Bartonella species in small mammals and their potential vectors in Asia
Tawisa Jiyipong, Sathaporn Jittapalapong, Serge Morand, Jean-Marc Rolain
1Research Unit on Infectious and Emerging Tropical Diseases (URMITE), CNRS-IRD-INSERM UMR 7278, IHU Méditerranée Infection, Faculty of Medicine and Pharmacy, Aix-Marseille-University, Marseille, France
2Department of Parasitology, Faculty of Veterinary Medicine, Kasetsart University, Bangkok, Thailand
3Institut Sciences of Evolution, UMR 5554 CNRS-IRD-UM2, CC65, Université de Montpellier 2, F-34095, Montpellier, France
4Center for Agricultural Biotechnology (AG-BIO/PEDRO-CHE), Kasetsart University, Kamphaeng Saen Campus, Nakhon Pathom, 73140, Thailand
5Center of Excellence on Agricultural Biotechnology (AG-BIO/PERDO-CHE), Bangkok 10900, Thailand
6Center of Advanced Studies for Agriculture and Food, KU Institute for Advanced Studies, Kasetsart University, Bangkok 10900, Thailand (CASAF, NRU-KU, Thailand)
7Walai Rukhavej Botanical Research Institute, Mahasarakham University, Mahasrakham, Thailand
Bartonella species in small mammals and their potential vectors in Asia
Tawisa Jiyipong1,4,5,6, Sathaporn Jittapalapong2,6, Serge Morand3,7, Jean-Marc Rolain1*
1Research Unit on Infectious and Emerging Tropical Diseases (URMITE), CNRS-IRD-INSERM UMR 7278, IHU Méditerranée Infection, Faculty of Medicine and Pharmacy, Aix-Marseille-University, Marseille, France
2Department of Parasitology, Faculty of Veterinary Medicine, Kasetsart University, Bangkok, Thailand
3Institut Sciences of Evolution, UMR 5554 CNRS-IRD-UM2, CC65, Université de Montpellier 2, F-34095, Montpellier, France
4Center for Agricultural Biotechnology (AG-BIO/PEDRO-CHE), Kasetsart University, Kamphaeng Saen Campus, Nakhon Pathom, 73140, Thailand
5Center of Excellence on Agricultural Biotechnology (AG-BIO/PERDO-CHE), Bangkok 10900, Thailand
6Center of Advanced Studies for Agriculture and Food, KU Institute for Advanced Studies, Kasetsart University, Bangkok 10900, Thailand (CASAF, NRU-KU, Thailand)
7Walai Rukhavej Botanical Research Institute, Mahasarakham University, Mahasrakham, Thailand
PEER REVIEW
Peer reviewer
Dr. Hossein Ghalehnoei, University of Medical Science, Tehran, Iran.
Tel: +989385518026, E-mail: hossein. ghalehnoei58@gmail.com
Co-reviewers: Misael Chinchilla, San José, Costa Rica,
Comments
This is a good research about the presence of various species of Bartonella and its role at causing different infections. The consideration of authors to various reservoirs of this bacteria in animals that a little have been already surveyed is interesting. Diagnostic tests referred in this article are very useful.
Details on Page 764
In this article, authors review the current knowledge of Bartonella infection in small mammals including rodents, insectivores, bats and exotic small mammal pets and their vectors in Asia. Species of Bartonella are Gram-negative intracellular bacteria that infect erythrocytes of various mammalian and non-mammalian animals and mainly transmitted by blood sucking arthropod vectors. The genus Bartonella includes several species of important human diseases with severe clinical signs. Several new Bartonella species were isolated from rodents and other small mammals, and from human patients in Asia. Bartonella species are identified using standard polymerase chain reaction amplification and a sequencing targeting two housekeeping genes (gltA and rpoB) and the internal transcribed spacer fragment. Authors also discuss the implications in term of potential emerging zoonotic diseases.
Bartonella, Small mammals, Rodents, Shrews, Asia
1. Introduction
Member of the genusBartonellaare fastidious, hemotropic, Gram-negative and aerobic bacilli bacteria belonging to the class Alphaproteobacteria. Several species have been implicated in causing human diseases, ranging with short-term fever to severe endocarditis. Five species are known to be causative of significant human diseases:Bartonella bacilliformis(B. bacilliformis) is the causative agent of Oroya fever and verruga peruana (Peruvian wart);Bartonella quintana(B. quintana) is the causative agent of trench fever;Bartonella henselae(B. henselae) is the causative agent of cat-scratch disease[1-3]. Additionally, several species have been described as coincident zoonotic infection agents includingBartonella alsatica(B. alsatica) [4],Bartonella koehlerae(B. koehlerae)[5],Bartonella vinsoniisubsp.berkhoffii(B. vinsoniisubsp.berkhoffii)[6,7],Bartonella tamiae(B. tamiae)[8],Bartonella rochalimae(B. rochalimae) [9],Bartonella washoensis(B. washoensis)[10] andCandidatus Bartonella mayotimonensis[11].
Bartonellaspecies parasitize the erythrocytes, endothelial cells, monocytes and dendritic cells of mammals[12,13]. Additionally, aBartonellaspecies has been described in loggerhead sea turtle[14]. Bacteria of the genusBartonellaare widespread in domestic and wild animals and are transmitted by a great variety of bloodsucking arthropods including fleas, mites, sand flies and ticks[15,16]. Since 1990s, numerous studies have demonstrated that several mammals such as cats, dogs, rabbits and rodents are potential reservoir hosts ofBartonella. Until now, new host species such as insectivores, bats and exotic pets are continually added in the reservoir hosts’ list. This review provides an update of reservoir host species focusing on small mammals, their potential vectors and case reports ofBartonellainfection in Asia, a hotspot of emerging infectious diseases[17,18]. Hosts such as cats, dogs, and rabbits have been excluded from this review since several review articles have been already written on these reservoir hosts[19,20].
2. Taxonomy and bacteriology of Bartonella
Bacteria of the genusBartonellabelong to the family Bartonellaceae, order Rhizobiales, class Alphaproteobacteria, and phylum Proteobacteria.Bartonellaare closely related withBrucellaspecies andAgrobacterium tumefaciens.Bartonellagenus includes a large diversity ofBartonellaspecies, which were identified from different host species. Since the development of more efficient molecular tools for detection, genetic criteria and species identification are greatly improved[21], and the description of newBartonellaspecies has rapidly increased over the last 10 years and is still continually growing. The genus currently contains more than 30 species and 3 subspecies includingB. alsatica,Bartonella australis,B. bacilliformis,Bartonella birtlesii(B. birtlesii),Bartonella bovis(B. bovis),Bartonella capreoli(B. capreoli),Bartonella chomelii,Bartonella coopersplainsensis(B. coopersplainsensis),Bartonella clarridgeiae(B. clarridgeiae),Bartonella doshiae(B. doshiae),Bartonella durdenii,Bartonella grahamii(B. grahamii),B. henselae,Bartonella japonica(B. japonica),B. koehlerae,Bartonella melophagi,Bartonella phoceensis(B. phoceensis),Bartonella queenslandensis(B. queenslandensis),Bartonella quintana(B. quintana),Bartonella rattaustraliani(B. rattaustraliani),Bartonella rattimassiliensis(B. rattimassiliensis),B. rochalimae,Bartonella schoenbuchensis,Bartonella silvatica(B. silvatica),Bartonella silvicola,B. tamiae, Bartonella taylorii(B. taylorii), Bartonella tribocorum(B. tribocorum), Bartonella vinsoniisubsp.arupensis(B. vinsoniisubsp.arupensis), B. vinsoniisubsp.berkhoffii,Bartonella vinsoniisubsp.vinsonii(B. vinsoniisubsp.vinsonii),B. washoensis,CandidatusBartonella antechini,Candidatus Bartonella mayotimonensis,CandidatusBartonella thailandensis(Table 1). Of these, several species may cause either asymptomatic or mild diseases or severe diseases. Bacteria from this genus are fastidious to growin vitro. The culture on blood agar requires 7 to 45 d in primary isolation. Microscopically, theBartonellaspecies are Gram-negative bacilli. SeveralBartonellaspecies are flagellate,B. bacilliformis,B. bovisandB. clarridgeiae[22,23].

