The Pael-R gene does not mediate the changes in rotenone-induced Parkinson’s disease model cells
Ting Zou, Xiangqi Tang, Zhiling Huang, Niangui Xu, Zhiping Hu
The Pael-R gene does not mediate the changes in rotenone-induced Parkinson’s disease model cells
Ting Zou, Xiangqi Tang, Zhiling Huang, Niangui Xu, Zhiping Hu
Department of Neurology, Second Xiangya Hospital, Central South University, Changsha, Hunan Province, China
In this study, we established cell models for Parkinson’s disease using rotenone. An RNA interference vector targeting Parkin-associated endothelin receptor-like receptor (Pael-R) was transfected into the model cells. The results of reverse-transcription polymerase chain reaction and western blot analysis showed that Pael-R expression was decreased after RNA interference compared with the control group (no treatment) and the model group (rotenone treatment), while the rate of apoptosis and survival of dopaminergic cells did not differ signi fi cantly between groups, as detected by fl ow cytometry and an MTT assay. These experimental fi ndings indicate that the Pael-R gene has no role in the changes in rotenone-induced Parkinson’s disease model cells.
nerve regeneration; neurodegeneration; Parkinson’s disease; rotenone; Pael-R; RNA interference; apoptosis; mitochondria; neural regeneration
Zou T, Tang XQ, Huang ZL, Xu NG, Hu ZP. The Pael-R gene does not mediate the changes in rotenone-induced Parkinson’s disease model cells. Neural Regen Res. 2014;9(4):402-406.
Introduction
Parkinson’s disease is a common neurodegenerative disease. It is mainly caused by the loss of dopaminergic innervation owing to degenerative necrosis of nigral dopaminergic neurons[1-2]. Currently, the pathogenesis of Parkinson’s disease is not entirely clear, but it has been generally considered to be the result of interactions among various genetic and environmental factors[3-6].
Rotenone is a suppressant of mitochondrial compound I, and is widely used in pesticides[7-8]. It was previously found that dopaminergic neurons become apoptotic if they are exposed to rotenone[9-11]. Sherer et al.[12-14]found that rotenone-treated rats showed behavioral and pathological features of Parkinson’s disease; thus, such rats are considered to be an ideal Parkinson’s disease model and have been widely used in research investigating the pathogenesis of Parkinson’s disease. Presently, the pathways and molecular mechanisms by which rotenone induces apoptosis of dopaminergic neurons remain unclear.
Accumulating evidence has shown that Parkin is a common Parkinson’s disease-causing gene and its mutation correlates with mitochondrial function through reducing mitochondrial complex activity[15-18]. Moreover, Parkin mutation causes Parkin-associated endothelin receptor-like receptor (Pael-R) protein deposition, and the associated cytotoxicity leads to dopaminergic neuronal apoptosis[19-21]. In this study, we speculated that the Pael-R gene is possibly involved in the action of rotenone on cells; therefore, this study aimed to investigate the role of the Pael-R gene in rotenone-induced Parkinson’s disease model cells using RNA interference.
Results
Pael-R gene expression was downregulated by RNA interference
Total RNA was extracted from cultured PC12 cells representing the control group (no treatment at all), model group (rotenone treatment) and RNA interference group (rotenone treatment plus RNA interference vector). Pael-R gene primers were used for reverse-transcription polymerase chain reaction (RT-PCR) ampli fi cation, and 599 bp of the Pael-R gene interior fragment was obtained (Figure 1A).
The gray scale values of RT-PCR bands were analyzed and the absorbance ratios between the Pael-R gene and GAPDH gene in each group were calculated and used for further comparison. There was no signi fi cant difference between the control group without rotenone treatment and the model group in terms of the level of Pael-R gene expression (P >0.05). This indicates that rotenone has no signi fi cant impact on the transcription of the Pael-R gene in induced dopaminergic neuron-like cells. The absorbance ratio from the RTPCR results in the RNA interference group was 14% less than that in the model group (P < 0.05; Figure 1A). This evidence confirmed that the pRNA-U6/Pael-R interference vector suppresses the expression of the Pael-R gene.
