Expression of a Lysine-rich Gene in Bacillus subtilis 168
Li Xue-lin, Liu Xiao-fei, Gao Xue-jun*, Qiao Bin, Pan Hong-bao, and Ao Jin-xia
1 College of Veterinary Medicine, Northeast Agricultural University, Harbin 150000, China;
2 College of Food Engineering, Harbin University of Commerce, Harbin 150076, China
Introduction
L-lysine is one of the eight essential amino acids for humans and other monogastric animals (Galili et al., 2000; He et al., 2009). It is mainly used as a feed additive in the animal feed industry, and mixed with various common livestock feed, such as cereals which do not contain suff i cient levels of L-lysine for the livestock's nutritional requirements, especially for single-stomach (monogastric)animals like broilers,bovine and swine (Leuchtenberger, 1996; Kircher and Pfefferle, 2001). Biotechnology has provided alternative methods to improve the lysine content in feed. On the one hand, many genetic engineering researches were devoted to: understanding the regulation of lysine metabolism and its exploitation for increasing free lysine level in seeds (Zhu and Galili, 2003; Frizzi et al., 2008; ILSI, 2008);using proteins that are enriched in lysine content(Beauregard and Hefford, 2006). On the other hand,the synthetic L-lysine is produced exclusively using bacterial fermentations. Bacterial fermentations were devoted to: increase of the microorganism's intrinsic productivity characteristics by classical mutagenesis and genetic engineering (Tosaka et al., 1979; Jose,1997); more recent developments of recombinant DNA technology and molecular biology also have been used in order to improve L-lysine producing strains (Becker et al., 2008; Ohnishi et al., 2002; Anastassiadis, 2007).But research on improvement of lysine content by transferring alien lysine-rich genes into bacterial is almost absent.
A cDNA encoding lysine-rich protein cloned from winged bean is expressed in plants (Li et al., 2006;Gao et al., 2001; Meng et al., 2001). The lysine-rich protein consists of 158 amino acids with a molecular weight of 16.8 ku. There are 17 lysine residues in the protein, and lysine is the most abundant amino acid in this protein, amounting to 10.8% (Sun, 1998).
Bacillus subtilis 168 is an attractive host for the expression of heterologous proteins, because the absence of exterior membrane simplifies the protein secretion pathways, therefore, allows higher secretion levels of extracellular proteins. Moreover,Bacillus subtilis 168 is regarded as an ideal expression system due to its nonpathogenic trait, wellestablished fermentation technology and ripened genetic manipulation as well as multiple regulators which possibly influence expression and secretion(Stragier and Losick, 1996). Previous reports paid much attention to the expression of lysine-rich gene in plants. However, studies about lysine-rich gene expression in Bacillus subtilis are seldom. In this study, the lys gene from winged bean was cloned and expressed in Bacillus subtilis 168.
Materials and Methods
Lysine-rich protein gene, strain, plasmid and medium
The winged bean (Psophocarpus tetragonolobus(L.)DC)was obtained from Shun Qiang Nong Ke Company (China). E. coli top10 and Bacillus subtilis 168 were provided by Laboratory of Dairy Science,Ministry of Education. Plasmid pHT43 was purchased from Molecular Biotechnology Company (Germany).
LB medium: 1% w/v peptone, 0.5% w/v yeast extract, 1% w/v NaCl , and autoclave to sterilize ( pH 7.0).
Bacillus subtilis 168 transformation medium:
10×SpⅠSalts (100 mL): 2% w/v (NH4)2SO4, 14%w/v K2HPO4, 6% w/v KH2PO4, 0.2% w/v MgSO4·7H2O,1% w/v sodium citrate.
CAYE (100 mL): 2% w/v trypton, 10% w/v yeast extraction.
SpⅠmedium: 10% v/v 10×SpⅠSalts, 1% v/v CAYE, 1% v/v 50% glucose.
SpⅡmedium: 10% v/v SpⅠmedium, 1% v/v 50 mmol · L-1CaCl2, 1% v/v 250 mmol · L-1MgCl2.
Fermentation medium: 1.5% w/v peptone, 2% w/v yeast extract, 0.3% w/v K2HPO4, 0.1% w/v sodium citrate, 1% w/v starch.