Table 1 List of the currently known described species of Bartonella.
3. Bartonella genomes
The genome sizes ofBartonellaspecies range from 1.5 to 2.5 Mb. SeveralBartonellaspecies includingB. bacilliformis,B. birtlesii,B. clarridgeae,B. grahamii,B. henselae,B. quintana,B. rattimassiliensis,B. rattaustralianiandB. tribocorumwere completely sequenced and two species (B. grahamiiandB. tribocorum) contain plasmid. The first genomes have been described forB. henselaeandB. quintanawith 1.9 and 1.6 Mb, respectively[24], followed by the sequencing of the genomes ofB. bacilliformis(1.4 Mb) (the Institute for Genomic Research, unpublished),B. clarridgeiae(1.5 Mb)[25],B. grahamii(2.3 Mb)[26],B. tribocorum(2.6 Mb)[27],B. rattismassiliensis(2.0 Mb)[28],B. rattaustraliani(2.1 Mb)[29],B. birtlesii(1.8 Mb)[30] andB.quintina(1.6 Mb)[31] among others. The guanine-cytosine content ofBartonellaspecies range from 38.5 mol% forB. quintanato 41.1 mol% forB. vinsonii[2]. Additionally, some species such asB. bacilliformis,B. henselae,B. quintanaandB. vinsoniisubsp.berkhoffiihave been shown to contain phage[32]. The phage particles fromB. bacilliformisandB. vinsoniisubsp. berkhoffiiwere tailed, whereas those fromB. henselaeandB. quintanalacked tails, but all contained 14 kb linear, double-stranded DNA, packaged in a round to head.
To date,Bartonellaspecies are identified using standard polymerase chain reaction (PCR) amplification and a sequencing targeting two housekeeping genes (gltA and rpoB) and the internal transcribed spacer fragment[2].
4. Hosts and reservoirs
Bartonellaspecies may show either low specificity with some species infecting several different host species, while some other species show high specificity by infecting a single host. For example,B. bovisgenerally infects only one ruminant species and is seldom associated with other animals. The prevalence ofBartonellain both wild and domestic mammals has been studied in many different countries. To date, several animal species have been reported as a potential reservoir hosts, such as cats, dogs, rabbits, ruminants, monkeys, marsupials, marine mammals, bats, insectivores and rodents[19,33].
4.1. Rodents
Wild rodents are known to be important reservoir hosts of various pathogens, which are causative of human illnesses including Bartonellosis. Of the current species of the genusBartonella, fifteen species includingB. birtlesii,Bartonella elizabethae (B. elizabethae),B. coopersplainsensis,B. doshiae,B. grahamii,B. japonica,B. phoceensis,B. queenslandensis,B. rattaustraliani,B. rattimassiliensis,B. silvatica,B. taylorii,B. tribocorum,B. vinsoniisubsp.arupensis, B. vinsoniisubsp.vinsonii,B. washoensishave been isolated from various rodent species[2]. Of these,B. birtlesii,B. elizabethae,B. grahamii, andB. washoensisare causative agent of human illnesses.
In Asia, the occurrence ofBartonellainfection in rodent populations have been reported in several countries including China, Japan, Taiwan, Lao PDR, Cambodia, Thailand, Indonesia, Bangladesh and Israel.Bartonellais present in most surveyed rodent populations, with an overall prevalence in the sites investigated ranging from 6% in Korea[34] to 47% in China[35]. Table 2 summarizesBartonellaspecies, host species and geographic distribution ofBartonellain Asia.Bartonellainfections are highly prevalent in rodents in China, Korea, Japan, Russia and Taiwan ranging from 8.6% to 82.3%. SeveralRattusspecies have been found as highly infected including the house ratRattus tanezumi (R. tanezumi). Several species of thegeneraApodemus,Eothenomys,MusandMyodeshave also been shown to hostBartonella. A high diversity ofBartonellaspecies was observed from rodents in this region includingB. elizabethae,B. grahamii,B. queenslandensis,B. phoceensis,B. rattimassiliensis,B. taylorii, B. tribocorumand unknown species.
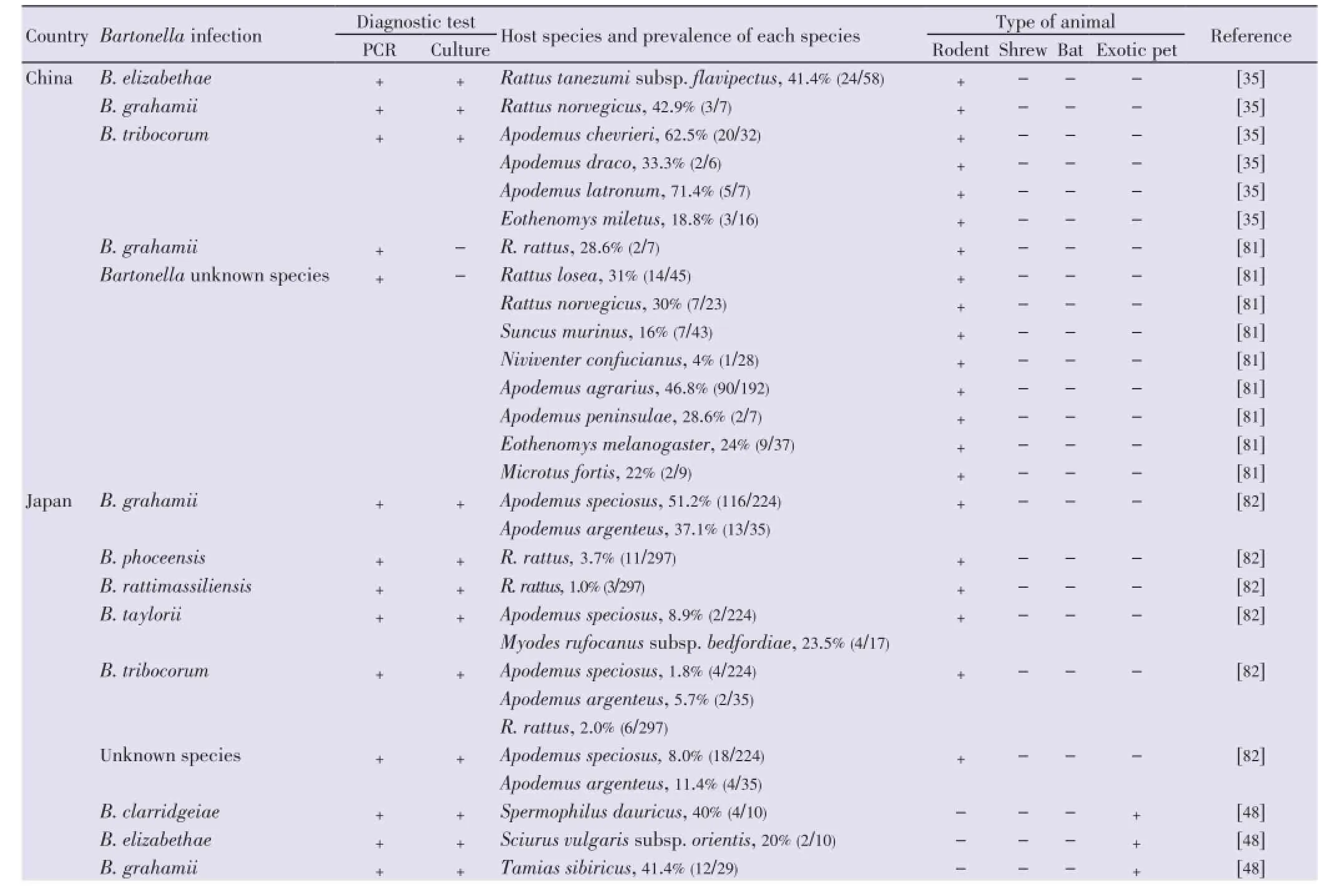
Table 2 Bartonella infection and host species in East Asia.