The Pael-R protein level in all three cell groups was determined by western blot analysis. A Pael-R band was obtained at 67 kDa (Figure 1B). The gray scale analysis of RT-PCR results revealed similar levels of Pael-R protein in model and control cells (P > 0.05), suggesting that rotenone also has no significant influence on Pael-R protein levels. In the RNA interference group, the Pael-R protein level was lower than those in the control and model groups (P < 0.05; Figure1B), indicating that transfection of cells with the pRNA-U6/ Pael-R vector could effectively down-regulate Pael-R protein expression.
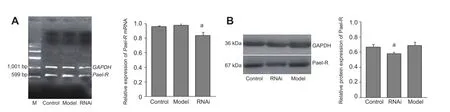
Figure 1 Effect of RNA interference (RNAi) on Pael-R gene and protein expression in Parkinson’s disease model cells.
RNA interference had no signi fi cant in fl uence on apoptosis of Parkinson’s disease model cells
Morphological changes to cells were observed under a fluorescence microscope with Hoechst 33258 staining. There were no apoptotic cells in the control group. In the model group, some cells showed nuclear condensation or nuclear division and the nuclei appeared light blue with an irregular shape. These fi ndings indicated the presence of apoptosis. In the RNA interference group, some cells showed nuclear condensation and division, which also con fi rmed the presence of apoptotic cells (Figure 2A–C).
We also examined the rates of apoptosis in the different cell groups by fl ow cytometry. The results were similar to those obtained with Hoechst 33258 staining. Cell apoptosis rates in the model and RNA interference groups were not statistically signi fi cantly different (P > 0.05; Figure 2D). This suggested that there is no signi fi cant in fl uence of down-regulation of Pael-R gene expression on rotenone-induced apoptosis.
RNA interference had no signi fi cant in fl uence on the viability of Parkinson’s disease model cells
According to the results of the MTT assay, the cell survival rates in the model group and RNA interference group were signi fi cantly lower than that in the control group (P < 0.05). There was no statistically signi fi cant difference between the model group and the RNA interference group (P > 0.05; Figure 2E). These fi ndings indicated that the cell survival rates decrease after rotenone treatment, but that this decrease in survival could not be improved by down-regulating Pael-R expression in cells.
Discussion
Widespread attention has been paid to rotenone as a potential cause of Parkinsonism. Recent studies have shown that long-term exposure to rotenone may cause changes at the cellular level in the form of injury to central dopaminergic neurons, obvious degeneration and apoptosis of substantia nigra dopaminergic neurons and the appearance of Lewy bodies in neurons, tremor, unsteady gait and other Parkinson’s disease symptoms[22-23]. Currently, the precise mechanisms underlying rotenone-induced apoptosis in dopaminergic neurons are unclear. In the cell injury induced by rotenone, the following processes are known to participate: increased levels of reactive oxygen species; lipid peroxidation; progressive glutathione deficiency; mitochondrial depolarization; release of cytochrome C; and activation of caspase-3[24-27]. These factors collectively lead to apoptosis, which, particularly when affecting dopaminergic neurons, is one of the important insults leading to Parkinson’s disease[28-29]. In 2006, Feng[30]reported that apoptosis of dopaminergic neurons induced by rotenone may be related to damage to the intracellular microtubule system, thus leading to release of dopamine in cells, which causes cell toxicity. In addition, Pan-Montojo et al.[31]found that rotenone causes alpha-synuclein deposition in the nervous system and further induces apoptosis of neuronal cells.
It has been found that some similar molecular mechanisms in the pathogenesis of Parkinson’s disease are mediated by environmental toxins and genetic flaws[32-33]. Pael-R protein deposition is an important factor during the course of apoptosis of dopaminergic neurons owing to Parkin gene mutations[34-36]. We discussed the relationship between down-regulation of Pael-R gene expression and apoptosis of dopaminergic neurons induced by rotenone in this study. Furthermore, because the function of Pael-R is not well understood, it may be a component of other signaling pathways; therefore, we performed transient transfection of cells with an RNA interference vector to partially down-regulate the expression of Pael-R, and maintained a lower level of Pael-R expression in cells to avoid in fl uences from other pathways.
