Molecular Dynamics Simulations of Ions Diffusion in Carbon Nanotubes Embedded in Cell Membrane
Qing Song Tu,Michelle Lee,Samuel Zhangand Shaofan Li
1 Introduction
A key function of biological membrane is to provide mechanisms for mediating exchange of ions,nutrients,metabolites,peptides and proteins across it[Sotomayor,M.and Schulten,K.(2007);Nelson,P.H.(2003);Hille,B.(2001)].Ion channels are found in cell membranes and are responsible for controlling ion transport and exchange through cells,and they also play a crucial role in electrical properties of neurons and muscle[Tytgat,J.and Hess,P.(1992);Noskov,S.Y.and Roux,B.(2006);Cannon,S.C.,Brown,R.H.,Jr and Corey,D.P.(1991)].Defects in ion channel function are associated with a number of diseases conditions including myasthenia,immune system diseases,central nervous system diseases and even cancer[Delpire,E.,Lu,J.,England,R.,Dull,C.and Thorne,T.(1999);Hübner,C.A.and Jentsch,T.J.(2002)].
Gating and selectivity are the two most important characteristics of biological ion channels.They are often highly selective for a particular ionic species,leading to a classification into sodium(Na+),potassium(K+),calcium(Ca2+),chloride(Cl−),and unspecific cation channels.In recent years,considerable researches have been carried out to investigate questions of how different ion transports across the cell membrane,whether they are active or passive,are achieved and,in particular,how each channel being selected under different physiology condition[Katz,A.M.,Messineo,F.C.,and Herbette,L.(1982);Stein,W.(1986);Yellen,G.A.R.Y.(1984)].
Since the 2resolution X-ray crystal structure of KcsA was reported,numerous MD(Molecular Dynamics)simulations have been performed and reported in the literature that studied the gating mechanism and cation selectivity[Liu,H.,Jameson,C.J.and Murad,S.(2008);Forrest,L.R.and Sansom,M.S.(2000);Khalili-Araghi,F.,Gumbart,J.,Wen,P.C.,Sotomayor,M.,Tajkhorshid,E.and Schulten,K.(2009);Lindahl,E.and Sansom,M.S.(2008)].Using this technique,we can directly investigate the microscopic details of permeation through ion channels of known structure[Berneche,S.and Roux,B.(2000);Feller,S.E.(2000)].Previous MD studies have been used to study the effective short-time local diffusion coefficients of K+and Cl−in a series of ion channel models[Marañón Di Leo,J.and Marañón,J.(2005);Sansom,M.S.,Shrivastava,I.H.,Ranatunga,K.M.and Smith,G.R.(2000)].Liu et al.studied the permeation of ions and water in a membrane consisting of single wall neutral CNTs revealed changes in the hydration shell of the ions upon confinement in tubes of up to 0.90 nm effective internal diameter[Liu,H.,Murad,S.and Jameson,C.J.(2006)].Peter and Hummer studied computationally the Na+ion transport through narrow hydrophobic pores in model membranes formed of hexagonally packed armchair CNT(10,10)[Peter,C.and Hummer,G.(2005)].
The present work is aimed at investigating diffusion and selectivity of carbon nanotube embedded in the cell membrane to Na+and K+ions,in order to find the minimum diameter for different ions to pass through,and compare with the socalled biological selectivity filters.The present work is a systematic study of ion and water transport through CNT embedded in cell membrane.This work shares various methodological details with other reported studies[Kalra,A.,Garde,S.and Hummer,G.(2003);Holt,J.K.,Noy,A.,Huser,T.,Eaglesham,D.and Bakajin,O.(2004);Sun,L.and Crooks,R.M.(2000)].There are,however,several modeling differences as follows:(1).the simulations are devoted to single CNTs embedded in the explicit bilayer lipid membranes with atomistic details,rather than to use the coarse-graining method or other mesoscale cell membrane models that may have the similar structure;(2).a two layer-membrane structure is used in the simulation to form a cellular chamber with only one side pierced by CNT,so that it ensures ions diffusion in only one direction.The paper is organized into four Sections.In Section 2,we first discuss some technical details of the molecular modeling procedure,including the CNT-Membrane model and the methodology of molecular dynamics simulations.Then in Section 3,we report the simulation results,and finally in Section 4,we present the conclusion by discussing the numerical results.