continued Table 2
In Southeast Asia,Bartonellaspecies have been investigated from rodents in Indonesia, Lao PDR, Cambodia and Thailand[36-40] (Table 3). Again, the genusRattusappears to be the more prevalent rodent genus followed by the generaBandicotaandMus. In South and West Asia, studies ofBartonellain rodents are more limited (Table 4).
Altogether, it appears thatB. coopersplainsensisis mainly found inBandicotaspecies [Bandicota indica(B. indica) andBandicota savilei(B. savilei)] andR. tanezumi;B. elizabethaeinApodemus agrariusandRattusspecies [Rattus rattus(R. rattus) andR. tanezumi];B. grahamiiinRattus norvegicus;B. queenslandensisinMusspecies [Mus cookii(M. cookii),B. phoceensisinR. tanezumi,B. rattimassiliensisinM. cookiiandR. tanezumi;B. rochalimaeinMus cervicolor;B. silvaticainClethrionomys rufocanus;B. tayloriiinC. rufocanus;B. tribocoruminApodemusspecies (A. chevrieri,A. latronum) and inRattusspecies (R. tanezumi,Rattus norvegicus);B. washoensisinPachyuromys duprasi.
4.2. Shrews
Several shrew species live in the domestic areas or peridomestic environments such as agricultural areas. Isolation ofBartonellafrom shrews has been carried out in Asia and North America. In Asia, mostBartonellainfections in shrews have been described in East Asia (Korea and Taiwan) and South Asia (Bangladesh and Nepal)[41-43]. In Southeast Asia, the prevalence ofBartonellain shrews has been reported inSattus murinusin Indonesia[37] (Table 3). Potential new species ofBartonellahas been recently observed inSattus murinusin Cambodia[36].
Finally, it appears thatSattus murinusis a main host forB. elizabethae,B. phoceensis,B. rattimassiliensisandB. tribocorum, althoughCrocidura attenuatais found infected with high prevalence byB. rattimassiliensisandB. tribocorum.
4.3. Bats
The knowledge ofBartonellain bats is still scarce, with only few studies published concerning bats from Asia[44]. Studies on bats and their arthropod vectors havedemonstrated that bats were infected with unknownBartonellaspecies[45]. Another study found new putative species in bats from Kenya[46]. In Asia, reports are even scarcer.Bartonellainfection in bat has been described in Taiwan[47]. Six blood samples ofMiniopterus schreibersiiwere positive with new putativeBartonellaspp. with high prevalence (42.9%, Table 2).
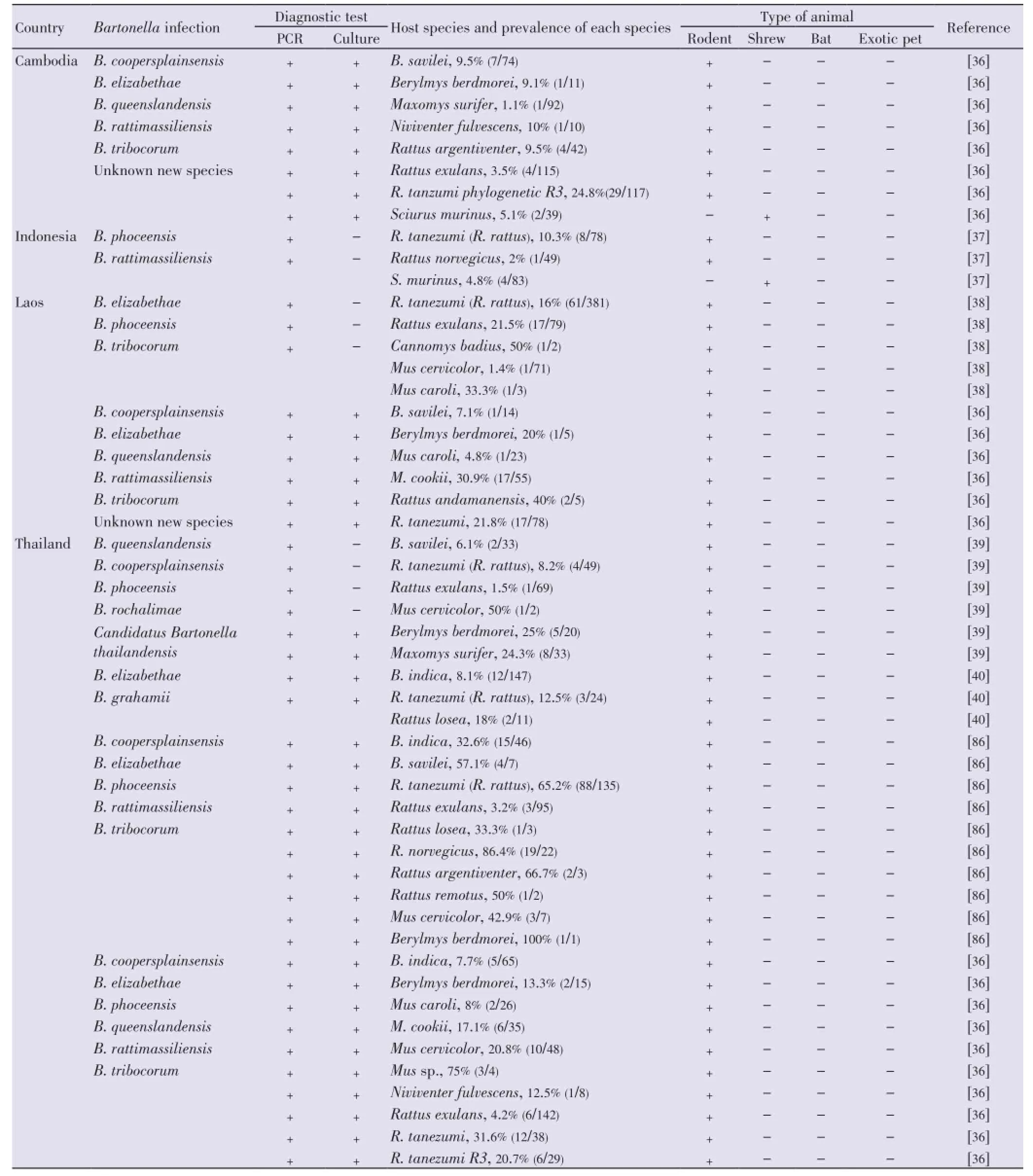
Table 3 Bartonella infection and host species in Southeast Asia.