Enzymes and chemicals
Trizol from Invitrogen; PrimeScript RT-PCR Kit,dNTP, Taq DNA polymerase, restriction enzymes and T4-DNA ligase were purchased from TaKaRa Biotechnology (Dalian, China)Company; DNA purification kit and plasmid extraction kit from Axygen;RNA easy mini kit from Qiagen; IPTG, chloromycetin and all chemicals used were of analytical grade and were obtained from Shanghai Sangon Biological Engineering Technology & Services Company.
Total RNA isolation and reverse transcription reaction
Total mRNA was isolated from developing seeds of winged beans using trizol reagent according to the instruction of the manufacturer (Invitrogen).Reverse transcription reaction (40 μL)consisted of the following components: 10 μg of total RNA, 1 μL of moloney murine leukemia virus reverse transcriptase,1 μL of RNase inhibitor, 4 μL of deoxynucleoside triphosphate, 2 μL of Oligo dT, 4 μL of dithiothreitol,and 8 μL of 5× reverse transcriptase buffer. The procedure of the reverse transcription was taken according to the instructions of the manufacturer.
Primer design and PCR amplif i cation
According to the published sequence of the winged beans mRNA from United States Patent with an accession number of US 6,184,437 B1B, and two DNA oligonucleotide primers were designed and synthesized by Invitrogen Biontechnology (Shanghai,China). The primers were designed to incorporate the restriction enzyme sites Xba I and Sma I at their 5'ends, allowing the directional cloning of PCR product into the plasmid expression vector pHT43.
Primers were as the followings:
Forward one: 5'- GCTCTAGACATTATGGGTGTT TTCACATATGAG-3' (Xba I)
Backward one: 5'- TCCCCCGGGATTGTATTCAG GATGGGCCAAAAGG-3' (Sma I)
The cDNA encoding lysine-rich protein was cloned using the total cDNA as template and the primers designed. The PCR condition was 94℃ denaturation for 5 min, 94℃ for 50 s, 52℃ for 60 s, 72℃ for 80 s,36 cycles, fi nally 72℃ for 30 min. The amplif i ed PCR products were analyzed by agarose gel electrophoresis.
Construction of expression plasmid
The PCR products were double-digested by Xba I and Sma I and ligated to plasmid pHT43 digested by the same restriction enzymes and the recombination plasmid pHT43/lys was constructed. The ligation reaction was transformed into E. coli Top 10 and plated on LB with ampicillin (100 μg · mL-1). Plasmid DNA was isolated and verif i ed by sequencing. Plasmid DNA was isolated from transformants by the alkaline lysis method in accordance with the manufacturers'instructions (Invitrogen). Clones were verified by DNA sequencing (TaKaRa Dalian, China).
Bacillus subtilis 168 transformation and selection
Plasmids carrying the lys gene were isolated from E. coli Top10 by miniprep. The preparation of Bacillus subtilis 168 competent cells and plasmid transformation were performed according to the methods described by Spizizen (1961).
Expression of lys gene in Bacillus subtilis 168
Bacillus subtilis 168 cells containing the lys gene were streaked on a nutrient agar plate supplemented with chloramphenicol (5 μg · mL-1), followed by incubation at 37℃ overnight. A single colony was inoculated into 3 ml of LB at 37℃ with shaking at 200 r · min-1for 12-13 h. The inoculated medium was then transferred to 20 mL of LB at a dilution factor of 1 : 20, followed by incubation at 37℃ with shaking at 200 r · min-1.When OD600reached 0.75-0.95, protein production was induced by IPTG (1 mmol · L-1)at 37℃ for 30 h.After incubation, the bacterial cells were harvested by centrifugation at 2 000 r · min-1and 4℃ for 20 min, and the supernatant (containing the lysine-rich protein)was collected by centrifugation and stored at 4℃ with the fi nal concentration 50% (NH4)2SO4for 12 h and then,the precipitate was collected by centrifugation. The precipitation was analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Detection of lys gene expression by SDSPAGE
SDS-PAGE of proteins was performed in 15% (wt/vol)polyacrylamide gels as described by Molecular Cloning (Joseph Sambrook, 2001).
RT-PCR
Total RNA extraction was performed at various phases of LB culture of induced Bacillus subtilis 168 with trizol reagent according to the instruction of the manufacturer (Invitrogen). The RNA was quantified spectrophotometrically and cDNA was synthesized by RNA. The cDNA was used as a template for PCR.