2 Model and Methods
In this section,we briefly outline the main method e molecular model used in the simulation.
2.1 CNT-Membrane model
The CNT-Membrane Model adopted in the present study consists of two POPE lipid bilayer membrane layers each with sizewhich form a cellular chamber with periodic boundary conditions as shown in Fig.1.A rigid nonpolar armchair-type CNT of lengthis embedded in the center of the right membrane layer.Water solution is modeled by TIP3P water model with two water reservoirs formed on outer side of cell chamber and one water reservoir is the cell chamber itself.Between the two membrane layers,solute ions including Na+,K+and Cl−with ionic concentration of approximately 1 M are located only in water solution of the cell chamber.The half sectional view of the whole model is shown in Fig.1.
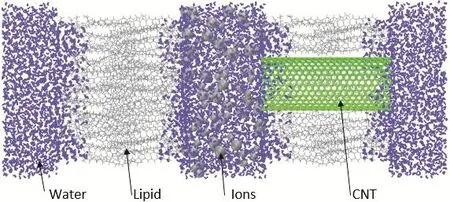
Figure 1:Snapshot(half sectional side view)of the model.
The POPE membrane is chosen as embedding material for the reason that it will not have any influence on water conduction in the nanotube,and results obtained can draw a direct comparison with the results of aquaporin.
Different sizes of CNT models are employed in the simulation in order to investigate the size effect for the ion diffusion under investigation.The different radii of CNTs and other structural details are specified in Tab.1.
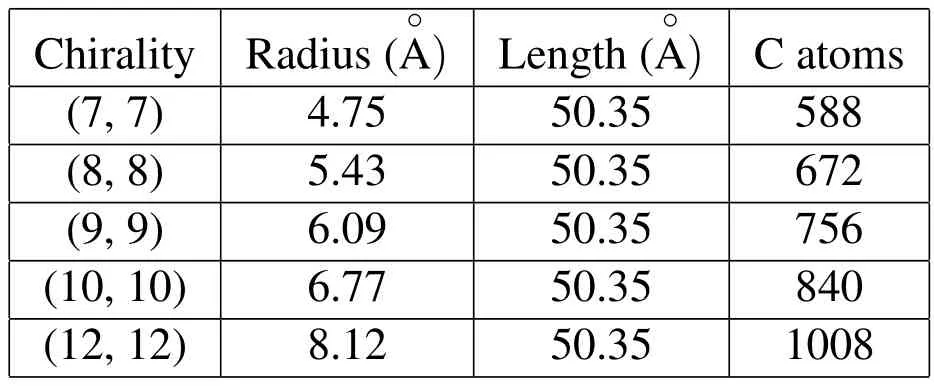
Table 1:Geometry of CNTs used
2.2 Molecular dynamics method
MD simulations are performed using the parallel molecular dynamics code NAMD[Phillips,J.C.,Braun,R.,Wang,W.,Gumbart,J.,Tajkhorshid,E.and Villa,K.(2005)],and the simulation results are processed and visualized by using a molecular visualization program VMD[Humphrey,W.,Dalke,A.and Schulten,K.(1996)].The CHARMM force fields are used in simulations to model the lipid bilayer membrane[Brooks,B.R.,Bruccoleri,R.E.,Olafson,B.D.,States,D.J.,Swaminathan,S.and Karplus,M.(1983)],the carbon nanotubes,as well as ions and water molecules,which have been well optimized for simulating bio-protein and bio-molecular systems.The equation of motion of each particle is described by Newton’s law:
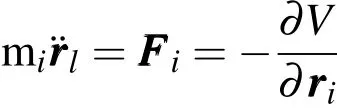
Where miis the atomic mass for i-th atom,andriis the spatial coordinate of the i-th atom or particle.V is defined as potential energy function of the particle,which is described by following equation in CHARMM force field:

Each term accounts for the energy caused by different parts of the system,including bond stretches(Vbond),bond angle(Vangle),torsion angle(Vdih),out of plane bending(Vimp),Van Der Waals energy(Vvdw)and the electrostatic energy(Vee).In order to run the MD simulations,we need to be sure that all parameters in the above equation are specified.More than ten parameters are used for each kind of ions,molecules,or atoms,which are listed in Tab.2.