4.4. Exotic pets
Exotic pets are wildlife that are traded around the world, with important illegal trafficking, and have been (and still being) imported into many countries. The illegal trade without quarantine may cause the dissemination of infectious diseases. However, there are little data onprevalence of infection withBartonellaspp. from exotic small mammal pets in Asia (Table 2). Among them, a study reported that several exotic species in Japan were infected withBartonellaspp. with quite high prevalence[48].

Table 4 Bartonella infection and host species in South and West Asia.

Table 5 Species of Bartonella detected on arthropod vectors from rodents and shrews in Asia.
4.5. Vectors
Several blood-sucking arthropods including fleas, mites, sandflies and ticks have been reported as potential vectors transmittingBartonellaamong animals and between animals and humans.
Of the several blood-sucking arthropods, fleas are the key vectors ofBartonellainfection. Several studies have suggested that fleas, especiallyXenopsylla cheopis, are potential vectors in rodents (Table 5). Ticks, mites and lice are also potential vectors forBartonellainfections inrodents. Epidemiological surveys ofBartonellainfection in ticks, mites and lice have been done in Korea, Taiwan and Thailand[16,34,42,49].
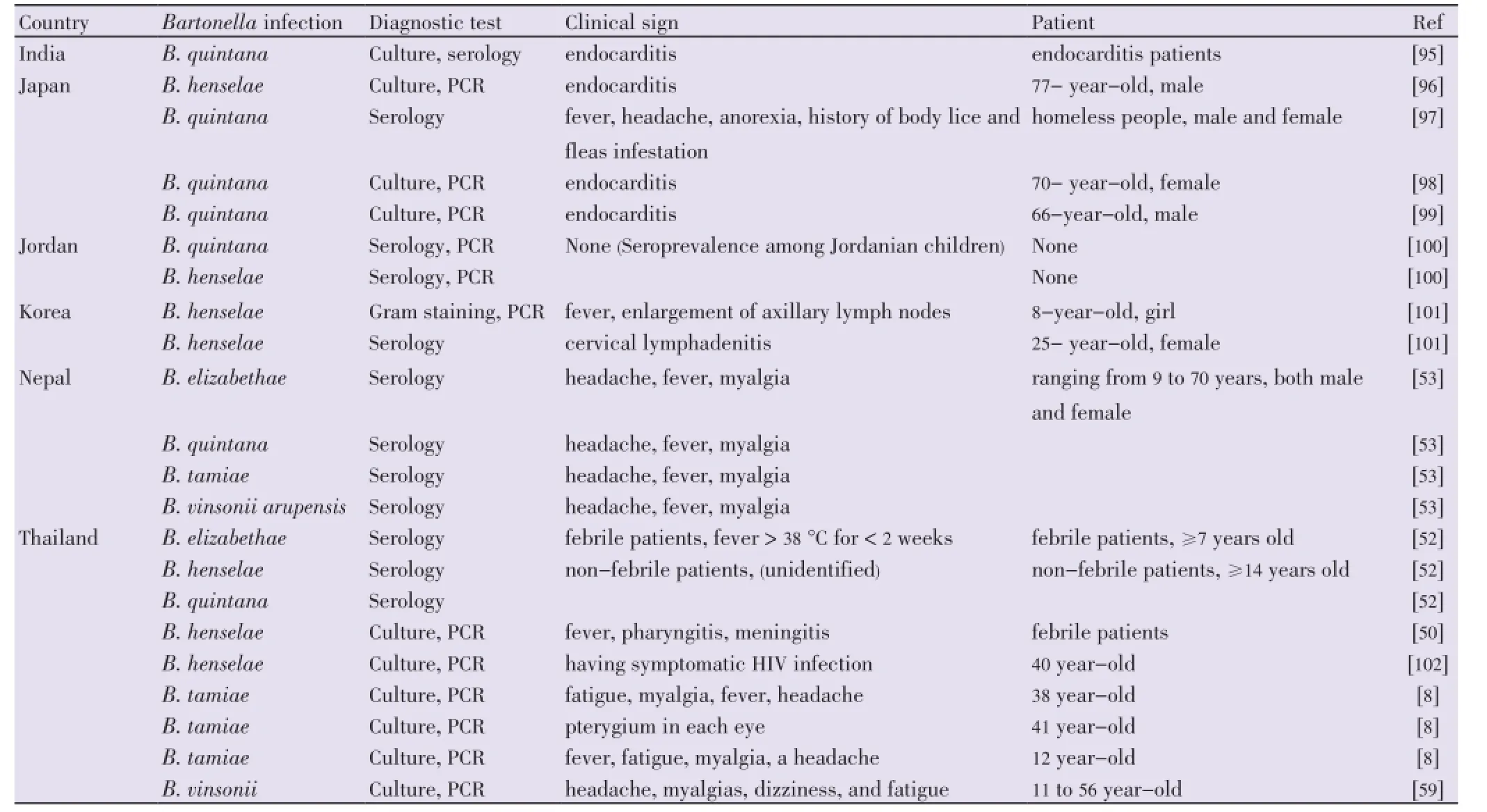
Table 6 Case reports of Bartonella infection in Asia.
6. Zoonotic diseases and case reports in Asia
Bartonellaspecies are important zoonotic bacteria with increasing array of emerging infections in humans and animals. Bartonellosis has been described in Asia. Possible routes of transmission are contacts with infected animals or blood-sucking arthropod via their feces. FourBartonellaspecies includingB. henselae,B. quintana,B. elizabethaeandB. tamiaehave been reported as causative agents of several human diseases in Asia (Table 6). However, few cases of human infections withBartonellabacteria of rodent origins have been reported. The first case ofBartonellaendocarditis in Thailand was found in a 57-year-old male poultry farmer in Khon Kaen province, where the source of pathogen was a cat.B. tamiaewas isolated from three febrile patients in Thailand[8]. In another report, several farmers were supposed to be exposed to infected rodents at home or fields[50]. Recently, four patients were found infected byB. vinsoniiin Northern and Northeastern Thailand, with dogs and rats supposed to be reservoirs[51]. Additional serological studies evidenced exposure to rodentBartonellaspp. in Thailand but also in Nepal[52,53]. However, to date, onlyB. elizabethaewas found in rodent surveys, where the most common rodents were murid fromApodemusspp. andRattusspp. (and particularly the commensalR. rattusandR. tanezumi).
7. Perspectives
The diversity ofBartonellaspecies is far from being known as the recent study of Jiyiponget al. emphasized on three new putative species[36]. Indeed, Southeast Asia is a hotspot of mammal species[54] and a diversification centre for several rodent families. Two-thirds of living rodent species belongs to the family Muridae[55], which also represents most of the rodents found in Southeast Asia with 35 species[56]. It appears that less than 50% of these murid species have been investigated for the presence ofBartonellaspecies (Table 3). The knowledge is even poorer for shrews and bats. This emphasizes the need to increase the screening, detection and characterization ofBartonellain small mammals[57].
SinceBartonellaspecies may be transmitted to humans by ticks, fleas and lice, surveys ofBartonelladistribution within arthropods are needed. In particular, the population changes of these vectors ofBartonellain relation to the impacts of habitat and land-use changes should be better investigated[57].
