Genetic polymorphisms of GSTM1, GSTP1 and GSTT1 genes and lung cancer susceptibility in the Bangladeshi population
Mir Muhammad Nasir Uddin, Maizbha Uddin Ahmed, Mohammad Safiqul Islam, Mohammad Siddiqul Islam, Muhammad Shahdaat Bin Sayeed, Yearul Kabir, Abul Hasnat*
1Department of Clinical Pharmacy and Pharmacology, Faculty of Pharmacy, University of Dhaka, Dhaka, Dhaka-1000, Bangladesh
2Department of Pharmacy, Noakhali Science and Technology University, Sonapur, Noakhali-3814, Bangladesh
3Department of Biochemistry and Molecular Biology, University of Dhaka, Dhaka-1000, Bangladesh
Genetic polymorphisms of GSTM1, GSTP1 and GSTT1 genes and lung cancer susceptibility in the Bangladeshi population
Mir Muhammad Nasir Uddin1, Maizbha Uddin Ahmed1, Mohammad Safiqul Islam2, Mohammad Siddiqul Islam1, Muhammad Shahdaat Bin Sayeed1, Yearul Kabir3, Abul Hasnat1*
1Department of Clinical Pharmacy and Pharmacology, Faculty of Pharmacy, University of Dhaka, Dhaka, Dhaka-1000, Bangladesh
2Department of Pharmacy, Noakhali Science and Technology University, Sonapur, Noakhali-3814, Bangladesh
3Department of Biochemistry and Molecular Biology, University of Dhaka, Dhaka-1000, Bangladesh
PEER REVIEW
Peer reviewer
Dr. Md. Kamrul Hossain, Professor and Chairman, Department of Pharmacy, University of Chittagong-4331, Chittagong, Bangladesh.
Tel: +880 31 2606001 4486, 621726
E-mail: mkhossain73@yahoo.com
Comments
This is an interesting study of the association ofGSTfamily genes and lung cancer in the Bangladeshi population asGSTM1, GSTTI and GSTP1 genotypes are well established risk factors for lung cancer. The interaction of GSTP1 gene and tobacco use on lung cancer risk is quite interesting in the current study. The finding about GSTP1 genotyping and it’s correlation to lung cancer is also quite interesting.
Details on Page 987
Article history:
Received 29 Aug 2014
Received in revised form 9 Sep 2014
Accepted 8 Oct 2014
Available online 17 Oct 2014
Objective:To verify possible associations between polymorphisms of glutathione S-transferase Mu (GSTM1), glutathione S-transferase θ (GSTT1) and glutathione S-transferase Pi (GSTP1) genes and susceptibility to lung cancer.
Lung cancer, Glutathione S-transferase, Genetic polymorphism, Smoking
1. Introduction
Lung cancer is currently one of the most common malignant diseases and is responsible for the leading cause of cancer related deaths worldwide[1,2]. It is considered to be the leading cancer site in males and accounts for 17% of the total new cancer cases and 23% of the total cancer deaths. Among females, it was the fourth most commonly diagnosed cancer and the second leading cause of cancer death[3]. Overall five-year survival rates remain poor and are in the range of 5% to10%[4]. According to the WorldHealth Organization data published in April 2011, lung cancers account 1.89% of total deaths in Bangladesh. Total number of lung cancer patients aged 30 years was estimated to be 196 000 in Bangladesh[5,6]. In the USA, the lifetime chance of developing lung cancer is 1 in 13 (men) and 1 in 16 (women)[7]. Approximately, half of all newly diagnosed cases in the US screening offers former smokers[1]. Smoking is believed to be the primary cause of cancer, not only smokers but also many non-smokers including passive smokers develop lung cancer[8,9]. One of thirteen lifetime smokers develop lung cancers, implying that the differential risk for lung cancer may be explained by genetic susceptibility factors[10,11]. Polymorphism of human genes that encodes the enzymes involved in metabolic activation and detoxification of pulmonary carcinogens such as polycyclic aromatic hydrocarbons and aromatic amines has been reported. Inter individual differences in the ability to activate and detoxify these pulmonary carcinogens are expected to affect the risk of developing lung cancer[12]. Glutathione S-transferases (GSTs) are phase II transformation enzymes involved in the detoxification of hazardous agents[13,14].GSTgene family encodes genes that are critical for certain life processes, as well as for detoxication and toxification mechanisms. The main role of GSTs is to detoxify xenobiotics by catalyzing the nucleophilic attack by glutathione synthetase on electrophilic carbon, sulfur, or nitrogen atoms and converts to nonpolar xenobiotic substrates, thereby preventing their interaction with crucial cellular proteins and nucleic acids[15]. Several studies performed in different populations that examined the role of genetic polymorphisms to lung cancer often showed contradictory results[16]. Glutathione S-transferase Mu (GSTM1), presents in human lung tissue is characterized by two active allelesGSTM1*A, GSTM1*Band a non-functional null allele which resulting from the entireGSTM1gene deletion mutation. UnlikeGSTM1, Glutathione S-transferase θ (GSTT1) is polymorphic and characterized by a functional (wild) allele and a non-functional (null) allele. This null allele results from total or partial deletion of the gene.GSTT1gene may have a diminished ability to metabolically eliminate carcinogenic compounds. Individuals who are carriers of such genotypes may therefore be at increased cancer risk[12,17-19]. Glutathione S-transferase P1 (GSTP1) is the most predominant GSTs in lung tissue also considered to be most important in determining risk for lung cancer[20]. Four GSTP1 alleles have been recognized the wild type allele (GSTP1*A) differs by an A:G transition at nucleotide 313 (Val 105-Ala114) from GSTP1*B and from GSTP1*C by this transition and a C:T transition at 341 (Val 105 -Val 114). A GSTP1*D allele (Ile 105 -Val 114) has also been identified. Most frequently observed single nucleotide polymorphisms in GSTP1 are rs1695 (formerly rs947894 which is due to an A313G substitution resulting in an Ile105Val amino acid change) and rs1138272 (formerly rs1799811 which is due to a C341T substitution resulting in an Ala114Val amino acid change reduce catalytic activity of the enzyme[18,19,21]. It has the highest specific activity towards the active benzo(a)pyrenediol epoxide metabolite of cigarette, and is almost exclusively active towards the (+)-enantiomer of anti-benzo pyrenediol epoxide, thought to be the ultimate mutagenic form of benzo(a)pyrene[22]. Accumulation of these two to have a direct relation to lung cancer[23]. thus, individual carrying val variant expected to have lower detoxication potential and greater risk for cancer because it has generally lower activity towards polycyclic aromatic hydrocarbon diol epoxides, especially benzo pyrenediol epoxide[24]. We conducted a case-control study to investigate the association between the risk of lung cancer andGSTM1,GSTT1and GSTP1 polymorphisms for first time in Bangladesh. No study ofGSTM1, GSTP1 andGSTT1as risk factors for lung cancer has been conducted on Bangladeshi lung cancer patients. A few case-control studies ofGSTM1andGSTT1have been conducted in Indian subcontinent populations that make up around one-sixth of the world’s population. The ancestry in Indian subcontinent is exceptional and the researchers showed that most Indian populations are genetic admixtures of two ancient, genetically divergent groups, which each contributed around 40%-60% of the DNA to most present-day populations. The researchers also found that Indian populations were much more highly subdivided than European populations but whereas European ancestry is mostly carved up by geography, Indian segregation was driven largely by caste[25]. Bangladesh which is situated in the south eastern region of Indian subcontinent supposed to have similar racial admixture and genetic diversity as populations in other part of Indian subcontinent still expected to have unique genetic characteristics.