Our results demonstrate that dopaminergic neuron-like cells show obvious apoptosis after the treatment with rotenone. The results of gene expression analyses indicate that there were no signi fi cant differences in Pael-R transcript and protein levels before and after rotenone treatment. This fi nding suggests that rotenone has no obvious effect on Pael-R gene expression, and that Pael-R probably does not participate in the course of apoptosis of dopaminergic neuron-like cells induced by rotenone. Further investigations revealed that after down-regulating Pael-R gene expression by RNAinterference in the rotenone-treated cells, there were no signi fi cant changes in cell viability and level of apoptosis. This result suggests that down-regulating Pael-R gene expression could not promote the survival of Parkinson’s disease model cells, and that down-regulation of Pael-R gene has no protective action in rotenone-induced Parkinson’s disease model cells.
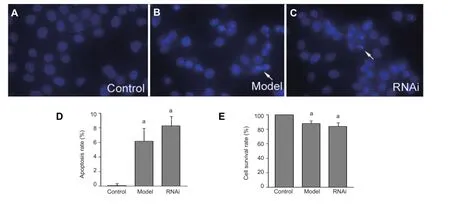
Figure 2 Effect of RNA interference (RNAi) on apoptosis and survival rate of Parkinson’s disease model cells.
Materials and Methods
Design
A controlled cytobiology study.
Time and setting
The experiments were performed at the Cancer Research Institute, Xiangya Medical School of Central South University, China from June 2007 to March 2008.
Materials
PC12 cells were purchased from the Cell Bank of Shanghai Institute of Biochemistry and Cell Biology, China, and rotenone was purchased from Sigma, St. Louis, MO, USA.
Methods
Induction of dopaminergic neuron-like cells
PC12 cells were incubated in culture plates covered with poly-L-lysine, and cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) including 10% fetal calf serum (Invitrogen) in an incubator with 5% CO2at 37°C. PC12 cells were plated at a density of 2.5–10 × 104cells/cm2during induction and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) containing 50 ng/mL nerve growth factor (Invitrogen). The medium was changed every 2–3 days. Neurites could be observed 1 week after induction and culture; the cells then became dopaminergic neuron-like cells at 10 days[37-38].
Establishment of a rotenone-induced Parkinson’s disease cell model
Induced dopaminergic neuron-like cells were plated in 6-well plates at a density of 5 × 105cells/well. Rotenone was added after 24 hours to a fi nal concentration of 1 µmol/L. The rotenone-induced Parkinson’s disease cell model was established after 24 hours of treatment[39-40].
Down-regulation of the Pael-R gene using RNA interference in rotenone-induced Parkinson’s model cells
The pRNA-U6/Pael-R interference vector was designed for rat Pael-R gene and constructed as previously described[41]. Parkinson’s disease model cells treated with rotenone were plated in 6-well plates at a density of 5 × 105cells/well for 24 hours. Then, the pRNA-U6/Pael-R vector was transfected into cells together with Lipofectin 2000 (Invitrogen) according to the manufacturer’s protocol. Speci fi cally, 4 µg of vector was mixed with 10 µL of Lipofectin 2000 reagent, and cells were incubated with the mixture for 24 hours. Cells can be collected for tests 48 hours after transfection.
Detection of Pael-R gene expression by RT-PCR
Differences in the levels of transcription of the Pael-R gene were detected by RT-PCR after extracting total RNA from cells. Pael-R gene primer was designed using Primer3 software (Howard Hughes Medical Institute, Chevy Chase, MD, USA) to obtain 599 bp of the Pael-R gene interior fragment. The upstream primer was 5′-AAC CGA CGC GTG AGACTG AA-3′, and the downstream primer was 5′-TGC GCC ATC ATA AGT GAG AGC-3′. The rat GAPDH gene served as an internal reference; the upstream primer was 5′-TGG TGA AGG TCG GTG TGA AC-3′, the downstream primer was 5′-TTA CTC CTT GGA GGC CAT GT-3′, and the product was 1,001 bp. The gray scale values for bands in the RT-PCR results were then analyzed by the Gel Doc XR+ System (Bio-Rad, Hercules, CA, USA) and results were calculated according to the formula: absorbance rate of Pael-R gene/GAPDH gene.