Rodents, shrews and bats live in a wide range of habitats that are frequented by humans[58], which warrants further investigations on the transmission ecology ofBartonellain order to improve prevention ofBartonellainfections. Identification of risky habitats for human transmission is needed to develop a surveillance strategy, which could be done only after the improvement of our knowledge on the diversity ofBartonellain small mammals and in their arthropod vectors.
This review by giving up-to-date list of reservoirs will help develop a strategy of reservoirs, vectors and habitatsprone to be sources of outbreaks and/or emergingBartonellainfections in humans.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgements
The study was supported by the French ANR CERoPath project (number ANR 07 BDIV 012) (www.ceropath.org), the French ANR BiodivHealthSEA project (number ANR 11 CPEL 002) (www.biodivhealthsea.org), the Infectiopole Sud and the Center of Excellence on Agricultural Biotechnology (Science and Technology Postgraduate Education and Research Development Office, Office of Higher Education Commission, Ministry of Education) (AG-BIO/PERDO-CHE). Part of this research was also funded by the Center of Advanced Studies for Agriculture and Food, Institute for Advanced Studies, Kasetsart University. We thank the editor and a reviewer for helpful comments.
Comments
Background
Bartonellais an important bacteria that can make various infections in different animals specially humans. This bacteria has various hosts and it is transmitted by several vectors. Species ofBartonellaare Gram-negative intracellular bacteria that infects erythrocytes of various mammalian and non-mammalian animals and mainly transmitted by blood-sucking arthropod vectors.
Research frontiers
This is a review article aboutBartonellainfection in small mammals including rodents, insectivores, bats and exotic small mammal pets and their vectors in Asia. This article dicusses the present species ofBartonellain animals cited.
Related reports
This is a review article and it is based on various reports that are already reported. Various articles mentioned role of different species ofBartonellain making infections in animals (mammals and non-mammals).Bartonellain different animals have been recognized.
Innovations and breakthroughs
This review article investigates various species ofBartonellain animals that already a little have been studied and can offer good information about the role ofBartonellaat causing infection in animals cited.
Applications
This review article surveys prevalence of various species ofBartonellain different areas of Asia. Informations offered in this article can be useful for more studies in future.
Peer review
This is a good research about the presence of various species ofBartonellaand its role at causing different infections. The consideration of authors to various reservoirs of this bacteria in animals that a little have been already surveyed is interesting. Diagnostic tests referred in this article are very useful.
[1] Boulouis HJ, Chang CC, Henn JB, Kasten RW, Chomel BB. Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet Res 2005; 36: 383-410.
[2] Saisongkorh W, Rolain JM, Suputtamongkol Y, Raoult D. Emerging Bartonella in humans and animals in Asia and Australia. J Med Assoc Thai 2009; 92: 707-731.
[3] Angelakis E, Billeter SA, Breitschwerdt EB, Chomel BB, Raoult D. Potential for tick-borne bartonelloses. Emerg Infect Dis 2010; 16: 385-391.
[4] Raoult D, Roblot F, Rolain JM, Besnier JM, Loulergue J, Bastides F, et al. First isolation of Bartonella alsatica from a valve of a patient with endocarditis. J Clin Microbiol 2006; 44: 278-279.
[5] Avidor B, Graidy M, Efrat G, Leibowitz C, Shapira G, Schattner A, et al. Bartonella koehlerae, a new cat-associated agent of culture-negative human endocarditis. J Clin Microbiol 2004; 42: 3462-3468.
[6] Roux V, Eykyn SJ, Wyllie S, Raoult D. Bartonella vinsonii subsp. berkhoffii as an agent of afebrile blood culture-negative endocarditis in a human. J Clin Microbiol 2000; 38: 1698-1700.
[7] Olarte L, Ampofo K, Thorell EA, Sanderson S, Doby E, Pavia AT, et al. Bartonella vinsonii endocarditis in an adolescent with congenital heart disease. Pediatr Infect Dis J 2012; 31: 531-534.
[8] Kosoy M, Morway C, Sheff KW, Bai Y, Colborn J, Chalcraft L, et al. Bartonella tamiae sp. nov., a newly recognized pathogen isolated from three human patients from Thailand. J Clin Microbiol 2008; 46: 772-775.
[9] Eremeeva ME, Gerns HL, Lydy SL, Goo JS, Ryan ET, Mathew SS, et al. Bacteremia, fever, and spleenomegaly caused by a newly recognized Bartonella species. N Engl J Med 2007; 356: 2381-2387.
[10] Kosoy M, Murray M, Gilmore RD Jr, Bai Y, Gage KL. Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J Clin Microbiol 2003; 41: 645-650.
[11] Lin EY, Tsigrelis C, Baddour LM, Lepidi H, Rolain JM, Patel R, et al. Candidatus Bartonella mayotimonensis and endocarditis. Emerg Infect Dis 2010; 16: 500-503.
[12] Dehio C. Bartonella interactions with endothelial cells and erythrocytes. Trends Microbiol 2001; 9: 279-285.
[13] Dehio M, Quebatte M, Foser S, Certa U. The transcriptional response of human endothelial cells to infection with Bartonella henselae is dominated by genes controlling innate immune responses, cell cycle, and vascular remodelling. Thromb Haemost 2005; 94: 347-361.
[14] Valentine KH, Harms CA, Cadenas MB, Birkenheuer AJ, Marr HS, Braun-McNeill J, et al. Bartonella DNA in loggerhead sea turtles. Emerg Infect Dis 2007; 13: 949-950.
[15] Tsai YL, Chang CC, Chuang ST, Chomel BB. Bartonella species and their ectoparasites: selective host adaptation or strain selection between the vector and the mammalian host? Comp Immunol Microbiol Infect Dis 2011; 34: 299-314.
[16] Billeter SA, Levy MG, Chomel BB, Breitschwerdt EB. Vector transmission of Bartonella species with emphasis on the potential for tick transmission. Med Vet Entomol 2008; 22: 1-15.
[17] Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature 2008; 451: 990-993
[18] Coker RJ, Hunter BM, Rudge JW, Liverani M, Hanvoravongchai P. Emerging infectious diseases in Southeast Asia: regional challenges to control. Lancet 2011; 377: 599-609.
[19] Chomel BB, Boulouis HJ, Maruyama S, Breitschwerdt EB. Bartonella spp. in pets and effect on human health. Emerg Infect Dis 2006; 12: 389-394.
[20] Breitschwerdt EB, Kordick DL. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin Microbiol Rev 2000; 13: 428-438.
[21] La Scola B, Zeaiter Z, Khamis A, Raoult D. Gene-sequencebased criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol 2003; 11: 318-321.
[22] Benson LA, Kar S, McLaughlin G, Ihler GM. Entry of Bartonella bacilliformis into erythrocytes. Infect Immun 1986; 54: 347-353.
[23] Rolain JM, Chanet V, Laurichesse H, Lepidi H, Beytout J, Raoult D. Cat scratch disease with lymphadenitis, vertebral osteomyelitis, and spleen abscesses. Ann N Y Acad Sci 2003; 990: 397-403.
[24] Alsmark CM, Frank AC, Karlberg EO, Legault BA, Ardell DH, Canbäck B, et al. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc Natl Acad Sci USA 2004; 101: 9716-9721.