2. Materials and methods
2.1. Study subjects and data collection
This study was conducted on 106 lung cancer patients and 116 healthy volunteers matched by age, sex and smoking status. Histologically confirmed lung cancer patients were recruited from three main cancer treatment based hospitals in Bangladesh (Ahsania Mission Cancer and General Hospital, Dhaka Medical College Hospital and Bangabandhu Sheikh Mujib Medical University) between the period of January 2009 and December 2011. Controls were selected after physical examination by matching age, sex and smoking status to lung cancer patients. No lung cancer case had a history or evidence of any other severe diseases like cardiovascular disease, kidney disease, previous cancer, and metastasized cancer and if preset they were excluded from the study. Controls were not relatives to the patients and no subject had a history or evidence of hepatic, renal, gastrointestinal or hematologic deviations or any acute or chronic diseases based on medical history, clinical examination and laboratory investigation (hematology, blood biochemistry andurine analysis). Other information like smoking status, demographic characteristics, and lifestyle factors were collected through interviews by trained nurses in the presence of expert physicians. Former smokers quit for >1 year before the recruitment, current and ex-smokers were considered as ever smokers. The guidelines of International Association of Lung Cancer were followed and the patients were histologically diagnosed with lung cancer[26]. The study protocol was approved by the ethical committees of the respective hospitals and the study was conducted in accordance with the declaration of Helsinki and its subsequent revisions[27]. Patients group (n=106) and the healthy volunteers had participated in a preliminary genotyping study of Islamet al[5].
2.2. Genotyping
Genomic DNA was extracted from blood samples of all subjects. Three milliliters of venous blood was collected from all patients and control subjects in ethylene diamine tetraacetic acid-Na2-containing sterile tubes and kept at 80 C until DNA extraction. Genomic DNA was extracted using Daly’s chemical method[5,28]. Genotyping was performed by PCR forGSTM1andGSTT1[18,19] (Figures 1 and 2), whereas genotyping of GSTP1 was performed by PCR-restriction fragment length polymorphism (RFLP) and primers were designed from the published paper[18,19] (Figure 3). Briefly, 25 µL PCR mixture consisted of 1 µL genomic DNA samples (50-70 ng/ µL), 2.5 µL of 10 standard Taq reaction buffer (with MgCl2), 0.5 µL dNTPs (10 mmol/L), 0.5 µL of each primer (10 mmol/L), 0.13 µL Taq DNA polymerase (5 IU/µL) (New England Biolabs, Ipswich, MA) and 20 µL nuclease free water. PCR products ofGSTM1andGSTT1were analyzed on a 2% agarose gel by staining with ethidium bromide whereas that of GSTP1 was analyzed with RFLP using the restriction enzymeBsmAI. The presence of oneGSTM1allele [GSTM1(+)] andGSTT1allele [GSTT (+)] was identified by the presence of 273 bp and 459 bp PCR amplified product respectively. PCR amplified product of breast canncer 2 (BRCA2) has been used as an internal control (marker) forGSTM1and PCR amplified product of cytochrome 3A5*3 (CYP3A5*3) has been used as an internal control (marker) forGSTT1. The required primers, PCR
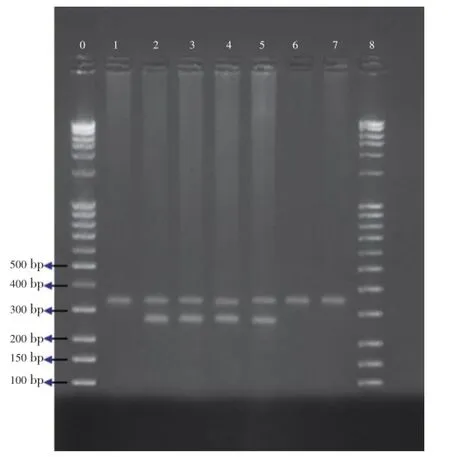
Figure 1.PCR assay forGSTM1gene polymorphism.Lanes 2, 3, 4, 5:GSTM1positive genotype (273 bp); Lanes 1, 6, 7:GSTM1null genotype; Lanes 0, 8: Marker;BRCA2gene was used as an internal positive control (346 bp).