Lysine content detected by HPLC in Bacillus subtilis 168
Lysine content was determined by measuring the ratio of the chromatographic peaks of the ions derived from the analytes (lysine)to those derived from the external standards.
Lysine determination was performed, the precipitate and concentrated supernatant liquid were hydrolyzed according to the GB/T18246-2000 method.
Samples were injected in a Diamonsil C18(2)5μ 250×4.6 mm column, maintained at 25℃, and the fl ow rate was 800 μL · min-1.
Results
Construction of expression vector in Bacillus subtilis 168
The lys gene encoding the lysine-rich protein was isolated from the seeds of winged beans (Fig. 1).Restriction enzyme analysis of the plasmid pHT43/lys showed that it was successfully constructed (Fig. 2).Plasmid extracts of recombinant clones were submitted to nucleotide sequencing and confirmed that they had inserted the actual sequence of the whole 478 bp fragment (Fig. 2).

Fig. 1 PCR products of lys gene of Psophocarpus tetragonolobus
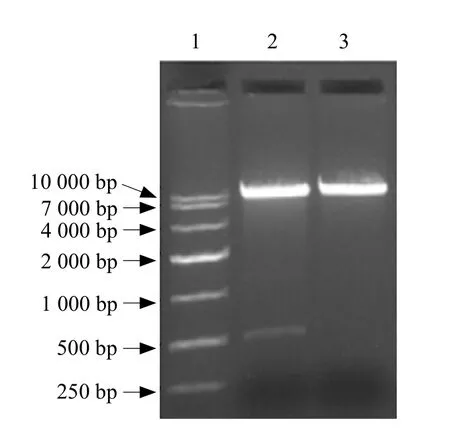
Fig. 2 Identif i cation of recombinant plasmid pHT43/lys
Detection of recombinant protein by SDSPAGE
The expression of the recombinant protein was induced by IPTG in supernatant. A single17 ku protein band was invisible in SDS-PAGE from the induced culture.
RT-PCR
Transcriptional analysis of lys was investigated by RTPCR in Bacillus subtilis 168 cells at different stages of cellular growth (Fig. 3). A 500 bp amplification product was detected corresponding to the expression of lys gene. The amplif i cation signal of lys gene was increasing during the lag phase till the end of the exponential growth phase and then decreased at the stationary phase till the recession phase. These results suggested that the expression of lys was enhanced during exponential growth phase of Bacillus subtilis 168.
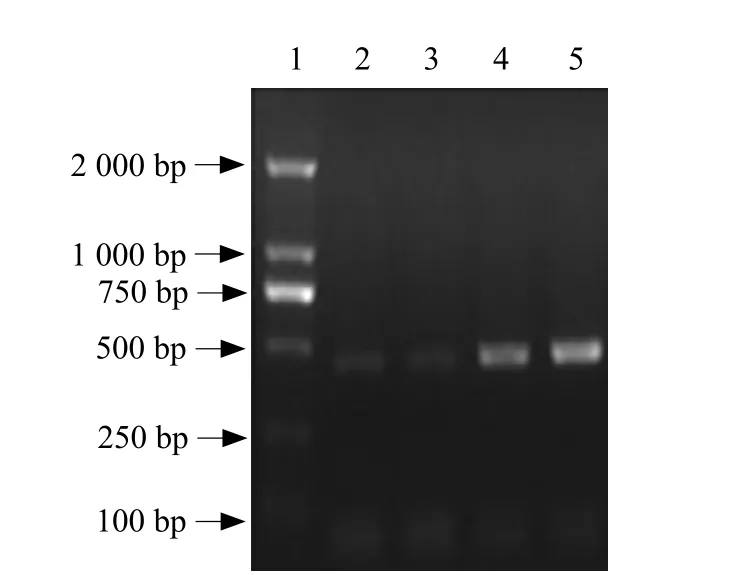
Fig. 3 RT-PCR products of lys gene from B. subtilis 168
Lysine content detected by HPLC
Linearity was assessed over concentrations varying from 2 μg · mL-1to 120 μg · mL-1for the lysine standard.The calibration plots for lysine were fit to linear equations of slope and intercept y =0.0003x+2.3725(Fig. 4). The correlation coeff i cient was in the excess of 0.99 in case.
The content of lysine in the induced pHT43/lys/Bacillus subtilis 168 increased by 9.85% compared with Bacillus subtilis 168.