[25] Engel P, Salzburger W, Liesch M, Chang CC, Maruyama S, Lanz C, et al. Parallel evolution of a type IV secretion system in radiating lineages of the host-restricted bacterial pathogen Bartonella. PLoS Genet 2011; 7: e1001296.
[26] Berglund EC, Frank AC, Calteau A, Pettersson OV, Granberg F, Eriksson AS, et al. Run-off replication of host-adaptability genes is associated with gene transfer agents in the genome of mouse-infecting Bartonella grahamii. PLoS Genet 2009; 5: e1000546.
[27] Saenz HL, Engel P, Stoeckli MC, Lanz C, Raddatz G, Vayssier-Taussat M, et al. Genomic analysis of Bartonella identifies type IV secretion systems as host adaptability factors. Nat Genet 2007; 39: 1469-1476.
[28] Merhej V, Croce O, Robert C, Rolain JM, Raoult D. Genome sequence of Bartonella rattimassiliensis, a bacterium isolated from European Rattus norvegicus. J Bacteriol 2012; 194: 7013.
[29] Merhej V, Croce O, Robert C, Rolain JM, Raoult D. Genome sequence of Bartonella rattaustraliani, a bacterium isolated from an Australian rat. J Bacteriol 2012; 194: 7012.
[30] Rolain JM, Vayssier-Taussat M, Gimenez G, Robert C, Fournier PE, Raoult D. Genome sequence of Bartonella birtlesii, a bacterium isolated from small rodents of the genus Apodemus. J Bacteriol 2012; 194: 4779.
[31] Li H, Tong YG, Huang Y, Bai JY, Yang H, Liu W, et al. Complete genome sequence of Bartonella quintana, a bacterium isolated from rhesus macaques. J Bacteriol 2012; 194: 6347.
[32] Chenoweth MR, Somerville GA, Krause DC, O’Reilly KL, Gherardini FC. Growth characteristics of Bartonella henselae in a novel liquid medium: primary isolation, growth-phasedependent phage induction, and metabolic studies. Appl Environ Microbiol 2004; 70: 656-663.
[33] Kosoy M, Hayman DT, Chan KS. Bartonella bacteria in nature: where does population variability end and a species start? Infect Genet Evol 2012; 12: 894-904.
[34] Chae JS, Yu DH, Shringi S, Klein TA, Kim HC, Chong ST, et al. Microbial pathogens in ticks, rodents and a shrew in northern Gyeonggi-do near the DMZ, Korea. J Vet Sci 2008; 9: 285-293.
[35] Bai Y, Kosoy MY, Maupin GO, Tsuchiya KR, Gage KL. Genetic and ecologic characteristics of Bartonella communities in rodents in southern China. Am J Trop Med Hyg 2002; 66: 622-627.
[36] Jiyipong T, Jittapalapong S, Morand S, Raoult D, Rolain JM. Prevalence and genetic diversity of Bartonella spp. in small mammals from Southeastern Asia. Appl Environ Microbiol 2012; 78: 8463-8466.
[37] Winoto IL, Goethert H, Ibrahim IN, Yuniherlina I, Stoops C, Susanti I, et al. Bartonella species in rodents and shrews in the greater Jakarta area. Southeast Asian J Trop Med Public Health 2005; 36: 1523-1529.
[38] Angelakis E, Khamphoukeo K, Grice D, Newton PN, Roux V, Aplin K, et al. Molecular detection of Bartonella species in rodents from the Lao PDR. Clin Microbiol Infect 2009; 15: 95-97. [39] Saisongkorh W, Wootta W, Sawanpanyalert P, Raoult D, Rolain JM. “Candidatus Bartonella thailandensis”: a new genotype of Bartonella identified from rodents. Vet Microbiol 2009; 139: 197-201.
[40] Castle KT, Kosoy M, Lerdthusnee K, Phelan L, Bai Y, Gage KL, et al. Prevalence and diversity of Bartonella in rodents of northern Thailand: a comparison with Bartonella in rodents from southern China. Am J Trop Med Hyg 2004; 70: 429-433.
[41] Bai Y, Montgomery SP, Sheff KW, Chowdhury MA, Breiman RF, Kabeya H, et al. Bartonella strains in small mammals from Dhaka, Bangladesh, related to Bartonella in America and Europe. Am J Trop Med Hyg 2007; 77: 567-570.
[42] Tsai YL, Chuang ST, Chang CC, Kass PH, Chomel BB. Bartonella species in small mammals and their ectoparasites in Taiwan. Am J Trop Med Hyg 2010; 83: 917-923.
[43] Kim CM, Kim JY, Yi YH, Lee MJ, Cho MR, Shah DH, et al. Detection of Bartonella species from ticks, mites and small mammals in Korea. J Vet Sci 2005; 6: 327-334.
[44] Bai Y, Kosoy M, Recuenco S, Alvarez D, Moran D, Turmelle A, et al. Bartonella spp. in bats, Guatemala. Emerg Infect Dis 2011; 17:1269-1272.
[45] Bai Y, Recuenco S, Turmelle A, Osikowicz LM, Gómez J, Rupprecht C, et al. Prevalence and diversity of Bartonella spp. in bats in Peru. Am J Trop Med Hyg 2012; 87: 518-523.
[46] Kosoy M, Bai Y, Lynch T, Kuzmin IV, Niezgoda M, Franka R, et al. Bartonella spp. in bats, Kenya. Emerg Infect Dis 2010; 16: 1875-1881.
[47] Lin JW, Hsu YM, Chomel BB, Lin LK, Pei JC, Wu SH, et al. Identification of novel Bartonella spp. in bats and evidence of Asian gray shrew as a new potential reservoir of Bartonella. Vet Microbiol 2012; 156: 119-126.
[48] Inoue K, Maruyama S, Kabeya H, Hagiya K, Izumi Y, Une Y, et al. Exotic small mammals as potential reservoirs of zoonotic Bartonella spp. Emerg Infect Dis 2009; doi: 10.3201/eid1504.081223.
[49] Kabeya H, Colborn JM, Bai Y, Lerdthusnee K, Richardson JH, Maruyama S, et al. Detection of Bartonella tamiae DNA in ectoparasites from rodents in Thailand and their sequence similarity with bacterial cultures from Thai patients. Vector Borne Zoonotic Dis 2010; 10: 429-434.
[50] Kosoy M, Bai Y, Sheff K, Morway C, Baggett H, Maloney SA, et al. Identification of Bartonella infections in febrile human patients from Thailand and their potential animal reservoirs. Am J Trop Med Hyg 2010; 82: 1140-1145.
[51] Bai Y, Kosoy MY, Diaz MH, Winchell J, Baggett H, Maloney SA, et al. Bartonella vinsonii subsp. arupensis in humans, Thailand. Emerg Infect Dis 2012; doi: 10.3201/eid1806.111750.
[52] Bhengsri S, Baggett HC, Peruski LF, Morway C, Bai Y, Fisk TL, et al. Bartonella seroprevalence in rural Thailand. Southeast Asian J Trop Med Public Health 2011; 42: 687-692.
[53] Myint KS, Gibbons RV, Iverson J, Shrestha SK, Pavlin JA, Mongkolsirichaikul D, et al. Serological response to Bartonella species in febrile patients from Nepal. Trans R Soc Trop Med Hyg 2011; 105: 740-742.
[54] Schipper J, Chanson JS, Chiozza F, Cox NA, Hoffmann M, Katariya V, et al. The status of the world’s land and marine mammals: diversity, threat, and knowledge. Science 2008; 322: 225-230.