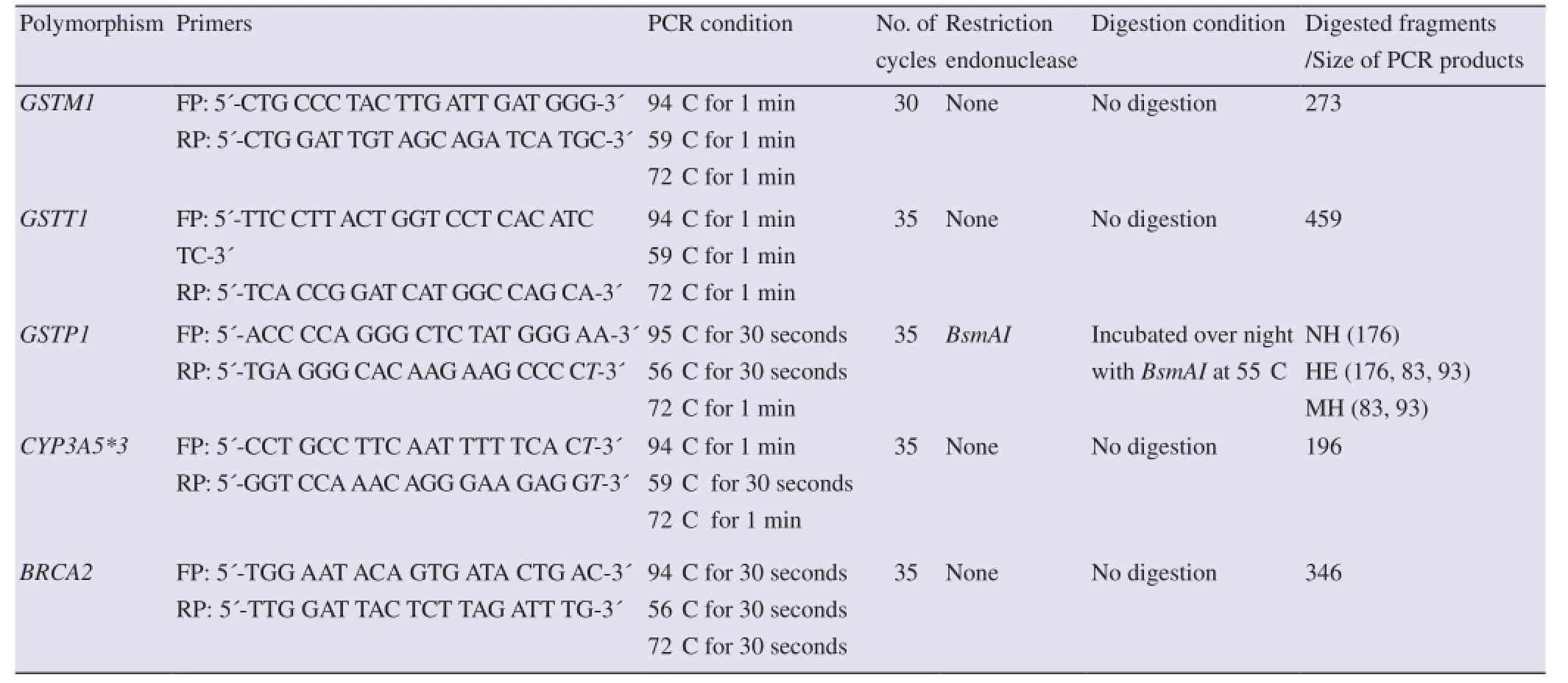
Table 1 Primers, PCR conditions, restriction enzymes and expected DNA fragments on digestion to genotype the selected polymorphisms.
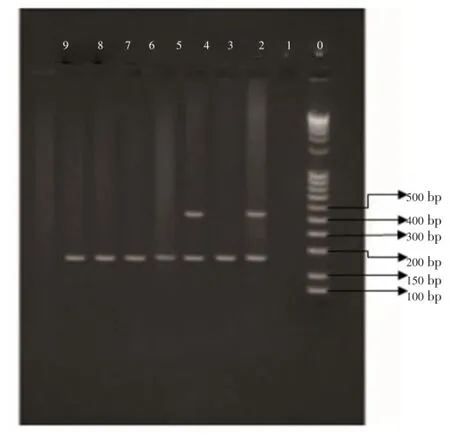
Figure 2.PCR assay forGSTT1gene polymorphism.Lanes 2, 4:GSTT1positive genotype; Lanes 3, 5, 6, 7, 8:GSTT1null genotype; Lane 0: Marker;CYP3A5*3gene was used as an internal positive control.
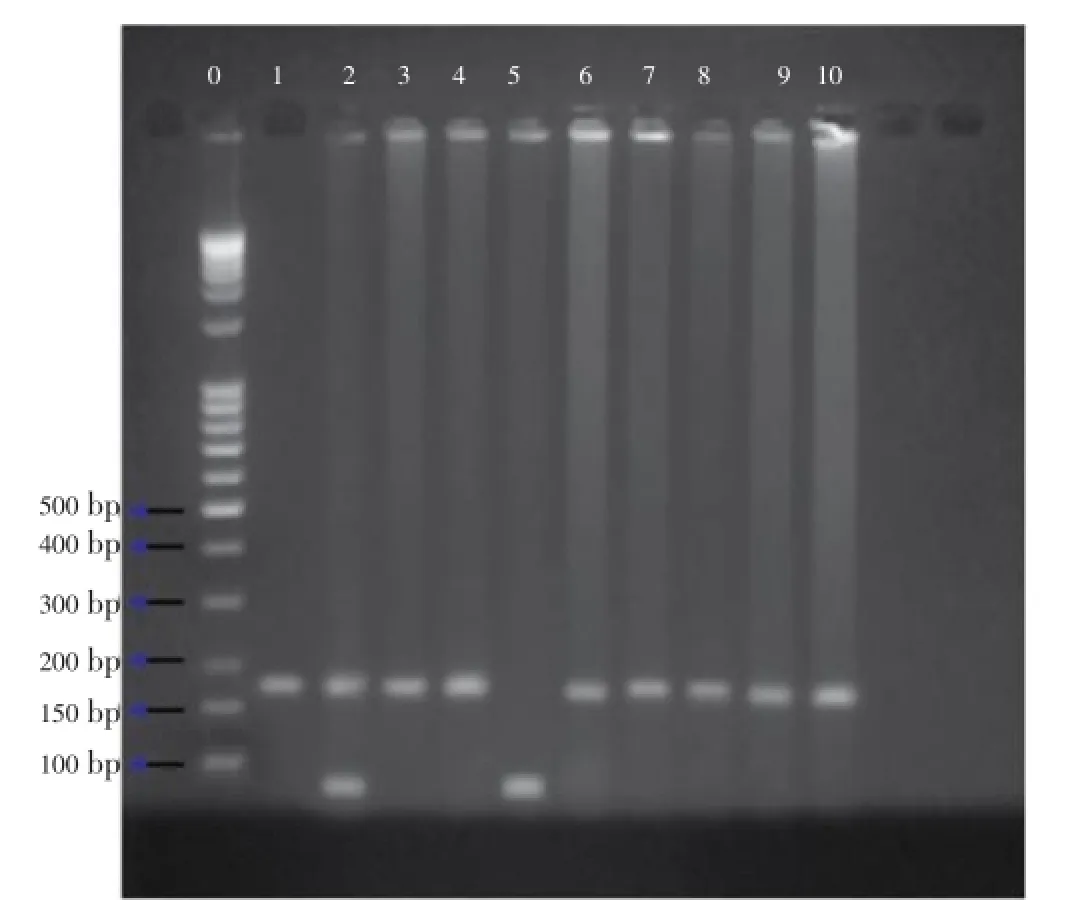
Figure 3.Gel electrophoresis of the digested PCR products showing individuals DNA for theGSTP1polymorphism.Heterozygous polymorphism: Lane 2; ile/val heterozygous; Homozygous polymorphism: Lanes 1, 3, 4, 6, 7, 8, 9, 10; val/val; Mutant homozygous: Lane 5; Electrophoresis of the digested PCR products showed individuals homozygous (ile/ile) for theGSTP1 BsmAIpolymorphism as one band of 176 bp. Heterozygous (ile/val, val) for the polymorphism resulted in three bands of 176, 91 and 85. Homozygotes mutant (val/val) showed two bands of 91 and 85 bp (which appear as one band due to close molecular size).