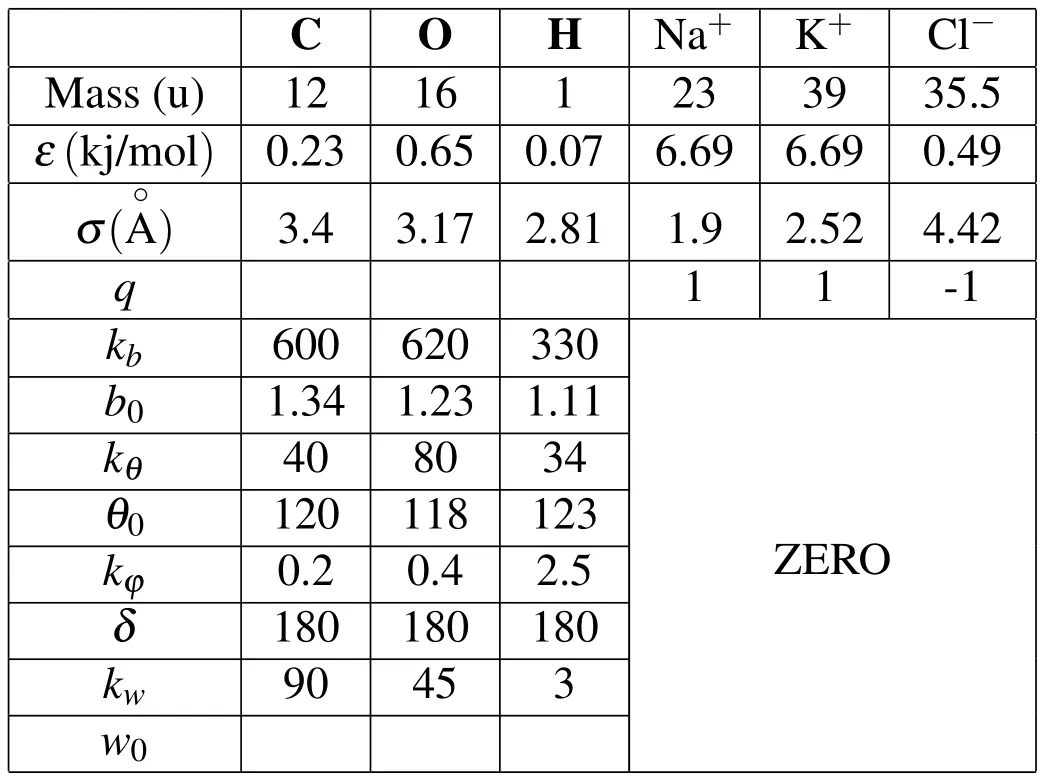
Table 2:Important atomic parameters
Parameters of bond energy for ions are all zero because ions are free in the solution and no bond formed between them.
In order to eliminate surface effects and to ensure a continuous flow of ions and water molecules,periodic boundary conditions in all three dimensions have been imposed;however,because of periodic boundary condition in the z direction,ions in one side can diffuse to the other size without go through the CNT,therefore,a two layer membrane structure is introduced in our model to form a closed cellular chamber.
Since the study of ion diffusion properties is the primary objective of this research,we employ a simple TIP3P model to describe water molecules,and we treat H2O molecule as a rigid body model,which will allow us to use larger time steps in computations,and thus it saves a lots of computational resources.
The simulation is carried out with a time step of 2fs.Each system is simulated for 5 ns followed by an equilibration for 0.2 ns at a constant temperature of 310 K;and the system is kept at a constant pressure of 1 bar.The PME(Particle-mesh Ewald)method has been employed in the simulation to account accurately for the long-range electrostatic interactions of the charges or ions and their periodic images[Hockney,R.W.and Eastwood,J.W.(1988)].