[55] Wilson DE, Reeder DA. Mammal species of the world: a taxonomic and geographic reference. 3rd ed. Maryland: Johns Hopkins University Press; 2005.
[56] Aplin KP, Brown PR, Jacob J, Krebs CJ, Singleton GR. Field methods for rodent studies in Asia and the Indo-Pacific. Canberra: ACIAR; 2003.
[57] Bordes F, Herbreteau V, Dupuy S, Chaval Y, Tran A, Morand S. The diversity of microparasites of rodents: a comparative analysis that helps in identifying rodent-borne rich habitats in Southeast Asia. Infect Ecol Epidemiol 2013; doi: 10.3402/iee. v3i0.20178.
[58] Boulouis HJ, Chang CC, Henn JB, Kssten RW, Chomel BB. Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet Res 2005; 36: 383-410.
[59] Heller R, Kubina M, Mariet P, Riegel P, Delacour G, Dehio C, et al. Bartonella alsatica sp. nov., a new Bartonella species isolated from the blood of wild rabbits. Int J Syst Bacteriol 1999; 49: 283-288.
[60] Fournier PE, Taylor C, Rolain JM, Barrassi L, Smith G, Raoult D, et al. Bartonella australis sp. nov. from kangaroos, Australia. Emerg Infect Dis 2007; 13: 1961-1963.
[61] Birtles RJ, Harrison TG, Fry NK, Saunders NA, Taylor AG. Taxonomic considerations of Bartonella bacilliformis based on phylogenetic and phenotypic characteristics. FEMS Microbiol Lett 1991; 67: 187-191.
[62] Bermond D, Heller R, Barrat F, Delacour G, Dehio C, Alliot A, et al. Bartonella birtlesii sp. nov., isolated from small mammals (Apodemus spp.). Int J Syst Evol Microbiol 2000; 50: 1973-1979.
[63] Bermond D, Boulouis HJ, Heller R, Van Laere G, Monteil H, Chomel BB, et al. Bartonella bovis Bermond et al. sp. nov. and Bartonella capreoli sp. nov., isolated from European ruminants. Int J Syst Evol Microbiol 2002; 52: 383-390.
[64] Maillard R, Riegel P, Barrat F, Bouillin C, Thibault D, Gandoin C, et al. Bartonella chomelii sp. nov., isolated from French domestic cattle (Bos taurus). Int J Syst Evol Microbiol 2004; 54: 215-220.
[65] Lawson PA, Clarridge JE, Collins MD. Description of Bartonella clarridgeiae sp. nov. isolated from the cat of a patient with Bartonella henselae septicemia. Med Microbiol Lett 1996; 5: 64-73.
[66] Gundi VA, Taylor C, Raoult D, La Scola B. Bartonella rattaustraliani sp. nov., Bartonella queenslandensis sp. nov. and Bartonella coopersplainsensis sp. nov., identified in Australian rats. Int J Syst Evol Microbiol 2009; 59: 2956-2961.
[67] Birtles RJ, Harrison TG, Saunders NA, Molyneux DH. Proposals to unify the genera Grahamella and Bartonella, with descriptions of Bartonella talpae comb. nov., Bartonella peromysci comb. nov., and three new species, Bartonella grahamii sp. nov., Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int J Syst Bacteriol 1995; 45: 1-8.
[68] Daly JS, Worthington MG, Brenner DJ, Moss CW, Hollis DG, Weyant RS, et al. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol 1993; 31: 872-881.
[69] Regnery RL, Anderson BE, Clarridge JE 3rd, Rodriguez-Barradas MC, Jones DC, Carr JH. Characterization of a novel Rochalimaea species, R. henselae sp. nov., isolated from blood of a febrile, human immunodeficiency virus-positive patient. J Clin Microbiol 1992; 30: 265-274.
[70] Inoue K, Kabeya H, Shiratori H, Ueda K, Kosoy MY, Chomel BB, et al. Bartonella japonica sp. nov. and Bartonella silvatica sp. nov., isolated from Apodemus mice. Int J Syst Evol Microbiol 2010; 60: 759-763.
[71] Droz S, Chi B, Horn E, Steigerwalt AG, Whitney AM, Brenner DJ. Bartonella koehlerae sp. nov., isolated from cats. J Clin Microbiol 1999; 37: 1117-1122.
[72] Chomel BB, McMillan-Cole AC, Kasten RW, Stuckey MJ, Sato S, Maruyama S, et al. Candidatus Bartonella merieuxii, a potential new zoonotic Bartonella species in canids from Iraq. PLoS Negl Trop Dis 2012; 6: e1843.
[73] Gundi VA, Davoust B, Khamis A, Boni M, Raoult D, La Scola B. Isolation of Bartonella rattimassiliensis sp. nov. and Bartonella phoceensis sp. nov. from European Rattus norvegicus. J Clin Microbiol 2004; 42: 3816-3818.
[74] Brenner DJ, O’Connor SP, Winkler HH, Steigerwalt AG. Proposals to unify the genera Bartonella and Rochalimaea, with descriptions of Bartonella quintana comb. nov., Bartonella vinsonii comb. nov., Bartonella henselae comb. nov. and Bartonella elizabethae comb. nov., and to remove the familyBartonellaceae from the order Rickettsiales. Int J Syst Bacteriol 1993; 43: 777-786.
[75] Dehio C, Lanz C, Pohl R, Behrens P, Bermond D, Piémont Y, et al. Bartonella schoenbuchii sp. nov., isolated from the blood of wild roe deer. Int J Syst Evol Microbiol 2001; 51: 1557-1565.
[76] Heller R, Riegel P, Hansmann Y, Delacour G, Bermond D, Dehio C, et al. Bartonella tribocorum sp. nov., a new Bartonella species isolated from the blood of wild rats. Int J Syst Bacteriol 1998; 48: 1333-1339.
[77] Welch DF, Carroll KC, Hofmeister EK, Persing DH, Robison DA, Steigerwalt AG, et al. Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis, from a cattle rancher: identity with isolates found in conjunction with Borrelia burgdorferi and Babesia microti among naturally infected mice. J Clin Microbiol 1999; 37: 2598-2601.
[78] Hofmeister EK, Kolbert CP, Abdulkarim AS, Magera JM, Hopkins MK, Uhl JR, et al. Cosegregation of a novel Bartonella species with Borrelia burgdorferi and Babesia microti in Peromyscus leucopus. J Infect Dis 1998; 177: 409-416.
[79] Breitschwerdt EB, Kordick DL, Malarkey DE, Keene B, Hadfield TL, Wilson K. Endocarditis in a dog due to infection with a novel Bartonella subspecies. J Clin Microbiol 1995; 33: 154-160.
[80] Kordick DL, Swaminathan B, Greene CE, Wilson KH, Whitney AM, O’Connor S, et al. Bartonella vinsonii subsp. berkhoffii subsp. nov., isolated from dogs; Bartonella vinsonii subsp. vinsonii; and emended description of Bartonella vinsonii. Int J Syst Bacteriol 1996; 46: 704-709.
[81] Liu Q, Sun J, Lu L, Fu G, Ding G, Song X, et al. Detection of Bartonella species in small mammals from Zhejiang Province, China. J Wildl Dis 2010; 46: 179-185.