2.3. Statistical analysis
χ2-tests and two-sided unpairedt-tests were used for comparing demographic variables, distribution of genotype between cases and controls. Unconditional logistic regression was used to estimate crude odds ratio (OR), adjusted OR and their 95% confidence intervals (CIs), with adjustment for age, sex and tobacco consumption status using the statistical software package SPSS version 20.0 (SPSS, Inc., Chicago, IL, USA).
3. Results
3.1. Cases and controls characteristics
The distributions of demographic characteristics and clinical data among study subjects are summarized in Table 2. Briefly, there were no significant differences in gender (P=0.505), mean age (P=0.576) and smoking status (P=0.228) between the two groups. The histological subtypes of lung cancer were squamous cell carcinoma (43.39%), adenocarcinoma (34.91%), small cell carcinoma (18.87%), large cell carcinoma (1.89%) and adenosquamous cell carcinoma (0.94%). Current smokers had been smoking regularly and non-smokers had never smoked during his/her lifetime. Those smokers who quit for more than 1 year before the recruitment were considered as former smokers. Current and ex-smokers were considered as ever smokers. Among the ever smokers 26.42% and 18.10% were former smokers in cases and controls, respectively whereas the observed ever smoking rate was 91.51% in the cases and 89.65% in controls. No significant difference of ever smokers and never smokers (P=0.637) was found between cases and controls. The distributions of demographic characteristics, clinical data, histological subtype of lung cancer and smoking status among study subjects was also summarized in Islamet al[5].
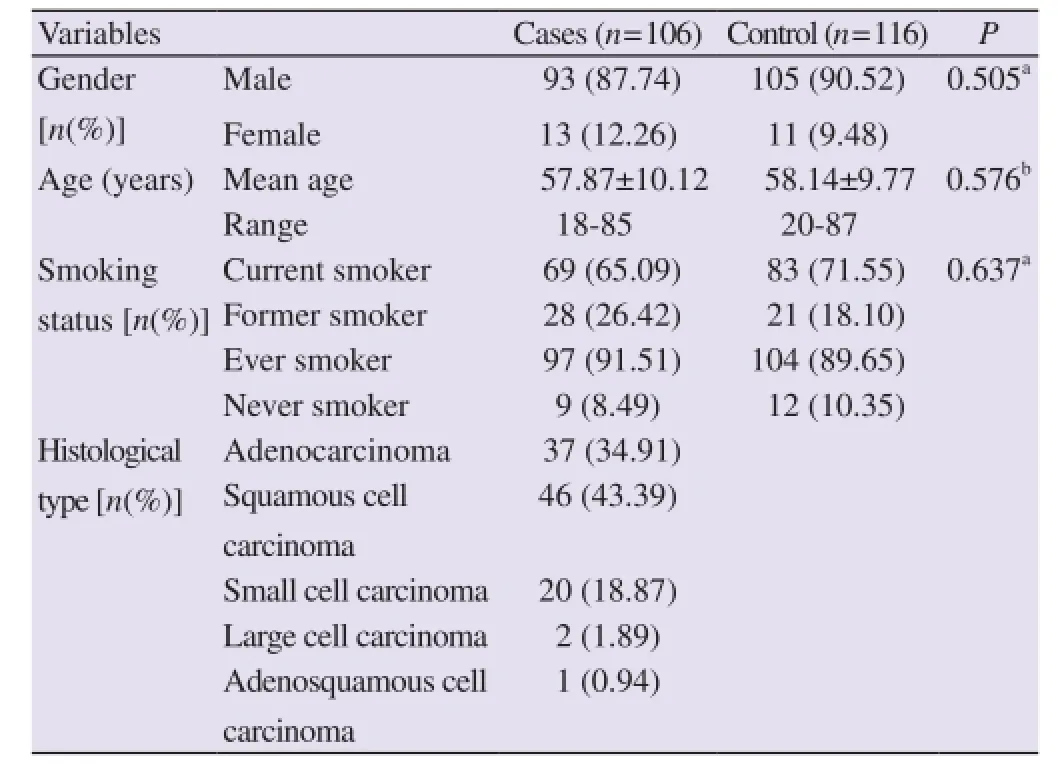
Table 2 Distribution of demographic variables of the lung cancer patients and controls.
3.2. GSTM1 and GSTT1 polymorphisms
The frequency of subjects carrying theGSTM1null genotype was slightly higher in the patient group (57.54%) compared with controls (56.03%). No significant difference was found between thegenotype frequency distribution of the two groups (P=0.934). Risk of lung cancer byGSTM1null genotype is not statistically significant (OR=1.06, 95%CI=0.62-1.81,P=0.820) (Table 3). Among the 106 cases 71.69% were carryingGSTT1null genotype, and 28.30% wereGSTT1positive whereas among the 116 controls 75.86% were carryingGSTT1null genotype, and 24.13% wereGSTT1positive. No significant difference was found between the genotype frequency distribution of the two groups (P=0.481). Risk of lung cancer byGSTT1null genotype is not statistically significant (Adjusted OR=0.84, 95%CI=0.46-1.55,P=0.573) (Table 3).