3 Simulations and Results
In this section,we present the main computation results that are recorded and obtained in the molecular dynamics simulation.
3.1 Average ion number inside the CNT
The average number of ions inside the CNTs of different sizes or chirality during the entire simulation process is shown in Fig.2.
No ions could pass through the CNT,if the radius of the CNT is less than 4.7which corresponded to the chirality(7,7).When the size of CTN reaches(8,8),one single Cl−ion begins to pass through the CNT,but no K+or Na+ion can be found during the whole simulation if CTN size is under(8,8).When the CTN size is at(9,9),the K+ion began to appear inside the CNT and the Na+only appear until the CTN size increases to(10,10).

Figure 2:Average number of ions inside CNT.
3.2 Dynamic distribution and diffusion of ions
To study the global picture of ion diffusion motion inside the CNT,we record all ion position changes inside CNT with time.A qualitative comparison between ions(current)position and CNT size(chirality size of(9,9)and(12,12))is displayed in Fig.3,which shows the instantaneous z-positions of the ions over typical simulation time intervals of 5 ns.
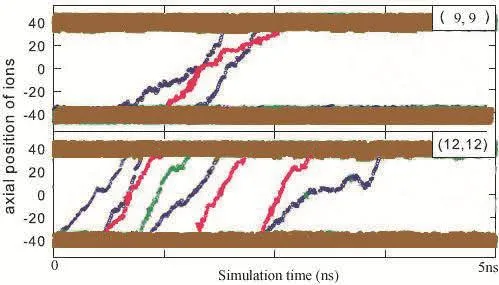
Figure 3:Snapshot of axial positions of ions with Cl−in blue,K+in red,and Na+in green;because the simulation is finished at time 5ns,some ions didn’t path through the entail CNT with partial path,which were not show.
From Fig.3,one can conclude that the ion diffusivity inside the CNT depends on the diameter of the CNTs;and it appears that the CNT with larger size has larger diffusivity.
3.3 Ion occupancy of Potassium
Moreover,we have tested the ion occupancy inside the carbon nanotubes of various diameters in the KCl solution,as shown in Fig.4.
Over the course of five nano-seconds,the ion occupancy of K+was observed largely between 2 and 4,the ions may enter the tube from either ends but most of them did not travel across the entire length of the nanotube,because some of them just enter and exit from the same end.
4 Discussions and Conclusions
In this section,we analyze the results obtained in the molecular dynamics simulation,and discuss the findings.
4.1 Minimum diameter for ion transport
In order to explain the simulation result,which indicates that different ions have different critical CNT diameters,we employ the so-called steered molecular dynamics(SMD)method[Wells,D.B.,Abramkina,V.,&Aksimentiev,A.(2007)]to calculate the energy barrier of ion permeation.A SMD program is provided by the NAMD software that can be used to calculate the axial potential of mean force(PMF)pro files,with formula:
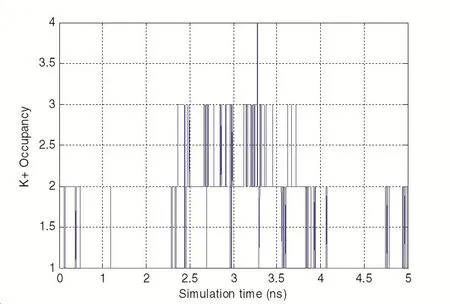
Figure 4:K+Occupancy vs.time in a(12,12)CNT
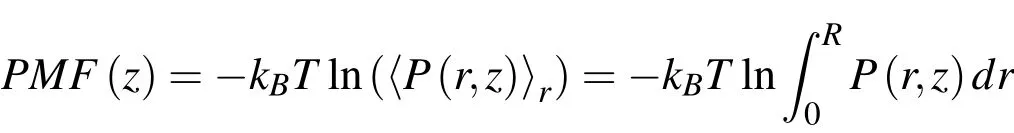
kBis the Boltzmann constant,and P(r,z)is the probability of transversely averaged position of atoms or ions.Thus it can be used to examine how a system’s energy changes as a function of axial distance.