[82] Inoue K, Maruyama S, Kabeya H, Yamada N, Ohashi N, Sato Y, et al. Prevalence and genetic diversity of Bartonella species isolated from wild rodents in Japan. Appl Environ Microbiol 2008; 74: 5086-5092.
[83] Lin JW, Chen CY, Chen WC, Chomel BB, Chang CC. Isolation of Bartonella species from rodents in Taiwan including a strain closely related to ‘Bartonella rochalimae’ from Rattus norvegicus. J Med Microbiol 2008; 57: 1496-1501.
[84] Hsieh JW, Tung KC, Chen WC, Lin JW, Chien LJ, Hsu YM, et al. Epidemiology of Bartonella infection in rodents and shrews in Taiwan. Zoonoses Public Health 2010; 57: 439-446.
[85] Samoylenko I, Raoult D, Malkova M, Tancev A, Yakimenko V, Rudakov N, et al. Detection of alpha-proteobacteria in rodents in a steppe-forest zone of Western Siberia. Clin Microbiol Infect 2009; 15: 127-129.
[86] Bai Y, Kosoy MY, Lerdthusnee K, Peruski LF, Richardson JH. Prevalence and genetic heterogeneity of Bartonella strains cultured from rodents from 17 provinces in Thailand. Am J Trop Med Hyg 2009; 81: 811-816.
[87] Gundi VA, Kosoy MY, Myint KS, Shrestha SK, Shrestha MP, Pavlin JA, et al. Prevalence and genetic diversity of Bartonella species detected in different tissues of small mammals in Nepal. Appl Environ Microbiol 2010; 76: 8247-8254.
[88] Harrus S, Bar-Gal GK, Golan A, Elazari-Volcani R, Kosoy MY, Morick D, et al. Isolation and genetic characterization of a Bartonella strain closely related to Bartonella tribocorum and Bartonella elizabethae in Israeli commensal rats. Am J Trop Med Hyg 2009; 81: 55-58.
[89] Morick D, Baneth G, Avidor B, Kosoy MY, Mumcuoglu KY, Mintz D, et al. Detection of Bartonella spp. in wild rodents in Israel using HRM real-time PCR. Vet Microbiol 2009; 139: 293-297.
[90] Marie JL, Fournier PE, Rolain JM, Briolant S, Davoust B, Raoult D. Molecular detection of Bartonella quintana, B. elizabethae, B. koehlerae, B. doshiae, B. taylorii, and Rickettsia felis in rodent fleas collected in Kabul, Afghanistan. Am J Trop Med Hyg 2006; 74: 436-439.
[91] Li DM, Liu QY, Yu DZ, Zhang JZ, Gong ZD, Song XP. Phylogenetic analysis of Bartonella detected in rodent fleas in Yunnan, China. J Wildl Dis 2007; 43: 609-617.
[92] Kim BJ, Kim SJ, Kang JG, Ko S, Won S, Kim H, et al. First report for the seasonal and annual prevalence of flea-borne Bartonella from rodents and soricomorphs in the Republic of Korea. Vector Borne Zoonotic Dis 2013; 13: 457-467.
[93] Morick D, Krasnov BR, Khokhlova IS, Shenbrot GI, Kosoy MY, Harrus S. Bartonella genotypes in fleas (Insecta: Siphonaptera) collected from rodents in the Negev desert, Israel. Appl Environ Microbiol 2010; 76: 6864-6869.
[94] Parola P, Sanogo OY, Lerdthusnee K, Zeaiter Z, Chauvancy G, Gonzalez JP, et al. Identification of Rickettsia spp. and Bartonella spp. in from the Thai-Myanmar border. Ann N Y Acad Sci 2003; 990: 173-181.
[95] Balakrishnan N, Menon T, Fournier PE, Raoult D. Bartonella quintana and Coxiella burnetii as causes of endocarditis, India. Emerg Infect Dis 2008; 14: 1168-1169.
[96] Tsuneoka H, Yanagihara M, Otani S, Katayama Y, Fujinami H, Nagafuji H, et al. A first Japanese case of Bartonella henselaeinduced endocarditis diagnosed by prolonged culture of a specimen from the excised valve. Diagn Microbiol Infect Dis 2010; 68: 174-176.
[97] Seki N, Sasaki T, Sawabe K, Sasaki T, Matsuoka M, Arakawa Y, et al. Epidemiological studies on Bartonella quintana infections among homeless people in Tokyo, Japan. Jpn J Infect Dis 2006; 59: 31-35.
[98] Yamada Y, Ohkusu K, Yanagihara M, Tsuneoka H, Ezaki T, Tsuboi J, et al. Prosthetic valve endocarditis caused by Bartonella quintana in a patient during immunosuppressive therapies for collagen vascular diseases. Diagn Microbiol Infect Dis 2011; 70: 395-398.
[99] Yoda M, Hata M, Sezai A, Unosawa S, Furukawa N, Minami K. First report of Bartonella quintana endocarditis in Japan. Circ J 2008; 72: 1022-1024.
[100] Al-Majali AM, Al-Qudah KM. Seroprevalence of Bartonella henselae and Bartonella quintana infections in children from Central and Northern Jordan. Saudi Med J 2004; 25: 1664-1669.
[101] Yoon HJ, Lee WC, Choi YS, Cho S, Song YG, Choi JY, et al. Cervical lymphadenitis in a patient coinfected with Toxoplasma gondii and Bartonella henselae. Vector Borne Zoonotic Dis 2010; 10: 415-419.
[102] Paitoonpong L, Chitsomkasem A, Chantrakooptungool S, Kanjanahareutai S, Tribuddharat C, Srifuengfung S. Bartonella henselae: first reported isolate in a human in Thailand. Southeast Asian J Trop Med Public Health 2008; 39: 123-129.
10.12980/APJTB.4.2014C742
*Corresponding author: Jean-Marc Rolain, Research unit on infectious and emerging tropical diseases (URMITE) CNRS-IRD-INSERM UMR 7278, IHU Méditerranée Infection, Faculty of Medicine and Pharmacy, Aix-Marseille-University, Marseille, France.
E-mail: jean-marc.rolain@univ-amu.fr
Foundation Project: Supported by the French ANR CERoPath project (number ANR 07 BDIV 012) and the French ANR BiodivHealthSEA project (number ANR 11 CPEL 002).
Article history:
Received 28 Dec 2013
Received in revised form 20 Feb, 2nd revised form 3 Apr, 3rd revised form 20 Apr 2014
Accepted 20 May 2014
Available online 14 Sep 2014
 Asian Pacific Journal of Tropical Biomedicine2014年10期
Asian Pacific Journal of Tropical Biomedicine2014年10期
- Asian Pacific Journal of Tropical Biomedicine的其它文章
- Antioxidant, antimicrobial, cytotoxic and analgesic activities of ethanolic extract of Mentha arvensis L.
- Efficacy of seed extracts of Annona squamosa and Annona muricata (Annonaceae) for the control of Aedes albopictus and Culex quinquefasciatus (Culicidae)
- In vitro antioxidant and anti-inflammatory activities of Korean blueberry (Vaccinium corymbosum L.) extracts
- In vitro antibacterial activity of leaf extracts of Zehneria scabra and Ricinus communis against Escherichia coli and methicillin resistance Staphylococcus aureus
- Production and purification of a bioactive substance against multi-drug resistant human pathogens from the marine-sponge-derived Salinispora sp.
- Calcinosis circumscripta in a captive African cheetah (Acinonyx jubatus)
