以 N,N′-二(4-甲氧基水杨基)丙烷-1,3-二胺为配体的三核钴ギ和镍ギ配合物的合成、结构及抗菌活性研究
周晓霜 程小珊 李英楠 田凤玉 徐芳元 由忠录
(辽宁师范大学化学化工学院,大连 116029)
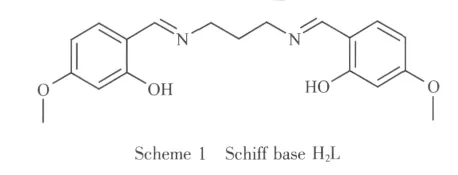
Multi-nuclear complexes containing Schiff bases have been attracted much attention for their versatile structures[1-4]and wide applications especially for their antibacterial,antifungal,and antitumor activities[5-7].The design and construction of such complexes largely depend on the nature of Schiff bases,as well as secondary bridging ligands,such as pseudohalide,4,4′-bipyridine,and multi-carboxylic acid[8-10].During search of literature,we noticed that acetate and nitrate anions come from metal salts are potential bridging ligands[11-12].Solvolthermal method is an efficient way to prepare single crystals of complexes,however,Schiff base complexes prepared by such way have seldom been reported.As an extension of work on the construction of polynuclear complexes as well as antimicrobial activities,in the present paper,a new cobaltギ complex[Co{CoL(μ2-CH3COO)}2](1)and a new nickelギ complex[Ni{NiL(μ2-NO3)(CH3OH)}2](2)with Schiff base N,N′-bis(4-methoxysalicylidene)propane-1,3-diamine(H2L;Scheme 1)have been synthesized under solvolthermal conditions and structurally characterized.It is notable that no complexes with H2L have been reported so far.
1 Experimental
1.1 General methods and materials
4-Methoxysalicylaldehyde and N,N′-propane-1,3-diamine were purchased from Lancaster.All other chemicals with AR grade were obtained as received.H2L wassynthesized according to the literature method[13].Elemental analyses for C,H and N were performed on a Perkin-Elmer 240C elemental analyzer.IR spectra were recorded on a Nicolet AVATAR 360 spectrometer as KBr pellets in the 4 000~400 cm-1region.
1.2 Preparation of[Co{CoL(μ2-CH3COO)}2](1)
H2L (0.1 mmol,34.2 mg)and Co(CH3COO)2·4H2O(0.1 mmol,24.9 mg)were mixed in methanol(8 mL).The mixture was stirred at room temperature for 10 min and then transferred to a stainless steel bomb,which was sealed,heated at 150℃for 12 h,and cooled gradually to room temperature.Red blockshaped crystals of the complex were collected,washed three times with methanol and dried in air.Yield:53%.Anal.Calcd.for C42H46Co3N4O12(%):C,51.7;H,4.8;N,5.7.Found(%):C,51.1;H,4.6;N,5.7.Selected IR data(KBr,cm-1):1 607(s),1 539(s),1 445(m),1 221(s),1 121(m),1 032(m),848(m),782(m),668(w),623(w),469(w).
1.3 Preparation of[Ni{NiL(μ2-NO3)(CH3OH)}2](2)
Complex 2 was prepared by a similar procedure as that described for 1,with Co(CH3COO)2·4H2O replaced by Ni(NO3)2·6H2O (0.1 mmol,29.1 mg),resulting green block-shaped crystals of the complex.Yield:37%.Anal.Calcd.for C40H48N6Ni3O16(%):C,46.0;H,4.6;N,8.0.Found(%):C,46.3;H,4.8;N,8.2.Selected IR data (cm-1):3 435 (b,w),1 618(s),1 599(s),1 543(s),1 446(m),1 418(s),1 394(s),1 350(m),1 303(s),1 215(s),1 176(w),1 128(m),1 029(w),978(m),845(m),624(w),592(w),469(w).
1.4 X-ray crystallography
Diffraction intensities for the complexes were collected at 298(2)K using a Bruker SMART ApexⅡCCD area-detector with Mo Kα radiation(λ=0.071 073 nm).The collected data were reduced using SAINT program[14],and empirical absorption corrections were performed using SADABS program[15].Structures of the complexes were solved by direct method and refined against F2by full-matrix least-squares method using SHELXTL[16].Allofthenon-hydrogenatomswererefined anisotropically.H8 atom of the methanol molecule in 2 was located from a difference Fourier map and refined isotropically,with O-H distance restrained to 0.085(1)nm.The remaining hydrogen atoms were placed in geometrically ideal positions and constrained to ride on their parent atoms.Crystallographic data for the complexes are summarized in Table 1.Selected bond lengths and angles are given in Table 2.