We apply a constant velocity of 20A/ns to the ions along the axial direction,and corresponding force at each time step is stored in the output file.The result is shown in Fig.5:
Fig.5 shows that free energy profiles of potassium ions under different radii have different energy barrier as the ions traverse across nanotubes.It can be seen that the potassium ions face an appreciable energy barrier to enter the tube,which significantly increases with decreasing pore radius.
Comparing the free energy different ions in CNT with diameter(7,7)to(12,12),we can see that the energy needed for ions to pass through diameter(7,7)and(8,8)are so large that it is almost impossible for ions to diffuse through;however,the energy inside the CNT is almost the same as that outside the CNT for(12,12),which indicates that the ions can move through almost frictionless.
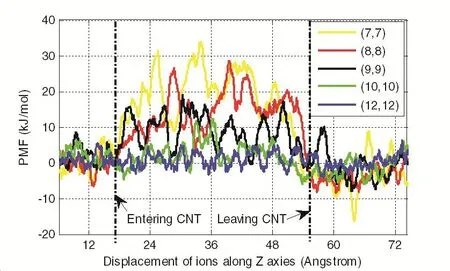
Figure 5:PMF for K+under different diameters
We may draw a conclusion that the free energy of K+determines the minimum diameter for K+to pass through the CNT.At diameter(9,9),the free energy is already small enough for K+permeation,which should be the critical diameter for K+ions.Similar analysis can be done for both Na+and Cl−ions,and our simulation results indicate that the critical diameter is(8,8)for Cl−and(10,10)for Na+,respectively.
4.2 Selectivity of ion hydration shell order
When ions are dissolved in water,they attract and hold several water dipoles around them.The negative(oxygen)side of a water molecule attracts positive ions.Because of this ion-dipole force,water molecules cluster around positive ions,as shown in Fig.6 and 7.Similarly,the positive(hydrogen)ends of water molecules are attracted to negative ions.
Comparing the size of hydration shells around ions inside and outside CNT shows that a denser shell forms in bulk compared to ions captured in CNT.
Therefore,we may conclude that the K+ion have to lose part of its surrounding water molecules inside the nanotube channel in order to pass through the CNT.This suggests that the stripping of coordination shell is critical for the high K+selectivity of ion channel.This observation is very familiar with those results found in the biological channel.This stripping effect induces the enthalpy and entropic changes that will affect the ion selectivity and fl ux inside the narrow nano pore.This observation from the MD simulation has improved our understanding on the hydration level of Cl−and K+and the mechanisms of ion transport and separation in the nanotube channels.In general carbon nanotubes have relatively smaller diameters so that they are only able to support a linear arrangement of water molecules passing along the axis of the tubes,which prevents the ions to enter the nanotube channel with water molecules together,because there is not enough space to accommodate the spatial dimension of hydration shells.Therefore the desolvation of ions due to CNT’s spatial or size constraint will impose a large energy barrier as well as entropic penalty.
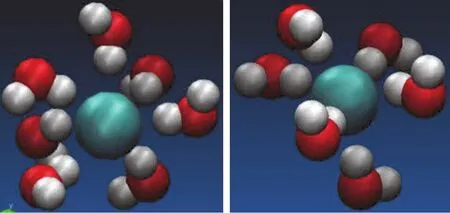
Figure 6:The hydration of Cl−ion for(a)in the bulk solvation;(b)in the CNT with diameter(9,9).