Detection of Pael-R protein by western blot analysis
Proteins from all three cell groups were prepared by cell lysis. Differences in the levels of Pael-R protein were assessed using goat-anti-rat Pael-R antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Total protein (20 µg) was taken from each group for electrophoresis and subsequent transfer of proteins onto a polyvinylidene fl uoride membrane. Membranes were blocked for 1 hour at room temperature, then incubated in primary antibody (1:1,000) overnight at 4°C, followed by incubation with rabbit anti-goat IgG (1:5,000; Sigma) for 1 hour at room temperature. Next, samples were treated for chemiluminescence with ECL fluid. Gray scale value analysis was then performed using the Gel Doc XR+ System (Bio-Rad) according to the formula: absorbance rate of Pael-R protein/GAPDH protein.
Cell morphological changes tested by Hoechst 33258 staining
Hoechst 33258 solution (Sigma) was added into the three groups of cultured cells to a fi nal concentration of 10 µg/mL; then, cells were incubated for 5 minutes at room temperature. Cell morphology was observed after PBS elution under an inverted fl uorescence microscope (Leica, Wetzlar, Germany).
Cell apoptosis test by flow cytometry
Cells in the three groups were collected and the level of cell apoptosis in each group was assessed by cell cytometry (BD, Franklin Lakes, NJ, USA) using annexin V-FITC/propidium iodide double staining[24].
Cell viability tested by MTT assay
Cells from the three groups were plated into 96-well plates at a density of 1 × 104cells/well. MTT solution (10 µL, 5 g/L, Sigma) was added into each well at 48 hours after RNA interference. Cells were incubated for 4 hours in 5% CO2at 37°C, then culture medium was removed and 150 µL of dimethyl sulfoxide (Sigma) was added into each well and cultured for 10 minutes until all particles were completely dissolved. Finally, the absorbance value at 490 nm was measured using a Microplate Reader (Bio-Rad) and the cell survival rate was calculated. Survival rate (%) = treated group absorbance/ untreated group absorbance × 100%.
Statistical analysis
Statistical analysis was performed using SPSS software (Version 14.0, SPSS, Chicago, IL, USA). Data are expressed as mean ± SD and were analyzed using one-way analysis of variance[35]. Statistical signi fi cance was de fi ned as P < 0.05.
Acknowledgments:We would like to thank the colleagues at Cancer Research Institute, Xiangya Medical School of Central South University, China for sharing their equipment.
Author contributions:Zou T and Hu ZP designed the study and contributed to manuscript development. Zou T, Tang XQ and Huang ZL conducted the experiments. Zou T and Xu NG participated in data integrity and analysis. All authors had approved the final version of the paper.
Con fl icts of interest:None declared.
Peer review:Using RT-PCR, western blot analysis, MTT and flow cytometry, we detected the effects of RNA interference-mediated Pael-R gene down-regulation in rotenone-induced Parkinson’s disease model cells. The experiment purpose could be achieved by the techniques.
[1] Gelb DJ, Oliver E, Gilman S. Diagnostic criteria for Parkinson disease. Arch Neurol. 1999;56(1):33-39.
[2] Mhyre TR, Boyd JT, Hamill RW, et al. Parkinson’s disease. Subcell Biochem. 2012;65:389-455.
[3] Piccini P, Burn DJ, Ceravolo R, et al. The role of inheritance in sporadic Parkinson’s disease: evidence from a longitudinal study of dopaminergic function in twins. Ann Neurol. 1999;45(5):577-582.
[4] Elbaz A, Grigoletto F, Baldereschi M, et al. Familial aggregation of Parkinson’s disease: a population-based case-control study in Europe. EUROPARKINSON Study Group. Neurology. 1999;52(9): 1876-1882.
[5] Tsuboi Y. Environmental-genetic interactions in the pathogenesis of Parkinson’s disease. Exp Neurobiol. 2012;21(3):123-128.
[6] Popat RA, Van Den Eeden SK, Tanner CM, et al. Coffee, ADORA2A, and CYP1A2: the caffeine connection in Parkinson’s disease. Eur J Neurol. 2011;18(5):756-765.
[7] Markham A, Cameron I, Bains R, et al. Brain-derived neurotrophic factor-mediated effects on mitochondrial respiratory coupling and neuroprotection share the same molecular signalling pathways. Eur J Neurosci. 2012;35(3):366-374.