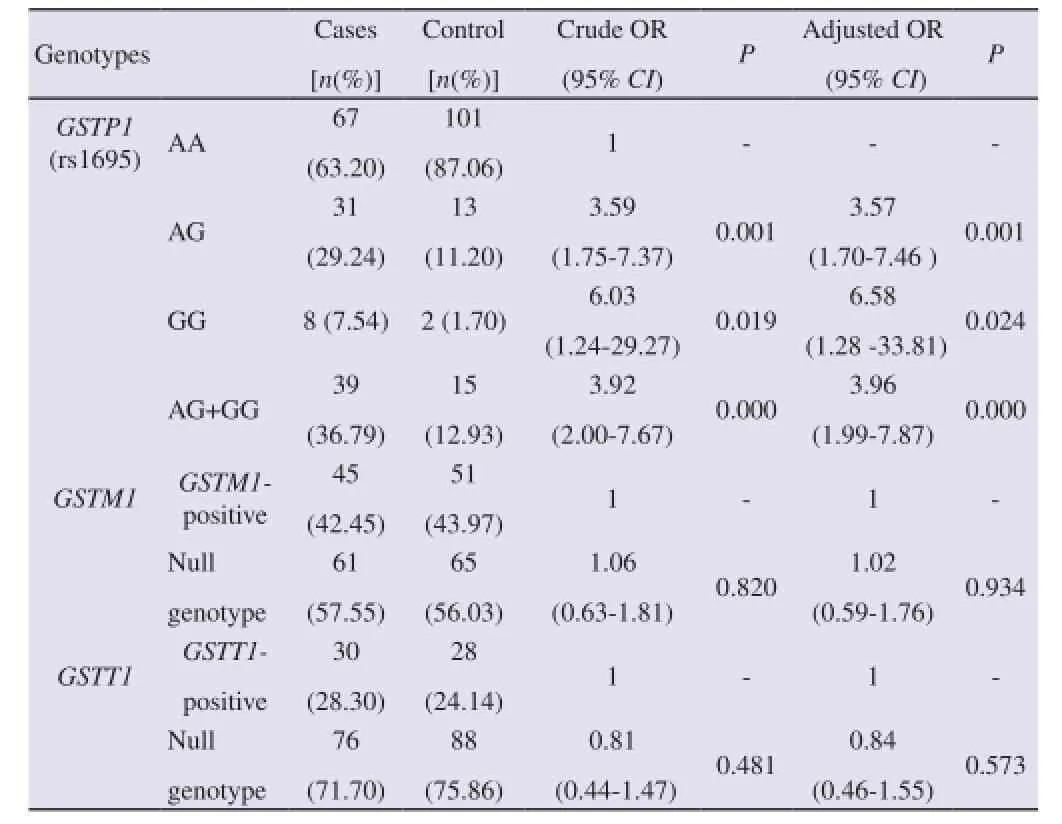
Table 3GSTM1,GSTP1(rs1695),GSTT1genotypes among lung cancer patients and controls.
3.3. GSTP1 polymorphisms
The frequency distribution of patients obeys the Hardy-Weinberg equilibrium (χ2=2.83,P=0.092), whereas that of controls deviates from the equilibrium (χ2=4.29,P=0.038). The distribution of the GSTP1 genotypes was significantly different between the cases and controls (AA, AG and GG genotypes; 63.20%, 29.24% and 7.54%vs87.06%, 11.20% and 1.7%,P=0.001). AG, GG and AG+GG genotypes increased the risk of lung cancer (Adjusted OR=3.56, 95%CI=1.70-7.46,P=0.001; adjusted OR=6.57, 95%CI=1.28-33.81,P=0.024; adjusted OR=3.95, 95%CI=1.98-7.87,P=0.0005, respectively) compared to the AA genotype.
3.4. Association between lung cancer risk and tobacco consumption
As tobacco consumption is the potential risk factors to lung cancer, we further calculated the modifying effect ofGSTM1,GSTT1and GSTP1 genotypes on the association of tobacco consumption with lung cancer (Table 4). Current, ex-smokers and chewing tobacco constitutes tobacco user. Never smoker is considered as tobacco nonuser. In tobacco userGSTM1null andGSTT1null genotypes do not increase the lung cancer risk significantly (OR=1.17, 95%CI=0.67-2.04,P=0.579; OR=0.73 95%CI=0.39-1.39,P=0.349) whereas AG, GG and AG+GG genotypes of GSTP1 increase the lung cancer risk significantly( OR=3.23, 95%CI=1.56 to 6.71,P=0.001; OR=8.71, 95%CI=1.02-74.14,P=0.048; OR=3.73, 95%CI=1.86-7.49,P=0.0005, respectively)
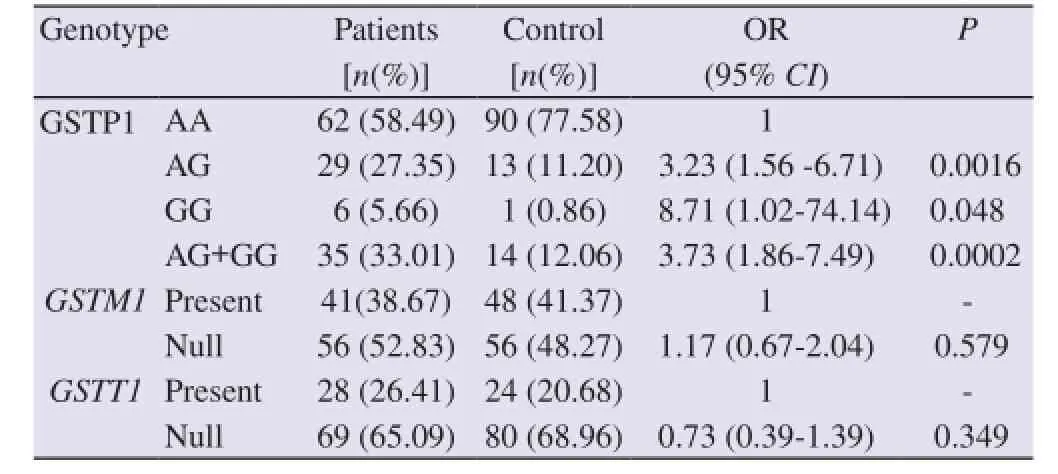
Table 4 Effect ofGSTM1, GSTP1andGSTT1genotypes on the association of tobacco use with lung cancer.