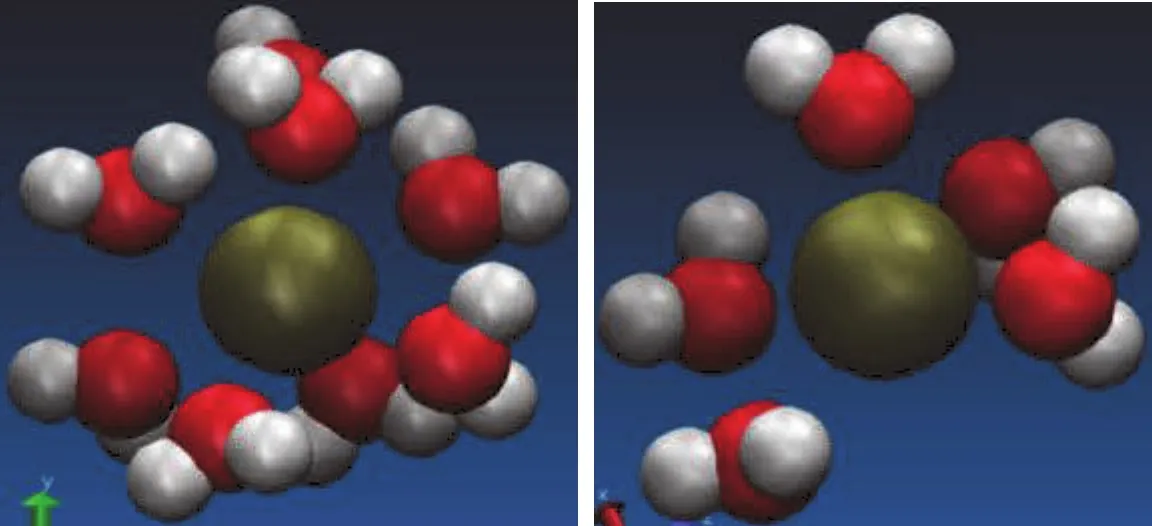
Figure 7:The hydration of K+ion for(a)in the bulk solvation;(b)in the CNT with diameter(9,9).
Acknowledgement:Mr.Qing-Song Tu is partially supported by a graduate fellowship from Chinese Scholarship Council(CSC),and this support is greatly appreciated.The authors would like to thank Ms.Hengameh Shams for many discussions and suggestions on this research.
Berneche,S.;Roux,B.(2000):Molecular Dynamics of the KcsA K+Channel in a Bilayer Membrane.Biophysical Journal,vol.78,no.6,pp.2900-2917.
Brooks,B.R.;Bruccoleri,R.E.;Olafson,B.D.;States,D.J.;Swaminathan,S.;Karplus,M.(1983):CHARMM:A program for macromolecular energy,minimization,and dynamics calculations.Journal of computational chemistry,vol.4,no.2,pp.187-217.
Cannon,S.C.;Brown,R.H.Jr;Corey,D.P.(1991):A sodium channel defect in hyperkalemic periodic paralysis:potassium-induced failure of inactivation.Neuron,vol.6,pp.619–626.
Delpire,E.;Lu,J.;England,R.;Dull,C.;Thorne,T.(1999):Deafness and imbalance associated with inactivation of the secretory Na–K–2Cl co-transporter.Nat.Genet.,vol.22,pp.192–195.
Feller,S.E.(2000):Molecular dynamics simulations of lipid bilayers.Current opinion in colloid&interface science,vol.5,no.3,pp.217-223.
Forrest,L.R.;Sansom,M.S.(2000):Membrane simulations:bigger and better?Current opinion in structural biology,vol.10,no.2,pp.174-181.
Hille,B.(2001):Ion channels of excitable membranes(Vol.507).Sunderland,MA:Sinauer.
Holt,J.K.;Noy,A.;Huser,T.;Eaglesham,D.;Bakajin,O.(2004):Fabrication of a carbon nanotube-embedded silicon nitride membrane for studies of nanometerscale mass transport.Nano letters,vol.4,no.11,pp.2245-2250.
Hockney,R.W.;Eastwood,J.W.(1988):Computer simulation using particles.CRC Press.
Humphrey,W.;Dalke,A.;Schulten,K.(1996):VMD:visual molecular dynamics.Journal of molecular graphics,vol.14,no.1,pp.33-38.
Hübner,C.A.;Jentsch,T.J.(2002).Ion channel diseases.Human molecular genetics,vol.11,no.20,pp.2435-2445.