CCDC:736569,1;736568,2.
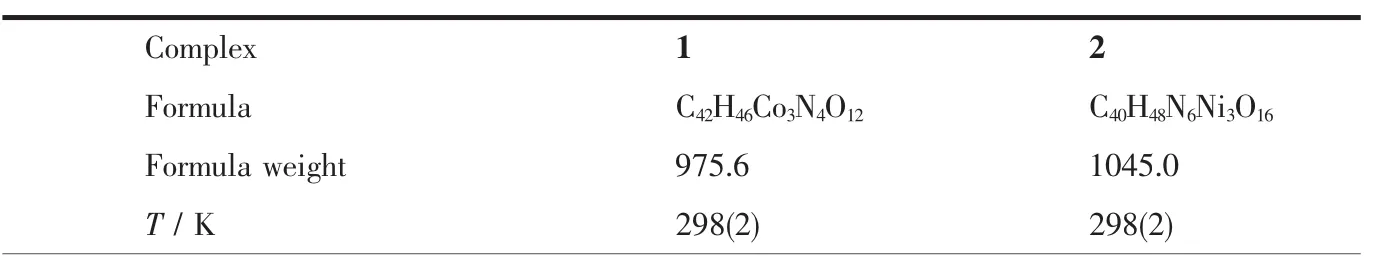
Table 1 Crystal data for the complexes
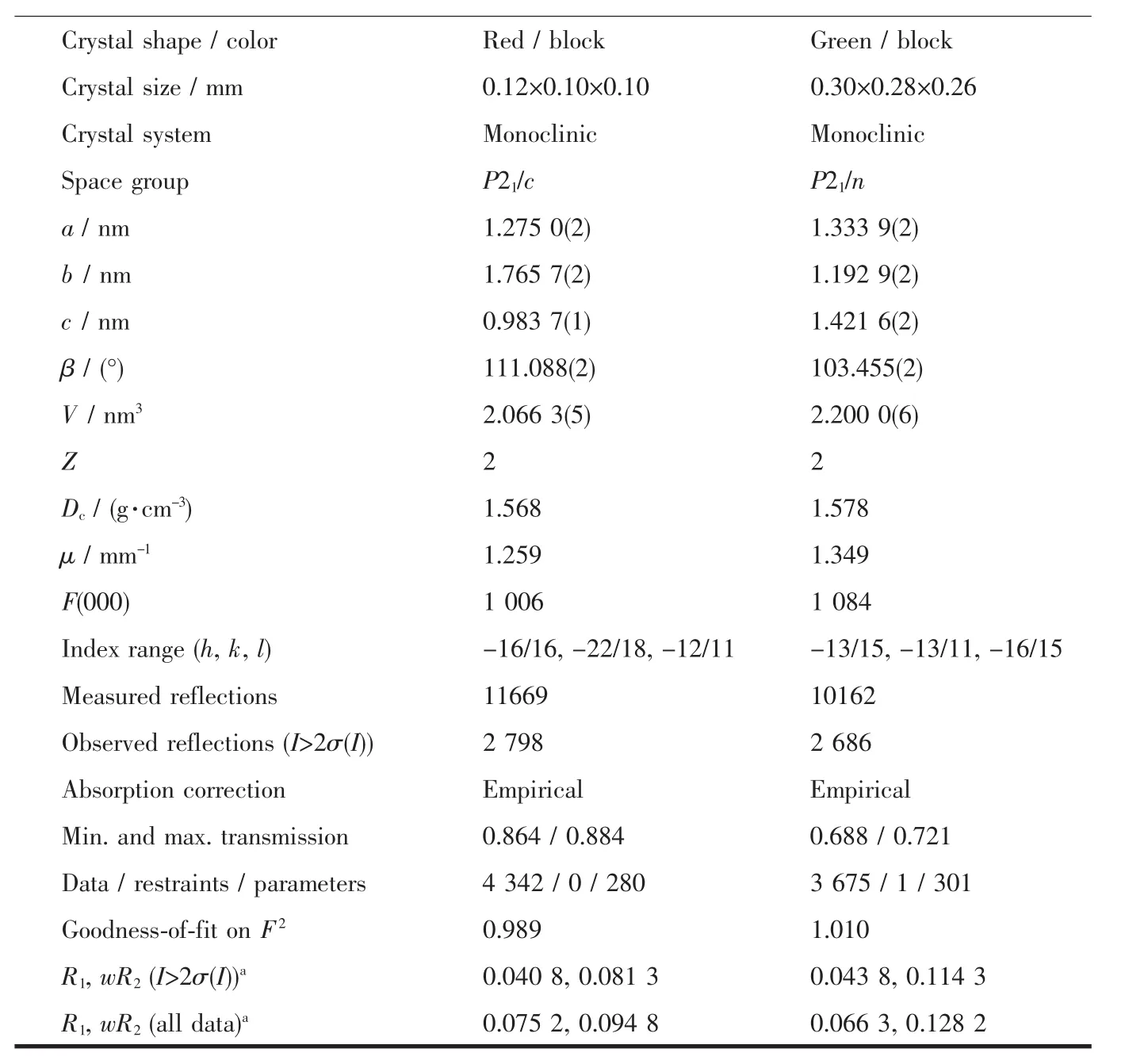
Continued Table 1
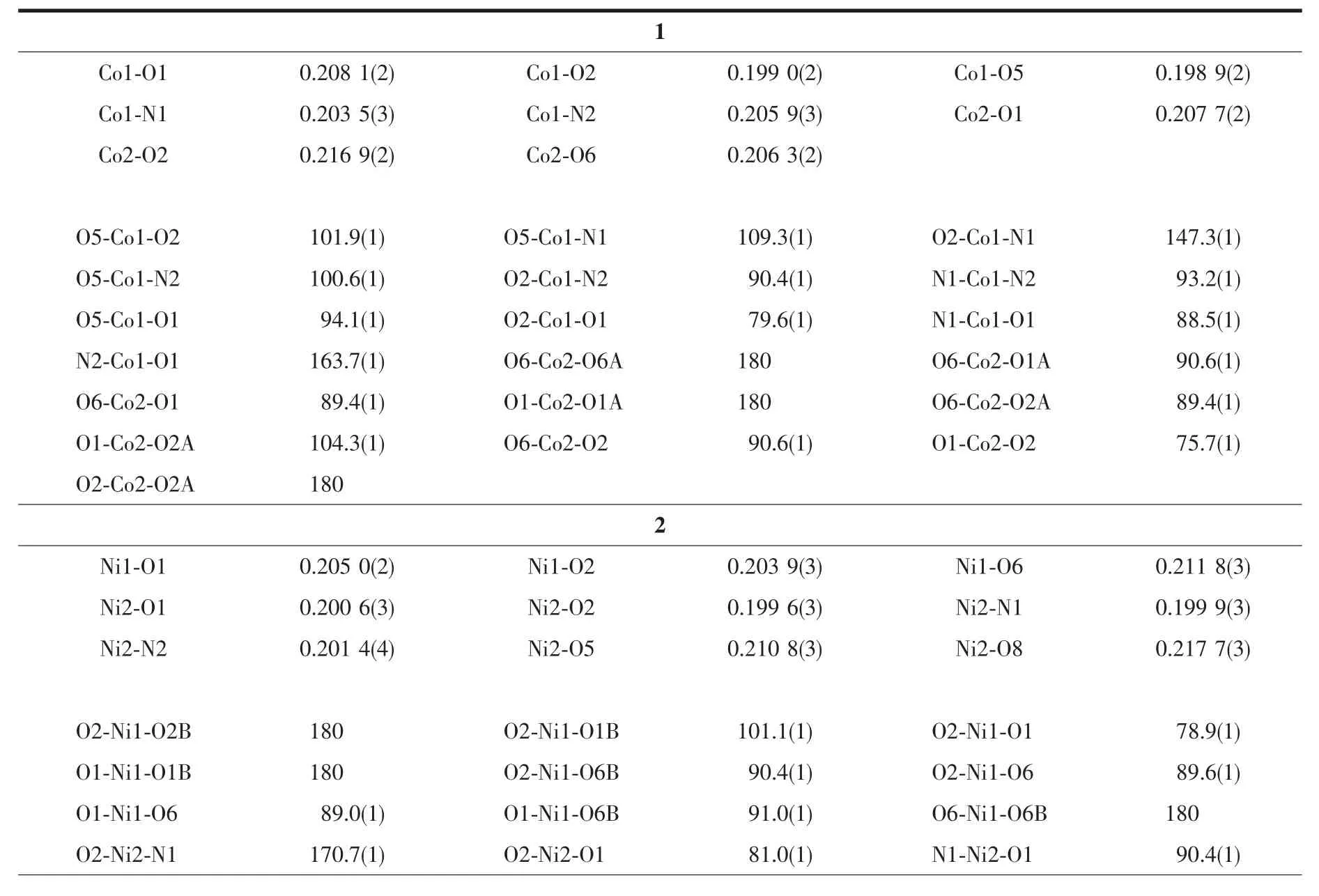
Table 2 Selected bond lengths(nm)and angles(°)for the complexes
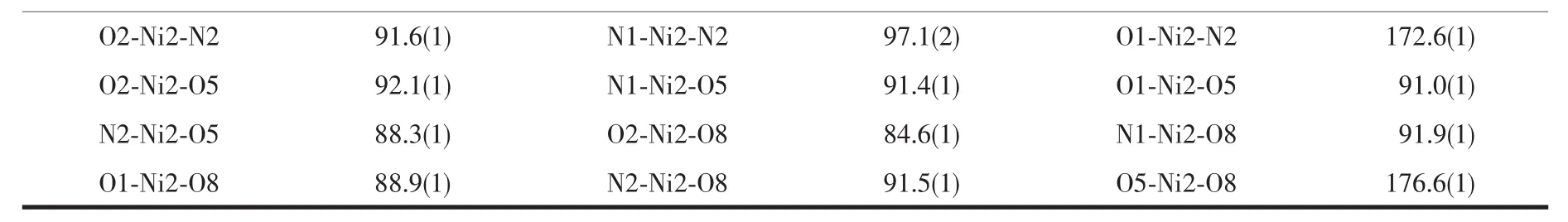
Continued Table 2
2 Results and discussion
2.1 Chemistry
Both complexes were prepared by reaction of H2L with cobalt acetate and nickel nitrate in methanol at solvolthermalcondition.The acetate and nitrate anions coordinate to the metal atoms through μ2-O,O′bridging mode.It should be noticed that we can not