基于RNAi技术解析美国白蛾HcAnk1和HcAnk2基因功能及对HcNPV的敏感性
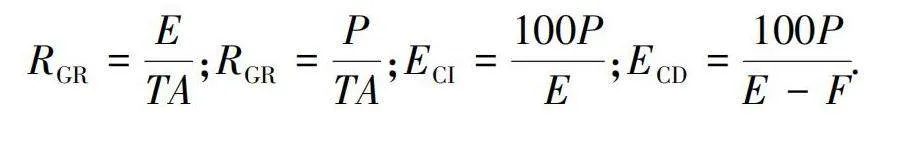

摘要:【目的】锚定蛋白是连接细胞骨架与质膜的衔接蛋白家族。但昆虫中关于锚定蛋白基因功能研究甚少。笔者克隆美国白蛾(Hyphantria cunea)锚定蛋白基因,阐明该基因在其各发育期和组织中的表达特征及对美国白蛾核型多角体病毒(Hyphantria cunea nucleopolyhedrosisvirus, HcNPV)的敏感性,为进一步开发HcNPV增效剂提供理论依据。【方法】通过转录组文库筛选出Ankyrin1(HcAnk1)和Ankyrin2(HcAnk2)基因,利用生物信息学分析基因特性,采用qRT-PCR检测不同发育阶段、不同组织以及不同HcNPV浓度胁迫下HcAnks的表达量,通过RNAi技术研究HcAnks基因沉默后美国白蛾对HcNPV的敏感性。【结果】HcAnk1和HcAnk2基因开放阅读框依次为1 392和1 866 bp,分别编码463和621个氨基酸,蛋白质分子质量分别为59.18和69.19 ku,理论等电点分别为5.74 和 8.66;进化树分析显示HcAnk1与粉纹夜蛾(Trichoplusia ni)亲缘关系最近而聚为一类。HcAnks在各个发育阶段均有表达,卵期表达量最高,6龄表达量最低;HcAnks分别在中肠和后肠中表达量最高;在卵巢和精巢中未检测到HcAnk1,而HcAnk2主要在中肠和精巢中表达。在不同浓度HcNPV胁迫下,HcAnks转录水平表现为低浓度诱导,高浓度抑制。沉默HcAnks美国白蛾4龄幼虫的相对生长率(RGR)和食物转化效率(ECD)显著降低。此外,沉默HcAnks的美国白蛾4龄幼虫对HcNPV敏感性显著增加。【结论】HcAnks在抵抗HcNPV侵染过程中发挥重要作用,HcAnks可作为HcNPV增效剂,用于美国白蛾无公害防治。
关键词:美国白蛾;锚定蛋白;美国白蛾核型多角体病毒;RNAi
中图分类号:S763.3 文献标志码:A开放科学(资源服务)标识码(OSID):
文章编号:1000-2006(2024)03-0181-10
HcAnk1 and HcAnk2 genes function and HcNPV susceptibility of Hyphantria cunea based on RNAi
PENG Mengmeng1, WU Hongqu1,2, ZHANG Jiawen1, YAN Liqiong1, CAO Chuanwang1, SUN Lili1*
(1. Key Laboratory of Sustainable Forest Ecosystem Management-Ministry of Education, Northeast Forestry University,
Harbin 150040, China; 2. Hubei Biopesticide Engineering Research Center, Wuhan 430070, China)
Abstract:【Objective】 Ankyrin is a family of adaptor proteins that connect the cytoskeleton to the plasma membrane. However, the functions of insect ankyrin are poorly understood. In this study, the Ankyrin genes of Hyphantria cunea were cloned and their expression characteristics were determined in different developmental stages and tissues of H. cunea. The mortality rate of H. cunea was measured under the stress of exposure to nucleopolyhedrosis virus, which provided a theoretical basis for further developing synergists for NPV-based control strategies. 【Method】 The Ankyrin1 (HcAnk1) and Ankyrin2 (HcAnk2) genes were screened by a transcriptome library, and the characteristics of the two Ankyrin genes were determined using bioinformatics. Using qRT-PCR, the expression of HcAnk1 and HcAnk2 genes were determined at different developmental stages and in different tissues, and under different HcNPV concentrations. The survival rate of H. cunea larvae under HcNPV stress was investigated after the gene silencing of HcAnk1 and HcAnk2 using an RNAi technique. 【Result】 The open reading frames of the HcAnk1 and HcAnk2 genes were 1392 and 1866 bp, encoding 463 and 621 amino acids, respectively. The molecular weights of the HcAnk1 and HcAnk2 proteins were 59.18 and 69.19 ku, respectively, and the theoretical isoelectric points were 5.74 and 8.66, respectively. A phylogenetic analysis showed that HcAnk1 was closely related to Trichoplusia ni and was clustered into one group. HcAnk1 and HcAnk2 were expressed at all developmental stages, with the highest expression in the egg stage and the lowest expression in the sixth instar larva. The highest expression of HcAnk1 and HcAnk2 was observed in the midgut and hindgut, respectively, but HcAnk1 was not detected in the ovary or testis, while HcAnk2 was mainly expressed in the midgut and testis. In tests with different levels of HcNPV stress, the transcription levels of HcAnk1 and HcAnk2 were induced at low concentrations and inhibited at high concentrations. The relative growth rate and food conversion efficiency (ECD) were significantly decreased after silencing HcAnks in H. cunea larvae. Additionally, the H. cunea with HcAnks silencing were significantly less resistant to HcNPV. 【Conclusion】 HcAnk1 and HcAnk2 play an important role in the resistance to HcNPV. HcAnk1 and HcAnk2 can be used as HcNPV synergists for the pollution-free control of H. cunea.