Kalra,A.;Garde,S.;Hummer,G.(2003):Osmotic water transport through carbon nanotube membranes.Proceedings of the National Academy of Sciences,vol.100,no.18,pp.10175-10180.
Katz,A.M.;Messineo,F.C.;Herbette,L.(1982):Ion channels in membranes.Circulation,vol.65,(1 Pt 2),I2-10.
Khalili-Araghi,F.;Gumbart,J.;Wen,P.C.;Sotomayor,M.;Tajkhorshid,E.;Schulten,K.(2009):Molecular dynamics simulations of membrane channels and transporters.Current opinion in structural biology,vol.19,no.2,pp.128-137.
Liu,H.;Murad,S.;Jameson,C.J.(2006):Ion permeation dynamics in carbon nanotubes.The Journal of chemical physics,vol.125,no.8,pp.084713.
Liu,H.;Jameson,C.J.;Murad,S.(2008):Molecular dynamics simulation of ion selectivity process in nanopores.Molecular Simulation,vol.34,no.2,pp.169-175.
Lindahl,E.;Sansom,M.S.(2008):Membrane proteins:molecular dynamics simulations.Current opinion in structural biology,vol.18,no.4,pp.425-431.
Marañón Di Leo,J.;Marañón,J.(2005):Hydration and diffusion of cations in nanopores.Journal of Molecular Structure:THEOCHEM,vol.729,no.1,pp.53-57.
Nelson,P.H.(2003):Modeling the concentration-dependent permeation modes of the KcsA potassium ion channel.Physical Review E,vol.68,no.6,061908.
Noskov,S.Y.;Roux,B.(2006):Ion selectivity in potassium channels.Biophysical chemistry,vol.124,no.3,pp.279-291.
Tytgat,J.;Hess,P.(1992):Evidence for cooperative interactions in potassium
channel gating.Nature,vol.359,no.6394,pp.420-423.
Peter,C.;Hummer,G.(2005):Ion transport through membrane-spanning nanopores studied by molecular dynamics simulations and continuum electrostatics calculations.Biophysical journal,vol.89,no.4,pp.2222-2234.
Phillips,J.C.;Braun,R.;Wang,W.;Gumbart,J.;Tajkhorshid,E.;Villa,K.(2005):Scalable molecular dynamics with NAMD.Journal of computational chemistry,vol.26,no.16,pp.1781-1802.
Sansom,M.S.;Shrivastava,I.H.;Ranatunga,K.M.;Smith,G.R.(2000):Simulations of ion channels–watching ions and water move.Trends in biochemicalsciences,vol.25,no.8,pp.368-374.
Stein,W.(1986):Transport and diffusion across cell membranes.Elsevier.
Sotomayor,M.;Schulten,K.(2007):Single-molecule experiments in vitro and in silico.Science,vol.316,pp.1144–1148.
Sun,L.;Crooks,R.M.(2000):Single carbon nanotube membranes:a welldefined model for studying mass transport through nanoporous materials.Journal of the American Chemical Society,vol.122,no.49,pp.12340-12345.
Wells,D.B.;Abramkina,V.;Aksimentiev,A.(2007):Exploring transmembrane transport throughα-hemolysin with grid-steered molecular dynamics.The Journal of chemical physics,vol.127,no.12,125101.
Yellen,G.A.R.Y.(1984):Ionic permeation and blockade in Ca2+-activated K+channels of bovine chromaffin cells.The Journal of general physiology,vol.84,no.2,pp.157-186.
 Computer Modeling In Engineering&Sciences2014年9期
Computer Modeling In Engineering&Sciences2014年9期
- Computer Modeling In Engineering&Sciences的其它文章
- Hybrid Simulation and Observation of Human Vertebral Endplate Morphology
- Variable Viscosity and Density Biofilm Simulations using an Immersed Boundary Method,Part I:Numerical Scheme and Convergence Results
- Activation Pattern of Nuclear Factor-kB in Skin after Mechanical Stretch–a Multiscale Modeling Approach
- A Computational Modeling Framework for Heat Transfer Processes in Laser-Induced Dermal Tissue Removal