Table 1 Lysine content analysis in Bacillus subtilis 168(g • L-1)

Fig. 4 Standard curve of lysine by HPLC
Discussion
The lys gene encoding the lysine-rich protein,containing 10.8 % of lysine and other essential amino acids, such as threonine, valine and isoleucine, etc.,was isolated from the seeds of winged bean. In the present study, lysine-rich gene was cloned and the corresponding protein was expressed in Bacillus subtilis 168. It was an effective method to enhance the lysine content by transferring alien lysine-rich genes in cereals. The results in this study were similar to the previous reports of maize, rice, wheat and lettuce (Li et al., 2006; Gao et al., 2001; Meng et al., 2001).
L-lysine is synthesized from L-aspartate as a part of the aspartate pathway in Bacillus subtilis. The first step in the common pathway, phosphorylation of L-aspartate, is catalyzed by aspartate kinase (AK).Bacillus subtilis has three independently regulated isozymic aspartate kinases. AKI (encoded by dapG)is allosterically inhibited by meso-diaminopimelate(meso-DAP), AKII (encoded by lysC)is inhibited by L-lysine, and AKIII (encoded by yclM, also designated thrD)is regulated by concerted feedback inhibition by L-lysine and L-threonine (Graves and Switzer, 1990). Although decreased feedback inhibition of AKI and AKII was demonstrated in Bacillus subtilis, improved L-lysine production was not demonstrated. Mutations in the 5' untranslated part of lysC mRNA decreased repression and increased L-lysine pro-duction in Bacillus subtilis 168, but significant L-lysine production by such mutants (more than 1 g · L-1)has not been reported.Until now, no mutation in any AKIII-encoding genes that leads to increased levels of L-lysine production in Bacillus species has been described.Dihydrodipicolinate synthase (DHDPS), encoded by dapA, has been proved to be another key enzyme for microbial L-lysine production at the end product of the pathway (Cremer et al., 1991; Eggeling and Sahm, 2003; Grundy et al., 2003). It is suggested that this enzyme is not feedback inhibited in Bacillus subtilis. Comparatively, DHDPS from plants is strongly inhibited by lysine. The last step in the L-lysine biosynthetic pathway is catalyzed by meso-DAP decar-boxylase which is encoded by lysA. In Bacillus subtilis, lysA is repressed by L-lysine (Belitsky, 2002).
Bacillus subtilis are gram-positive soil bacteria that produce and export a variety of hydrolytic enzymes that enable them to utilize complex carbohydrates in their natural habitats (Huang, 2005). Many genes have been successful expressed in Bacillus subtilis, as early as 1984, Ohmura achieved α-amylase gene signal peptide sequence and promoter E. coli β-lactamase expressed in Bacillus subtilis.
The sample of induced culture was detected by SDSPAGE and the recombinant protein band had not been found. It might have been degraded by extra cellular enzymes secreted by Bacillus subtilis. In conclusion,lysine content was increased by transferring alien lysine-rich gene in Bacillus subtilis 168 from 49.32 mg · L-1up to 54.18 mg · L-1.
Conclusions
In this study, lys gene was cloned from winged bean and the recombinant plasmid pHT43/lys was constructed, then it was expressed significantly in Bacillus subtilis 168. Through data analysis, the content of lysine increased by 9.85% in Bacillus subtilis 168.
Anastassiadis S. 2007. L-Lysine fermentation. Recent Patent Biontechnol, 1: 11-24.
Beauregard M, Hefford M A. 2006. Enhancement of essential amino acid contents in crops by genetic engineering and protein design.Plant Biotechnol J, 4: 561-574.
Becker J, Klopprogge C, Wittmann C. 2008. Metabolic responses to pyruvate kinase deletion in lysine producing Corynebacterium glutamicum. Microb Cell Fact, 5(13): 7-8.
Belitsky B R. 2002. Biosynthesis of amino acids of the glutamate and aspartate families, alanine and polyamines. Sonenshein A L, Hoch J A,Losick R. Bacillus subtilis 168 and its closest relatives: from genes to cells. ASM Press, Washington, DC.
Cremer J, Eggeling L, Sahm H. 1991. Control of the lysine biosynthesis sequence in Corynebacterium glutamicum as analyzed by overexpression of the individual corresponding genes. Appl Environ Microbiol, 57: 1746-1752.
Eggeling L, Sahm H. 2003. New ubiquitous translocators: amino acid export by Corynebacterium glutamicum and Escherichia coli. Arch Microbiol, 180: 155-160.