Keywords:Hyphantria cunea; Ankyrins; Hyphantria cunea nucleopolyhedrovirus; RNAi
Ankyrins are a family of intracellular structural proteins forming a fundamental component of the cytoskeleton and associated with several trans-sarcolemmal skeleton and membrane proteins[1-2]. Some Ank genes has been identified including one Caenorhabditis elegans (unc-44), two Drosophila (Dank1, Dank2), Ank1, which is expressed ubi-quitously, and the neuron-specific Ank2[3-4], and three mammalian (Ank1, Ank2, Ank3) genes. However, little is known about the ankyrins in Hyphantria cunea.
Ankyrins provide the linkage between the cytoskeleton and the plasma membrane, as well as the intracellular organelle membranes.Ankyrins were also involved in the signal transduction and protein sorting[5-6]. Ankyrins evolved within the Metazoa as an adaptation for organizing membrane microstructure and directing membrane traffic. The mutations of Ankyrin genes in human lead to severe genetic diseases such as fatal cardiac arrhythmias and hereditary spherocytosis[7]. Mutations of Ankyrin gene, unc-44, in the C. elegans affect axonal guidance and result in uncoordinated movement[8], indicating the importance of Ankyrins in animal development. The Ankyrin gene is ubiquitously expressed and enriched within the postsynaptic muscle membranes of the Drosophila neuromuscular junction (NMJ)[9-10]. RNAi-mediated depletion of Ankyrin from the muscle does not alter the stability or function of the NMJ[10]. In contrast, expression of Ankyrin2 is restricted to the nervous system[5-6]. In Drosophila, Ankyrin2-L (Ank2-L) is necessary to generate the membrane constrictions that normally separate individual synaptic boutons and achieve the normal spacing of sub-synaptic protein domains including the normal organization of synaptic cell adhesion molecules. Expression of a Ankyrin2 fragment disrupts the structure of the axon initial segment and causes axonal degeneration in Drosophila[11]. Studies have shown that viral Ankyrin (vAnkyrin) gene has certain homology with IKB (cactus) in D. melanogaster, which is an inhibitor of NF-kB transcriptional regulator family[12-15]. Gueguen et al.[16] found that polydnaviral Ankyrin proteins aid parasitic wasp survival by selective inhibition of hematopoietic and immune NF-kappa B signaling in insect hosts. However, there is limited information on the molecular mechanisms underlying the response of insect Ankyrin to nucleopolyhedrovirus. Baculoviruses are highly pathogenic to major insect pest species and used as biological control agents in agriculture and forestry[17]. The virus evolves to optimize host infection and its dispersion, but the host attempts to enhance protection against the pathogen.
The H. cunea is a notorious quarantine pest affecting hundreds of agricultural and forestry plants. The nucleo polyhedron virus is widely used in the biological control of H. cunea. However, the Hyphantria cunea nucleopolyhedrovirus (HcNPV) is slow to take effect, and also H. cunea has developed resistance to HcNPV gradually. Therefore, it is of great significance to find a synergist for the nuclear polyhedrosis virus. The previous research found Ankyrins in H. cunea (HcAnks) were induced by HcNPV and expected to be a synergist of HcNPV[18]. In this study, we cloned the full-length HcAnk1 and HcAnk2 genes of H. cunea, and evaluated the expression level of Anks genes in H. cunea larvae under HcNPV stress by RT-qPCR technology. Moreover, the effects of HcAnk1 and HcAnk2 on its nutrient utilization and survival were investigated under HcNPV stress.