4. Discussion
Several molecular studies have proved few significant piece of information regarding the relationship ofGSTM1andGSTT1with cancer susceptibility. First, the frequencies of homozygousGSTM1andGSTT1deletion carriers are very high (i.e., 20%-50%) in most populations studied to date[18,19,29-32]. Second,GSTM1and possibly,GSTT1may be involved in the etiology of cancer at more than one site[33-36]. Third, the risk conferred to individuals who carry homozygous deletions inGSTM1orGSTT1appears to be small in magnitude (e.g., OR<2). However, the magnitude of risk is larger (e.g., OR=3-5) when interactions ofGSTM1orGSTT1with other factors (e.g., cigarette smoking) are considered[37]. In our current study we investigated three high risk genetic polymorphisms ofGSTM1,GSTT1and GSTP1 as a genetic modifier of risk for individuals with lung cancer as susceptible genotypes. The mu and theta classes ofGSTisozymes (GSTM1andGSTT1respectively) have a common and broad range of substrate specificities, and they detoxify the reactive metabolites of benzo-a-pyrene and other polycyclic aromatic hydrocarbons[19,38-40]. Carriers of homozygous deletion inGSTM1andGSTT1genes have an absence of GST-m and GST-q enzyme activity respectively[19, 41]. These deletion variants are very useful in epidemiological studies of cancer because they divide individuals in two well-defined susceptibility classes: those who are and those who are not able to detoxify potential carcinogens by the metabolic pathways regulated byGSTM1andGSTT1genes. GSTP1 is the most abundant isoform in the lung and is also involved in response to oxidative stress[41,42]. A number of studies have tried to establish links between polymorphic expression of different GSTs and lung cancer risk in different ethnic populations and the results have been conflicting[16]. Among studies investigatingGSTM1null genotype and risk of developing lung cancer in different populations, some of them found significantly increased risk[43-45]. In few studies it was also observed thatGSTM1null genotypes appeared to play a protective role for cancer[46]. A meta-analysis of 11 studies found an OR of 1.6 (95%CI=1.26-2.04) for an association between theGSTM1null genotype and lung cancer risk. In another meta-analysisstudy, it was reported that there was no statistically significant relationship between the individuals carryingGSTM1null genotype and susceptibility to lung cancer but the number of patients carrying this genotype was greater in the lung cancer group.GSTM1null allele in the present study is 57.54% in patient group, which is not similar to the frequencies reported in Indian subcontinent. Additionally, the rate ofGSTM1null genotype was higher in the control group with compare to other control group in different Indian ethnic population[31,32,35,47], but results are consistent with studies conducted in Indian subcontinent which did not show any significant association betweenGSTM1null genotype and lung cancer risk. The incidence of theGSTT1null allele differs among global populations[31,48]. Significant differences inGSTT1null allele frequencies were observed between Caucasian, Asian, African and African American populations[30,31,48]. The prevalence ofGSTT1null allele in the present study in cases is 71.69%, which is not similar to the frequencies reported in Indian subcontinent[30-32,35,47,48]even our result is not consistent with studies conducted in Indian subcontinent[29,30]. Among world population, Korean population showed higher frequency ofGSTT1null allele compared with the white Americans, African Americans, Mexican-Americans and Turkish populations[18,30-31,48]. Although, some researchers reported a significant increase on lung cancer risk withGSTT1null genotype in various populations[29]. In our study, we found no significant relation betweenGSTM1and GTTT1 null genotype and susceptibility to lung cancer. Our results are in sequence with previous case-control studies and a meta-analysis study which observed no significant relationship betweenGSTT1deletion and lung cancer for Caucasians[18,49]. Earlier studies carried out in different populations which analyzed GSTP1 exon 5 polymorphism and lung cancer risk did not expose any significant association[49]. These apparently indicate that lack of involvement between GSTP1 exon 5 genotypes and the risk of developing lung cancer might be steady. Our findings suggest that GSTP1 exon 5 polymorphism (Ile105Val) is associated with increase risk in developing overall lung cancer and statistical analysis also found significance to support such finding. Thus, our results are in line up with the study of Wanget al.which found a significant increase in lung cancer risk with the GSTP1 exon 5 polymorphism. GSTP1 exon 5 polymorphism (Ile105Val) results inactive proteins with different enzyme activity. GSTP1 enzymes with 105Val allele showed decreasedGSTdetoxification capacity resulting in an increased concentration of carcinogens in their lung tissue[19]. Individuals with the 105Val allele have a higher risk of developing lung cancer than Individuals with the 105Ile allele.
The present work thus provides probably the first study of this nature from Bangladesh. We believe that further investigation ofGSTM1,GSTT1and GSTP1 allelic variants in Bangladesh should provide useful information for identification of founder mutations and ethnic predisposition alleles that results various cancerous disease phenotypes.
Our observations showed that carrying theGSTM1andGSTT1null genotype is not a risk factor alone for lung cancer. Our findings also suggest that GSTP1 exon 5 polymorphism (Ile105Val) is associated with high risk of lung cancer and especially in tobacco users. As otherGSTpolymorphisms play important overlapping roles in detoxifying tobacco carcinogens and because risk might be associated with these polymorphisms, further larger populations studies of risk associated with multiple polymorphisms are needed to fully understand the genetic interactions underlying risk susceptibility. Large scale multicenter studies are necessary to obtain more reliable and correct results.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgements
The research project was partially supported by the Ministry of National Science, Information and Communication Technology of People’s Republic of Bangladesh (Grant No.: 39.012.002.01.03.018.2012-721). The authors have no other relevant affiliations or financial involvement with any organization. The authors thank physicians and nurses of Ahsania Mission Cancer Hospital, Dhaka Medical College Hospital and Bangabandhu Sheikh Mujib Medical University, Dhaka, Bangladesh for their technical assistance during blood collection and patient counseling.
Comments
Background
Lung cancer is a major cause of cancer-related death in the developed countries and the overall survival rate has still an extremely poor. The study of genetic polymorphisms has touched every aspect of pulmonary and critical care medicine. The very nature of such studies promises to help in defining pathophysiologic mechanisms, to identify individuals who are at risk for developing disease and to suggest novel targets for drug treatment.
Research frontiers
The present research work depicts role ofGSTM1,GSTT1and GSTP1 polymorphisms as susceptible genotypes for lung cancer especially in relation to tobacco use. TheGSTM1andGSTT1were analyzed using PCR while GSTP1 was analyzed by using PCR and also by RFLP.
Related reports
The relationship betweenGSTM1polymorphism and lung cancer has already been observed in two large studies from Japan (Hayashiet al., 1992; Kiyoharaet al., 2002b) and two from China. The prevalence ofGSTT1null allele in the present study is 72%, which is also not similar to the frequencies reported in Indian subcontinent (Konwaret al., 2010). Genetic polymorphism of GSTP1 has been observed in studies of Anttilaet al., 1993 and Hayeset al., 1995.
Innovations and breakthroughs
A few case-control studies have been conducted in Indian subcontinent but the results are quite conflicting. In the present study the prevalence of genetic polymorphism in GSTP1,GSTM1andGSTT1genes and their association with risk to lung cancer has beenobserved. This is the first of its kind in Bangladesh.
Applications
It is possible that genetic susceptibility to lung cancer may in part be determined by the genetic factors associated with interindividual variations in carcinogen metabolizing enzymes. This article points out criteria that should be applied to design large scale multicenter studies.
Peer review
This is an interesting study of the association ofGSTfamily genes and lung cancer in the Bangladeshi population asGSTM1, GSTTI and GSTP1 genotypes are well established risk factors for lung cancer. The interaction of GSTP1 gene and tobacco use on lung cancer risk is quite interesting in the current study. The finding about GSTP1 genotyping and it’s correlation to lung cancer is also quite interesting.
[1] Tota JE, Ramanakumar AV, Franco EL. Lung cancer screening: review and performance comparison under different risk scenarios.Lung2014;192(1): 55-63.