[8] Chen G, Chen Z, Hu Y, et al. Inhibition of mitochondrial respiration and rapid depletion of mitochondrial glutathione by β-phenethyl isothiocyanate: mechanisms for anti-leukemia activity. Antioxid Redox Signal. 2011;15(12):2911-2921.
[9] Marella M, Seo BB, Matsuno-Yagi A, et al. Mechanism of cell death caused by complex I defects in a rat dopaminergic cell line. J Biol Chem. 2007;282(33):24146-24156.
[10] Sherer TB, Betarbet R, Testa CM, et al. Mechanism of toxicity in rotenone models of Parkinson’s disease. J Neurosci. 2003;23(34):10756-10764.
[11] Parameshwaran K, Irwin MH, Steliou K, et al. Protection by an antioxidant of rotenone-induced neuromotor decline, reactive oxygen species generation and cellular stress in mouse brain. Pharmacol Biochem Behav. 2012;101(3):487-492.
[12] Sherer TB, Kim JH, Betarbet R, et al. Subcutaneous rotenone exposure causes highly selective dopaminergic degeneration and alpha-synuclein aggregation. Exp Neurol. 2003;179(1):9-16.
[13] Alam M, Schmidt WJ. Rotenone destroys dopaminergic neurons and induces parkinsonian symptoms in rats. Behav Brain Res. 2002;136(1):317-324.
[14] Li BY, Yuan YH, Hu JF, et al. Protective effect of Bu-7, a fl avonoid extracted from Clausena lansium, against rotenone injury in PC12 cells. Acta Pharmacol Sin. 2011;32(11):1321-1326.
[15] Grünewald A, Voges L, Rakovic A, et al. Mutant Parkin impairs mitochondrial function and morphology in human fi broblasts. PLoS One. 2010;5(9):e12962.
[16] Flinn L, Mortiboys H, Volkmann K, et al. Complex I de fi ciency and dopaminergic neuronal cell loss in parkin-de fi cient zebra fi sh (Danio rerio). Brain. 2009;132(Pt 6):1613-1623.
[17] Sai Y, Zou Z, Peng K, et al. The Parkinson’s disease-related genes act in mitochondrial homeostasis. Neurosci Biobehav Rev. 2012;36(9): 2034-2043.
[18] Cookson MR. Parkinsonism due to mutations in PINK1, parkin, and DJ-1 and oxidative stress and mitochondrial pathways. Cold Spring Harb Perspect Med. 2012;2(9):a009415.
[19] Wang HQ, Imai Y, Inoue H, et al. Pael-R transgenic mice crossed with parkin de fi cient mice displayed progressive and selective catecholaminergic neuronal loss. J Neurochem. 2008;107(1):171-185.
[20] Kitao Y, Imai Y, Ozawa K, et al. Pael receptor induces death of dopaminergic neurons in the substantia nigra via endoplasmic reticulum stress and dopamine toxicity, which is enhanced under condition of parkin inactivation. Hum Mol Genet. 2007;16(1):50-60.
[21] Lundius EG, Stroth N, Vukojević V, et al. Functional GPR37 trafficking protects against toxicity induced by 6-OHDA, MPP+or rotenone in a catecholaminergic cell line. J Neurochem. 2013;124(3):410-417.
[22] Ayala A, Venero JL, Cano J, et al. Mitochondrial toxins and neurodegenerative diseases. Front Biosci. 2007;12:986-1007.
[23] Gomez C, Bandez MJ, Navarro A. Pesticides and impairment of mitochondrial function in relation with the parkinsonian syndrome. Front Biosci. 2007;12:1079-1093.
[24] Liu HQ, Zhu XZ, Weng EQ. Intracellular dopamine oxidation mediates rotenone-induced apoptosis in PC12 cells. Acta Pharmacol Sin. 2005;26(1):17-26.
[25] Giordano S, Lee J, Darley-Usmar VM, et al. Distinct effects of rotenone, 1-methyl-4-phenylpyridinium and 6-hydroxydopamine on cellular bioenergetics and cell death. PLoS One. 2012;7(9):e44610.
[26] Manjunath MJ , Muralidhara. Withania somnifera prophylaxis abrogates Rotenone-induced oxidative impairments and mitochondrial dysfunctions in striatum and cerebellum of mice: relevance to Parkinson’s disease. Cent Nerv Syst Agents Med Chem. in press.