obtain trinuclear complexes under ambient temperature during the preparation.Methanol was proved to be the best solvent for the preparation of the complexes.
2.2 Structure description of the complexes
Fig.1 and Fig.2 give perspective views of complexe 1 and 2,respectively,together with atomic labeling system.Both complexes are structurally similar centrosymmetric trinuclear compounds.Complex 1 is a phenolate O and μ2-acetato-bridged trinuclear cobaltギcompound,while 2 is a phenolate O and μ2-nitratobridged trinuclear nickelギcompound.
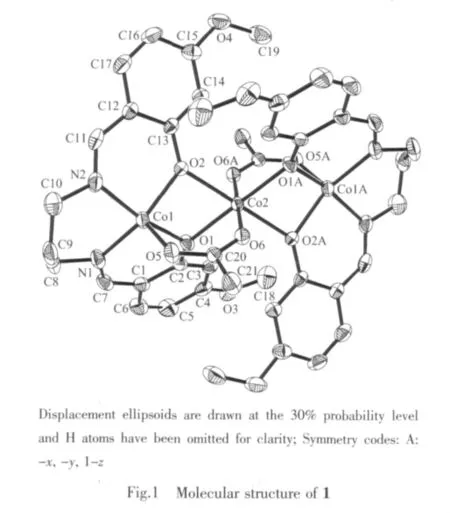
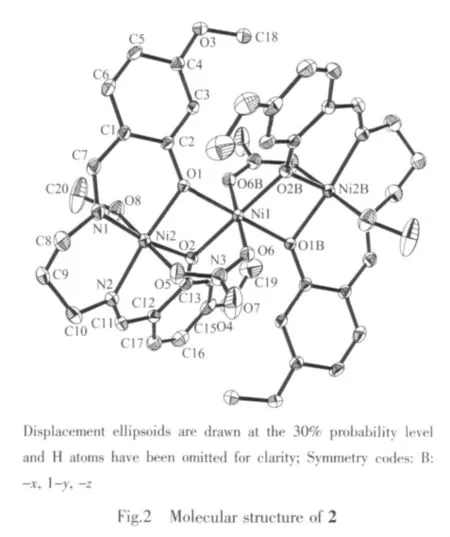
Each complex consists of two ML units (M=Co for 1,and M=Ni for 2)connected to each other by a completely encapsulated third metal atom M,which is located on a crystallographic inversion center.The cage of each central metal atom is formed by four phenolate O bridges from two ML moieties and by two other bridging functions(acetate for 1 and nitrate for 2)that furthermore connect the central metal with the two outer metal atoms resulting in an octahedral coordination.The coordination around each central metal atom displays only slight distortions,which can be observed from the coordinate bond lengths and angles.