[2] Haws L Jr, Haws BT. Aerodigestive cancers: lung cancer.FP Essent2014;424: 32-47.
[3] Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics.CA Cancer J Clin2011;61(2): 69-90.
[4] Van Meerbeeck JP, Fennell DA, De Ruysscher DK. Small-cell lung cancer.Lancet2011;378(9804): 1741-1755.
[5] Islam MS, Ahmed MU, Sayeed MS, Maruf AA, Mostofa AG, Hussain SM,et al.Lung cancer risk in relation to nicotinic acetylcholine receptor,CYP2A6andCYP1A1genotypes in the Bangladeshi population.Clin Chim Acta2013;416: 11-19.
[6] Palipudi KM, Sinha DN, Choudhury S, Zaman MM, Asma S, Andes L,et al.Predictors of tobacco smoking and smokeless tobacco use among adults in Bangladesh.Indian J Cancer2012;49(4): 387-392.
[7] Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010.CA Cancer J Clin2010;60(5): 277-300.
[8] Lam TK, Ruczinski I, Helzlsouer KJ, Shugart YY, Caulfield LE, Alberg AJ. Cruciferous vegetable intake and lung cancer risk: a nested case-control study matched on cigarette smoking.Cancer Epidemiol Biomarkers Prev2010;19(10): 2534-2540.
[9] Tang L, Zirpoli GR, Jayaprakash V, Reid ME, McCann SE, Nwogu CE,et al.Cruciferous vegetable intake is inversely associated with lung cancer risk among smokers: a case-control study.BMC Cancer2010;10: 162.
[10] DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL,et al.Cancer treatment and survivorship statistics, 2014.CA Cancer JClin2014;64(4): 252-271.
[11] Risch A, Plass C. Lung cancer epigenetics and genetics.Int J Cancer2008;123(1): 1-7.
[12] López-Cima MF, Alvarez-Avellón SM, Pascual T, Fernández-Somoano A, Tardón A. Genetic polymorphisms inCYP1A1,GSTM1,GSTP1andGSTT1metabolic genes and risk of lung cancer in Asturias.BMC Cancer2012;12: 433.
[13] Shukla RK, Kant S, Mittal B, Bhattacharya S. Polymorphism of cytochrome p450, glutathione-s-transferase and N-acetyltransferases: influence on lung cancer susceptibility.Niger J Med2010;19(3): 257-263.
[14] Dekant W. The role of biotransformation and bioactivation in toxicity.EXS2009;99: 57-86.
[15] Josephy PD. Genetic variations in human glutathione transferase enzymes: significance for pharmacology and toxicology.Hum Genomics Proteomics2010; doi: 10.4061/2010/876940.
[16] Altinisik J, Balta ZB, Aydin G, Ulutin T, Buyru N. Investigation of glutathione S-transferase M1 and T1 deletions in lung cancer.Mol Biol Rep2010;37(1): 263-267.
[17] Dzian A, Halasova E, Matakova T, Kavcova E, Smolar M, Dobrota D,et al.Lung adenocarcinoma and squamous cell carcinoma in association with genetic polymorphisms of GSTs in Slovak population.Neoplasma2012;59(2): 160-167.
[18] Ada AO, Kunak SC, Hancer F, Soydas E, Alpar S, Gulhan M,et al.Association betweenGSTM1,GSTT1, andGSTP1polymorphisms and lung cancer risk in a Turkish population.Mol Biol Rep2012;39(5): 5985-5993.
[19] Ramzy MM, Solliman MEDM, Abdel-Hafiz HA, Salah R. Genetic polymorphism ofGSTM1andGSTP1in lung cancer in Egypt.Int J Collab Res Intern Med Public Health2011;3: 41-51.
[20] Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases.Annu Rev Pharmacol Toxicol2005;45: 51-88.
[21] Harris MJ, Coggan M, Langton L, Wilson SR, Board PG. Polymorphism of the Pi class glutathione S-transferase in normal populations and cancer patients.Pharmacogenetics1998;8(1): 27-31.
[22] Robertson IG, Guthenberg C, Mannervik B, Jernström B. Differences in stereoselectivity and catalytic efficiency of three human glutathione transferases in the conjugation of glutathione with 7 beta,8 alphadihydroxy-9 alpha,10 alpha-oxy-7,8,9,10-tetrahydrobenzo(a)pyrene.Cancer Res1986;46(5): 2220-2224.
[23] Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53.Science1996;274(5286): 430-432.
[24] Coles B, Yang M, Lang NP, Kadlubar FF. Expression ofhGSTP1alleles in human lung and catalytic activity of the native protein variants towards 1-chloro-2,4-dinitrobenzene, 4-vinylpyridine and (+)-anti benzo[a]pyrene-7,8-diol-9,10-oxide.Cancer Lett2000;156(2): 167-175.
[25] Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history.Nature2009;461(7263): 489-494.
[26] Travis WD. Pathology of lung cancer.Clin Chest Med2011;32(4): 669-692.
[27] [The Helsinki Declaration of the World Medical Association (WMA). Ethical principles of medical research involving human subjects].Pol Merkur Lekarski2014;36(215): 298-301. Polish.
[28] Daly AK, Monkman SC, Smart J, Steward A, Cholerton S. Analysis of cytochrome P450 polymorphisms.Methods Mol Biol1998;107: 405-422.
[29] Yang X, Qiu MT, Hu JW, Wang XX, Jiang F, Yin R,et al.GSTT1null genotype contributes to lung cancer risk in Asian populations: a meta-analysis of 23 studies.PLoS One2013;8(4): e62181.
[30] Sharma A, Pandey A, Sardana S, Sehgal A, Sharma JK. Genetic polymorphisms ofGSTM1andGSTT1genes in Delhi and comparison with other Indian and global populations.Asian Pac J Cancer Prev2012;13(11): 5647-5652.
[31] Kumar A, Yadav A, Giri SK, Dev K, Gulati S, Gautam SK,et
al.Allelic variation ofGSTM1andGSTT1genes in Haryana population.Genomic Med Biomark Health Sci 2012;4(3): 98-102.
[32] Senthilkumar KP, Thirumurugan R.GSTM1andGSTT1allele frequencies among various Indian and non-Indian ethnic groups.Asian Pac J Cancer Prev2012;13(12): 6263-6267.
[33] Zhang BL, Sun T, Zhang BN, Zheng S, Lü N, Xu BH,et al.Polymorphisms ofGSTP1is associated with differences of chemotherapy response and toxicity in breast cancer.Chin Med J2011;124(2): 199-204.