Frizzi A, Huang S, Gilbertson L A, et al. 2008. Modifying lysine biosynthesis and catabolism in corn with a single bifunctional expression/silencing transgene cassette. Plant Biotechnol J, 6: 13-21.Galili S, Guenoune D, Wininge S, et al. 2000. Enhanced levels of free and protein2bound threonine in transgenic alfalfa (Medicago sativa L.)expressing a bacterial feedback-insensitive aspartate kinase gene.Transgenic Res, 9(4): 137-144.
Gao Y F, Jing Y X, Shen S H, et al. 2001. Transfer of lysine-rich protein gene into rice and production of fertile transgenic plants. Acta Botanica Sinica, 43(5): 506-511.
Graves, L M, Switzer R L. 1990. Aspartokinase III, a new isozyme in Bacillus subtilis 168. J Bacteriol, 172: 218-223.
Grundy, F J, Lehman S C, Henkin T M. 2003. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc Natl Acad Sci USA, 100: 12057-12062.
He X Y, Tang M Z, Luo Y B, et al. 2009. A 90-day toxicology study of transgenic lysine-rich maize grain (Y642)in rague-Dawley rats. Food Chem Toxicol, 47(2): 425-32.
Huang J, Wang G, Xiao L. 2005. Cloning, sequencing, and expression of the xylanase gene from a Bacillus subtilis strain B10 in Escherichia coli. Bioresour Technol, 97: 802-808.
ILSI. 2008. Maize with increased lysine (lysine maize-LY038). In nutritional and safety assessments of foods and feeds nutritionally improved through biotechnology: case studies. Compr Rev Food Sci Food Safety, 7: 99-106.
Jose M, Gasent-Ramirez, Benitez T. 1997. Lysine-overproducing mutants of saccharmyces cerevisiae baker's yeast isolated in continuous culture. Appl Environ Microbiol, 12: 4800-4806.
Kircher M, Pfefferle, W. 2001. The fermentative production of L-lysine as an animal feed additive. Chemosphere, 43: 27-31.
Leuchtenberger W. 1996. Amino acids: technical production and use.Rehm H J, Reed G, Puhler A, et al. Biotechnology: a multi volume comprehensive treatise. Products of Primary Metabolism. Weinheim,New York. pp: 465-502.
Li X T, Li X, Zhang J W, et al. 2006. Expression and inheritance of lysine-rich protein gene in lettuce. Chin J Appl Environ Biol, 12(4):472-475.
Meng C M, Chen X Q, Liang R Q, et al. 2004. Expression of lysinerich protein gene and analysis of lysine content in transgenic wheat.Chinese Science Bulletin, 49(19): 2053-2058.
Ohnishi J, Mitsuhashi S, Hayashi M, et al. 2002. A novel methodology employing Corynebacterium glutamicum genome information to generate a new L-lysine-producing mutant. Appl Microbiol Biotechnol, 58(2): 217-223.
Stragier P, Losick R. 1996. Molecular genetics of sporulation in Bacillus subtilis 168. Annu Rev Genet, 30: 297-41.
Sun S S M, Xiong L W, Jing Y X, et al. 1998. Lysine rich protein from winged bean: U.S.A. patent application no, 08/964,722.
Tosaka O, shihaya M, Moninaga Y. 1979. Mode of concersion of asparato B semialdehyde to L-threonine and L-lysine in B-revibacterium lactofermentum. Agri Biol Chem, 43(2): 265.
Zhu X, Galili G. 2003. Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also trans regulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell, 15: 845-853.
 Journal of Northeast Agricultural University(English Edition)2013年1期
Journal of Northeast Agricultural University(English Edition)2013年1期
- Journal of Northeast Agricultural University(English Edition)的其它文章
- Ecuador Export Potential Agricultural Products to China
- Effects of Sub-chronic Aluminum Exposure on Renal Pathologic Structure in Rats
- Effects of Low-temperature and Herbicide on Membrane Stability,Antioxidant Capacity, and Product of Metabolism in Barley Seedlings
- SWOT Analysis and Development Strategies of Maize Industry in Heilongjiang Province
- Research on DSP-Based Automatic Excitation Regulator in Small Rural Hydropower Station
- Isolation, Purif i cation and Cryopreservation of Cells from Neonatal Bovine Testis