The coordination around the inversion-related terminal metal atoms is square pyramidal for 1 and octahedral for 2.The square pyramid of each terminal Co atom in 1 is formed by two O and two N atoms of the Schiff base ligand in the equatorial plane,and by one O atom of the acetate group in the apical position.Substantialdistortion ofthe square pyramid is revealed by bond angles between apical and equatorial donor atoms.Co1 atom displaced out of the corresponding least-squares plane defined by equatorial donor atoms in the direction of the oxygen of the acetate bridge by 0.041 1(2)nm.The octahedron of each terminal Ni atom in 2 is formed by two O and two N atoms of the Schiff base ligand in the equatorial plane,and by two O atoms from one bridging nitrate group and one coordinated methanol molecule in the axial positions.The coordination around the terminal metal atom also displays slight distortion, as evidenced by coordinate bond lengths and angles.
The ML units in the complexes are butterflyshaped,with dihedral angles formed by the two aromatic rings of 52.4(3)°in 1 and 51.1(3)°in 2.The four-membered chelate rings M1-O1-M2-O2 in the complexes are not planar but are roof-shaped.The chelate rings formed by the terminal metal atom,N1,C8-C10,and N2 in the complexeshave chair conformations.
2.3 IR spectra
IR spectra of H2L and the complexes provide information about the metal-ligand bonding.The strong absorption band at 1 632 cm-1for H2L is assigned to the azomethine groups,ν(C=N)[17].The bands are shifted to lower wave numbers in the complexes,1 607 cm-1for 1,and 1 618 cm-1for 2,what can be attributed to the coordination of the nitrogen atoms of the azomethine groups to the metal atoms.The phenolic ν(Ar-O)in the free ligands exhibit strong bands at 1 206 cm-1[18].However,in the complexes,the bands appear in the region 1 215~1 221 cm-1,which may be assigned to the skeletal vibrations related to the phenolic oxygen of the Schiff base ligands[19].Other weak bands at 469 cm-1for the complexes may be assigned to ν(M-O)[20],and provides further evidence for coordination through the deprotonated phenolic oxygen atoms.For 1,the band indicative of the bridging acetate groups are at about 1 539 and 1 445 cm-1.For 2,the bands indicative of the bridging nitrate groups are at 1 446 and 1 303 cm-1.The close resemblance of the shapes and the positions of these bands suggest similar coordination modes for the complexes,in accordance with the structural features.
2.4 Antimicrobial activity
Antimicrobial assay was carried out as that described in our previous paper[21].The compounds were screened for antibacterial activity against two Gram(+)bacterial strains(Bacillus subtilis and Staphylococcus aureus)and two Gram(-)bacterial strains(Escherichia coli and Pseudomonas fluorescence)by MTT method.MIC (minimum inhibitory concentration)values of the compounds against four bacteria are listed in Table 3.Kanamycin and Penicillin G were used as standard drugs.The free Schiff base H2L showed moderate activities against B.subtilis and S.aureus,and no or very weak activities against E.coli and P.fluorescence.The two complexes showed obvious stronger activities against the bacteria than the Schiff base ligand.Complex 1 showed the most effective activity against S.aureus.In general,complex 1 has stronger activities against B.subtilis,S.aureus,and E.coli,but lower activity against P.fluorescence than complex 2.The antifungalactivitiesofthe complexeswere also evaluated against two fungal strains(Candida albicans and Aspergillus niger)by MTT method (Table 3).Ketoconazole was used as a standard drug.It is disappointed that only complex 1 showed weak activity against C.albicans.
