang,and P.Y.Wang,Nucleic.Acids Res.37, 1400(2009).
[14]Z.G.Liu,S.N.Tan,Y.G.Zu,Y.J.Fu,R.H.Meng, and Z.M.Xing,Micron.41,833(2010).
[15]V.A.Bloomf i eld,Biopolymers 44,269(1997).
[16]G.Felsenfeld and M.Groudine,Nature 421,448(2003).
[17]Y.Y.Liu,S.X.Dou,P.Y.Wang,P.Xie,and W.C. Wang,Acta.Phys.Sin.54,622(2005).
[18]Y.Y.Liu,P.Y.Wang,S.X.Dou,W.W.Zhang,X.J. Wang,and H.Y.Sang,Chin.Sci.Bull.56,1080(2011).
[19]A.Thastrom,P.T.Lowary,and J.Widom,Methods 33,33(2004).
[20]G.R.Schnitzler,C.L.Cheung,J.H.Hafner,A.J. Saurin,R.E.Kingston,and C.M.Lieber,Mol.Cel. Biol.21,8504(2001).
[21]Y.Y.Liu,P.Y.Wang,S.X.Dou,P.Xie,W.C.Wang, and H.W.Yin,Chin.Sci.Bull.50,731(2005).
[22]J.Zlatanova and S.H.Leuba,J.Muscle.Res.Cell. Motil.23,377(2002).
[23]N.A.Filenko,D.B.Palets,and Y.L.Lyubchenko,J. Amino.Acids.650840(2012).
ceived on July 30,2013;Accepted on December 26,2013)
∗Author to whom correspondence should be addressed.E-mail:liuyuying@cau.edu.cn,Tel.:+86-10-62736711
 CHINESE JOURNAL OF CHEMICAL PHYSICS2014年1期
CHINESE JOURNAL OF CHEMICAL PHYSICS2014年1期
- CHINESE JOURNAL OF CHEMICAL PHYSICS的其它文章
- Experimental and Theoretical Investigation on Excited State Intramolecular Proton Transfer Coupled Charge Transfer Reaction of Baicalein
- Theoretical Study of Reagent Rotational Excitation effect on the Stereodynamics of H+LiF→HF+Li Reaction
- Infrared Spectroscopy of COIsolated in Solid Nitrogen Matrix
- effects of Rotational Isomerism and Bond Length Alternation on Optical Spectra of FTC Chromophore in Solution
- High Pressure Structural Instability and Thermal Properties of Rutile TiO2from First-principles
- Kinetic Implication from Temperature effect on Hydrogen Evolution Reaction at Ag Electrode