1 Materials and Methods
1.1 Rearing and stress treatment of H. cunea
The eggs of H. cunea and artificial diets were purchased from Ecology and Nature Conservation Institute, Chinese Academy of Forestry (Beijing, China). The artificial diets were stored at 4 ℃ before hatching and usage. Briefly, H. cunea larvae were reared in 250 mL transparent plastic bottles at (25 ± 1)℃ with a photoperiod of 16∶8 hour light (L)∶dark (D), and fed an artificial diet.
Four different HcNPV stocks (3 × 105, 3 × 106, 3 × 107 and 3 × 108 PIBs/mL polyhedra, PIB is polyhedral inclusion body) were suspended in the distilled water, which were obtained from the Research Institute of Forest Ecology, Environment and Protection, Chinese Academy of Forestry (Beijing, China). A 10 μL of viral suspension was spread evenly on 0.50 g (5 mm × 5 mm × 1 mm) of artificial diets. These HcNPV treated diets were fed to 30 newly molted third instar H. cunea larvae after they had been previously starved for 12 h. Control H. cunea larvae were fed on the same quantity of artificial diets treated with the distilled water. Six replicates (30 individuals per replicate) were used for each treatment, and eight to ten larvae were randomly selected at 6, 12, 24, 48, 72, 96 and 120 h, and then stored for RNA extraction at -80 ℃ after being rapidly frozen in liquid nitrogen.
1.2 RNA isolation and first-strand cDNA synthesis
The RNA was extracted from H. cunea eggs, 1st-to 7th-instar larvae, pupae, adults, tissue samples (the head, silk glands, foregut, midgut, hindgut, epidermis, testis, ovary, Malpighian tubules, and fat body of day 1 of the 7th instar H. cunea larvae) and HcNPV treated larvae using the RNeasy Mini kit (Qiagen, Valencia, CA, USA). The cDNA was synthesized using the total RNA (0.5 μg) and the PrimeScript®RT Reagent Kit with gDNA Eraser (Perfect Real Time; TaKaRa, Japan) according to the manufacturer’s protocol.
1.3 Characteristics of HcAnk genes and phylogenetic analysis
The H. cunea transcriptome was analyzed by Solexa sequencing at GENEWIZ Company (Suzhou, China)[18]. The HcAnk genes were identified on the basis of functional annotation and confirmed using RT-PCR and sequencing. The phylogenetic tree was created using the neighbor-joining (NJ) method in MEGA 6.0 with 1 000 bootstrap replicates[19].
1.4 Quantitative RT-PCR analysis
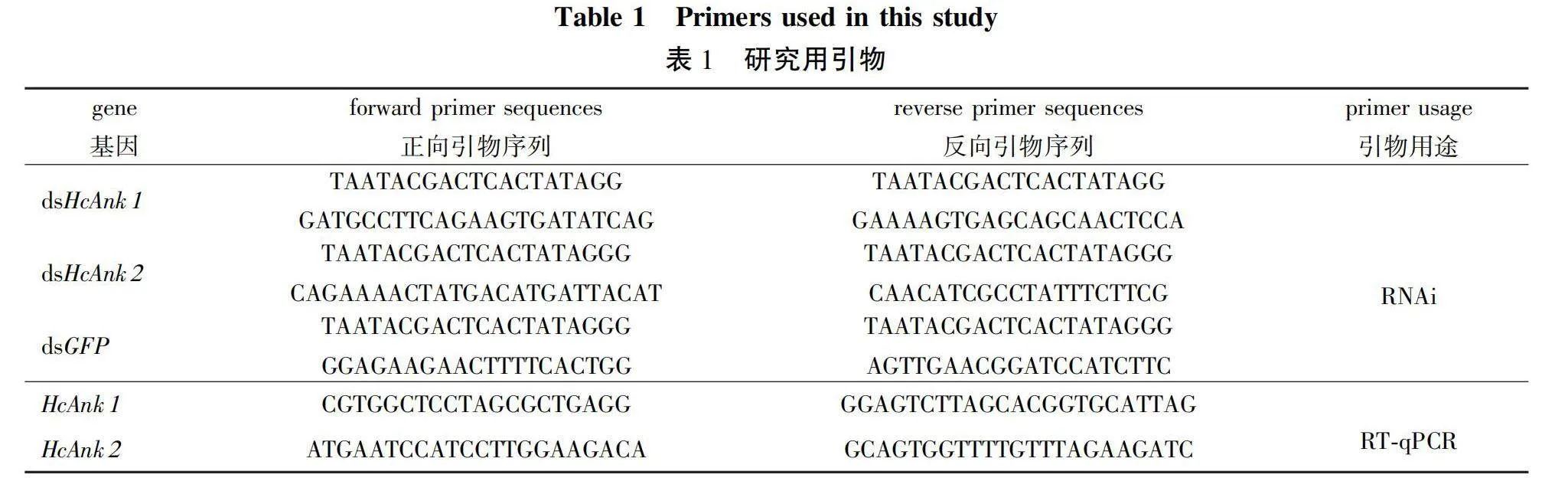
Total RNA of tissues of H. cunea at developmental stages were treated with DNaseI to remove the genomic DNA. Approximately 0.5 μg of total RNA was reverse transcribed to cDNA using 1 μmol/L of oligo d(T) primer. 10 μL of the synthesized cDNAs was diluted to 100 μL with sterile water and used as the template for RT-qPCR in an MJ OpticonTM2 machine (BioRad, Hercules, CA, USA). The most stable reference genes, EF1-α and RPL13, in H. cunea were chosen as internal controls to normalize the amount of total RNA present in each reaction. The reaction mixture (20 μL) contained 10 μL of SYBR Green RT-qPCR master mix (Toyobo), gene-specific primers (1 μL each; 0.5 μmol/L, Table 