[27] Verma R, Nehru B. Effect of centrophenoxine against rotenone-induced oxidative stress in an animal model of Parkinson’s disease. Neurochem Int. 2009;55(6):369-375.
[28] Exner N, Lutz AK, Haass C, et al. Mitochondrial dysfunction in Parkinson’s disease: molecular mechanisms and pathophysiological consequences. EMBO J. 2012;31(14):3038-3062.
[29] Hauser DN, Hastings TG. Mitochondrial dysfunction and oxidative stress in Parkinson’s disease and monogenic parkinsonism. Neurobiol Dis. 2013;51:35-42.
[30] Feng J. Microtubule: a common target for parkin and Parkinson’s disease toxins. Neuroscientist. 2006;12(6):469-476.
[31] Pan-Montojo F, Anichtchik O, Dening Y, et al. Progression of Parkinson’s disease pathology is reproduced by intragastric administration of rotenone in mice. PLoS One. 2010;5(1):e8762.
[32] Ritz BR, Manthripragada AD, Costello S, et al. Dopamine transporter genetic variants and pesticides in Parkinson’s disease. Environ Health Perspect. 2009;117(6):964-969.
[33] Hill-Burns EM, Singh N, Ganguly P, et al. A genetic basis for the variable effect of smoking/nicotine on Parkinson’s disease. Pharmacogenomics J. 2013;13(6):530-537.
[34] Imai Y, Soda M, Inoue H, et al. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell. 2001;105(7):891-902.
[35] Zou T, Xiao B, Tang J, et al. Downregulation of Pael-R expression in a Parkinson’s disease cell model reduces apoptosis. J Clin Neurosci. 2012;19(10):1433-1436.
[36] Dusonchet J, Bensadoun JC, Schneider BL, et al. Targeted overexpression of the parkin substrate Pael-R in the nigrostriatal system of adult rats to model Parkinson’s disease. Neurobiol Dis. 2009;35(1):32-41.
[37] Genchi GG, Ciofani G, Polini A, et al. PC12 neuron-like cell response to electrospun poly(3-hydroxybutyrate) substrates. J Tissue Eng Regen Med. in press.
[38] Akashi S, Shirai K, Okada T, et al. Neoechinulin a imparts resistance to acute nitrosative stress in PC12 cells: a potential link of an elevated cellular reserve capacity for pyridine nucleotide redox turnover with cytoprotection. Biol Pharm Bull. 2012;35(7):1105-1117.
[39] Akashi S, Kimura T, Takeuchi T, et al. Neoechinulin a impedes the progression of rotenone-induced cytotoxicity in PC12 cells. Biol Pharm Bull. 2011;34(2):243-248.
[40] Konishi K, Watanabe N, Arai T. SIN-1 cytotoxicity to PC12 cells is mediated by thiol-sensitive short-lived substances generated through SIN-1 decomposition in culture medium. Nitric Oxide. 2009;20(4):270-278.
[41] Zou T, Xiao B, Tang JG, et al. Construction of shRNA interference vector for rat Pael- R gene and functional identi fi cation. Shengwu Jishu. 2011;21(4):12-16.
Copyedited by McGowan D, Zhang XL, Ding XH, Yu J, Yang Y, Li CH, Song LP, Zhao M
10.4103/1673-5374.128245
Zhiping Hu, M.D., Department of Neurology, Second Xiangya Hospital, Central South University, Changsha 410011, Hunan Province, China, huzhiping_csu@hotmail. com.
http://www.nrronline.org/
Accepted: 2014-01-19
- 中国神经再生研究(英文版)的其它文章
- Posterior quadrantic disconnection maintains the activity of isolated temporal-parietal-occipital nerve tissue: neuroprotective measures in the surgical treatment of epilepsy
- Examination of Huntington’s disease in a Chinese family
- Circadian fl uctuations in three types of sensory modules in healthy subjects
- 7.0T nuclear magnetic resonance evaluation of the amyloid beta (1–40) animal model of Alzheimer’s disease: comparison of cytology veri fi cation
- Local inhibition of GABA affects precedence effect in the inferior colliculus
- Compound Formula Rehmannia alleviates levodopainduced dyskinesia in Parkinson’s disease