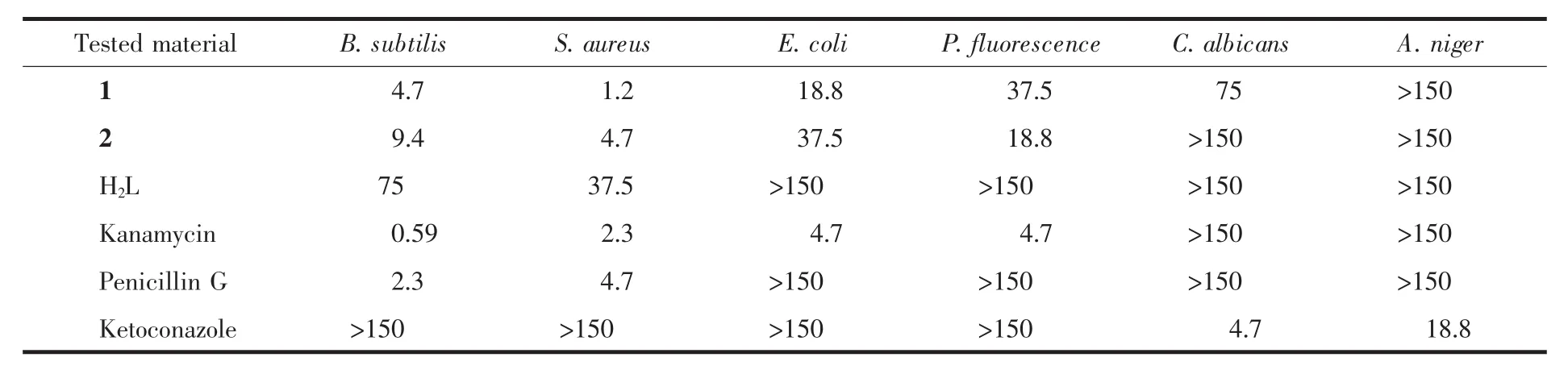
Table 3 MIC values of H2L and the complexes(mol·L-1)
[1]Salassa G,Castilla A M,Kleij A W.Dalton Trans.,2011,40:5236-5243
[2]You Z L,Zhou P.Transition Met.Chem.,2008,33:453-457
[3]ZHANG Qi-Long(张奇龙),ZHU Xing-Cheng(朱兴城),XU Hong(徐红),et al.Chinese J.Inorg.Chem.(Wuji Huaxue Xuebao),2012,28(6):1151-1156
[4]LUO Xin-Ze(罗新泽),ZHOU Hong-Bo(周红波).Chinese J.Inorg.Chem.(Wuji Huaxue Xuebao),2012,28(6):1183-1189
[5]Rosu T,Pahontu E,Maxim C,et al.Polyhedron,2011,30:154-162
[6]Bagihalli G B,Avaji P G,Patil S A,et al.Eur.J.Med.Chem.,2008,43:2639-2649
[7]QIU Xiao-Yang(仇晓阳),LIU Qi-Feng(刘起峰),ZHANG Ping(张平),et al.Chinese J.Inorg.Chem.(Wuji Huaxue Xuebao),2012,28(2):362-366
[8]Novitchi G,Wernsdorfer W,Chibotaru L F,et al.Angew.Chem.Int.Ed.Engl.,2009,48:1614-1619
[9]Hou P,You Z L,Zhang L,et al.Transition Met.Chem.,2008,33:1013-1017
[10]Sutradhar M,Mukherjee G,Drew M G B,et al.Inorg.Chem.,2006,45:5150-5161
[11]Dong W K,Li L,Sun Y X,et al.Transition Met.Chem.,2010,35:787-794
[12]Banu K S,Chattopadhyay T,Banerjee A,et al.Inorg.Chem.,2008,47:7083-7093
[13]Kabak M,Elmali A,Elerman Y,et al.Z.Naturforsch.B,2003,58:1141-1146
[14]Bruker,SMART and SAINT.Bruker AXS Inc.,Madison,2002.
[15]Sheldrick G M.SADABS.University of Göttingen,Germany,1996.
[16]Sheldrick G M.SHELXTL V5.1,Software Reference Manual,Bruker AXS Inc,Madison,1997.
[17]Kovacic J E.Spectrochim.Acta Sect.A,1963,23:183-187
[18]Percy G C,Thornton D A.J.Inorg.Nucl.Chem.,1972,34:3357-3363
[19]Tokii T,Muto Y,Imai K,et al.J.Inorg.Nucl.Chem.,1973,35:1539-1545
[20]Bekheit M M,Ibrahim K M.Synth.React.Inorg.Met.-Org.Nano-Met.Chem.,1986,16:1135-1147
[21]Zhang M,Xian D M,Li H H,et al.Aust.J.Chem.,2012,65:343-350