[34] Safarinejad MR, Shafiei N, Safarinejad SH. Glutathione S-transferase gene polymorphisms (GSTM1,GSTT1,GSTP1) and prostate cancer: a case-control study in Tehran, Iran.Prostate Cancer Prostatic Dis2011;14(2): 105-113.
[35] Konwar R, Manchanda PK, Chaudhary P, Nayak VL, Singh V, Bid HK. Glutathione S-transferase (GST) gene variants and risk of benign prostatic hyperplasia: a report in a North Indian population.Asian Pac J Cancer Prev2010;11: 1067-1072.
[36] Sailaja K, Surekha D, Rao DN, Rao DR, Vishnupriya S. Association of theGSTP1gene (Ile105Val) polymorphism with chronic myeloid leukemia.Asian Pac J Cancer Prev2010;11(2): 461-464.
[37] Rebbeck TR. Molecular epidemiology of the human glutathione S-transferase genotypesGSTM1andGSTT1in cancer susceptibility.Cancer Epidemiol Biomarkers Prev1997;6(9): 733-743.
[38] Fedets’ OM. [Structure and functions of glutathione transferases].Ukr
Biokhim Zh2014; 86(3): 23-32. Ukrainian.
[39] Board PG, Menon D. Glutathione transferases, regulators of cellular metabolism and physiology.Biochim Biophys Acta2013;1830(5): 3267-3288.
[40] Wu B, Dong D. Human cytosolic glutathione transferases: structure, function, and drug discovery.Trends Pharmacol Sci2012;33(12): 656-668.
[41] Zimniak P, Nanduri B, Pikula S, Bandorowicz-Pikula J, Singhal SS, Srivastava SK,et al.Naturally occurring human glutathione S-transferase GSTP1-1 isoforms with isoleucine and valine in position 104 differ in enzymic properties.Eur J Biochem1994;224(3): 893-899.
[42] Manevich Y, Hutchens S, Tew KD, Townsend DM. Allelic variants of glutathione S-transferase P1-1 differentially mediate the peroxidase function of peroxiredoxin VI and alter membrane lipid peroxidation.Free Radic Biol Med2013;54: 62-70.
[43] Liu D, Wang F, Wang Q, Guo X, Xu H, Wang W,et al.Association of glutathione S-transferase M1 polymorphisms and lung cancer risk in a Chinese population.Clin Chim Acta2012;414: 188-190.
[44] Liu X, Li Z, Zhang Z, Zhang W, Li W, Xiao Z,et al.Meta-analysis ofGSTM1null genotype and lung cancer risk in Asians.Med Sci Monit2014;20: 1239-1245.
[45] Langevin SM, Ioannidis JP, Vineis P, Taioli E, Genetic Susceptibility to Environmental Carcinogens group (GSEC). Assessment of cumulative evidence for the association between glutathione S-transferase polymorphisms and lung cancer: application of the Venice interim guidelines.Pharmacogenet Genomics2010;20(10): 586-597.
[46] Yadav DS, Devi TR, Ihsan R, Mishra AK, Kaushal M, Chauhan PS,et al.Polymorphisms of glutathione-S-transferase genes and the risk of aerodigestive tract cancers in the Northeast Indian population.Genet Test Mol Biomarkers2010;14(5): 715-723.
[47] Vettriselvi V, Vijayalakshmi K, Solomon FDP, Venkatachalam P. Genetic variation ofGSTM1,GSTT1andGSTP1genes in a South Indian population.Asian Pac J Cancer Prev2006;7(2): 325-328.
[48] Piacentini S, Polimanti R, Porreca F, Martínez-Labarga C, De Stefano GF, Fuciarelli M.GSTT1andGSTM1gene polymorphisms in European and African populations.Mol Biol Rep2011;38(2): 1225-1230.
[49] Honma HN, De Capitani EM, Perroud MW Jr, Barbeiro AS, Toro IF, Costa DB,et al.Influence of p53 codon 72 exon 4,GSTM1,GSTT1andGSTP1*Bpolymorphisms in lung cancer risk in a Brazilian population.Lung Cancer2008;61(2): 152-162.
10.12980/APJTB.4.2014APJTB-2014-0476
*Corresponding author: Dr. Abul Hasnat, Professor, Department of Clinical Pharmacy and Pharmacology, Faculty of Pharmacy, University of Dhaka, Dhaka-1000, Bangladesh.
Tel: +88-02-9667850, Ext.8164
Fax: +880-2-8615583
E-mail: ahasnat@du.ac.bd
Foundation Project: Partially supported by the Ministry of National Science, Information and Communication Technology (NSICT) of People’s Republic of Bangladesh (Grant No.: 39.012.002.01.03.018.2012-721).
Methods:A total of 106 lung cancer patients and 116 controls were enrolled in a case-control study. TheGSTM1andGSTT1were analyzed using PCR while GSTP1 was analyzed using PCR-restriction fragment length polymorphism. Risk of lung cancer was estimated as odds ratio at 95% confidence interval using unconditional logistic regression models adjusting for age, sex, and tobacco use.
Results:GSTM1null andGSTT1null genotypes did not show a significant risk for developing lung cancer. A significantly elevated lung cancer risk was associated with GSTP1 heterozygous, mutant and combined heterozygous+mutant variants of rs1695. When classified by tobacco consumption status, no association with risk of lung cancer was found in case of tobacco smokers and nonsmokers carrying null and present genotypes ofGSTM1andGSTT1. There is a three-fold (approximately) increase in the risk of lung cancer in case of both heterozygous (AG) and heterozygous+mutant homozygous (AG+GG) genotypes whereas there is an eightfold increase in risk of lung cancer in cases of GG with respect to AA genotype in smokers.
Conclusions:Carrying theGSTM1andGSTT1null genotype is not a risk factor for lung cancer and GSTP1Ile105Val is associated with elevated risk of lung cancer.
 Asian Pacific Journal of Tropical Biomedicine2014年12期
Asian Pacific Journal of Tropical Biomedicine2014年12期
- Asian Pacific Journal of Tropical Biomedicine的其它文章
- Changing trends of cardiovascular risk factors among Indians: a review of emerging risks
- An unusual cause of optic neuritis: rickettsiosis disease
- Seroprevalence of syphilis in patients attending a tertiary care hospital in Southern India
- Histopathological and molecular study of Neospora caninum infection in bovine aborted fetuses
- Expression of p-PPARγ in the aging thoracic aorta of spontaneously hypertensive rat and inhibitory effect of rosiglitazone
- In vitro germination and propagation of a threatened medicinal orchid, Cymbidium aloifolium (L.) Sw. through artificial seed