1), and 2 μL of cDNA template (equivalent to 100 ng of total RNA), and 7 μL nuclease-free water. The amplification was conducted with the following cycling parameters: 94 ℃ for 30 s followed by 45 cycles at 94 ℃ for 12 s, 60 ℃ for 30 s, 72 ℃ for 40 s and 1 s at 82 ℃ for plate reading. A melting curve was generated for each sample at the end of each run to assess the purity of the amplified products. RT-qPCR was carried out in triplicate biological repeats to ensure the reproducibility of the results. The clone expression levels were calculated from the threshold cycle according to the 2-△△Ct method[20].
1.5 RNAi analysis and bioassay in H. cunea larvae
Gene specific primers with T7 promoter (Table 1) were designed to synthesize the dsRNAs targeting 463 or 461 bp nucleotides of the ORF region of the HcAnk1 and HcAnk2 using the MEGAscript T7 High Yield Transcript Kit (Ambion) according to the manufacturer’s instructions. The dsRNA of a green fluorescent protein (GFP) gene was generated using a pMW1650 plasmid as template (a generous gift from Dr. Nannan Liu, Auburn University, Alabama, USA). The synthesized dsRNAs were diluted in diethyl pyrocarbonate (DEPC)-treated water with a concentration of 1 μg/μL. This dsRNA solution was microinjected into the 4th instar H. cunea larvae. The dsRNA (1μg) was microinjected into the penultimate posterior abdominal section of each 3rd instar H. cunea larvae with an injection needle (MICROLITERTM No.65 with 33-gauge needle, Hamilton Co., Reno, NV, USA). Controls were microinjected with the dsGFP. Ten H. cunea larvae were used for each replicate, and three replicates were performed. After 24, 48 or 72 h, the larvae injected with dsHcAnk1, dsHcAnk2 and dsGFP were collected and stored at -80 ℃ for RNA extraction.
The 4th instar H. cucea larvae were fed on the artificial diet treated with HcNPV (3×108 PIBs/mL) two hours after microinjection of dsGFP or dsHcAnk1 or dsHcAnk2. After HcNPV treatment, the cumulative mortality of the microinjected H. cucea larvae was determined during a period of 192 h.
1.6 Nutrition utilization
The procedures were carried out according to method developed by Sun et al.[21]. The silenced and normal 3rd instar larvae were reared for 72 h on the artificial diets in 250 mL transparent plastic bottles. Initial dry weight of each larva and every piece of feed were estimated by setting aside aliquots at the beginning of the experiment and then a fresh/dry conversion ratio was obtained. Ten H. cunea larvae were used in each replicate and each treatment was repeated three times. The following growth indices of larvae were calculated: relative consumption rate (RCR,RCR), relative growth rate (RGR,RGR), efficiency of conversion of ingested food (ECI,ECI), efficiency of conversion of digested food(ECD,ECD).
In the preceding formulae: A is mean of the dry weight of larvae during T;
E is dry weight of food eaten;
F is dry weight of feces produced;
P is dry weight gain of insect;
T is duration of the experimental period.
1.7 Statistical analysis
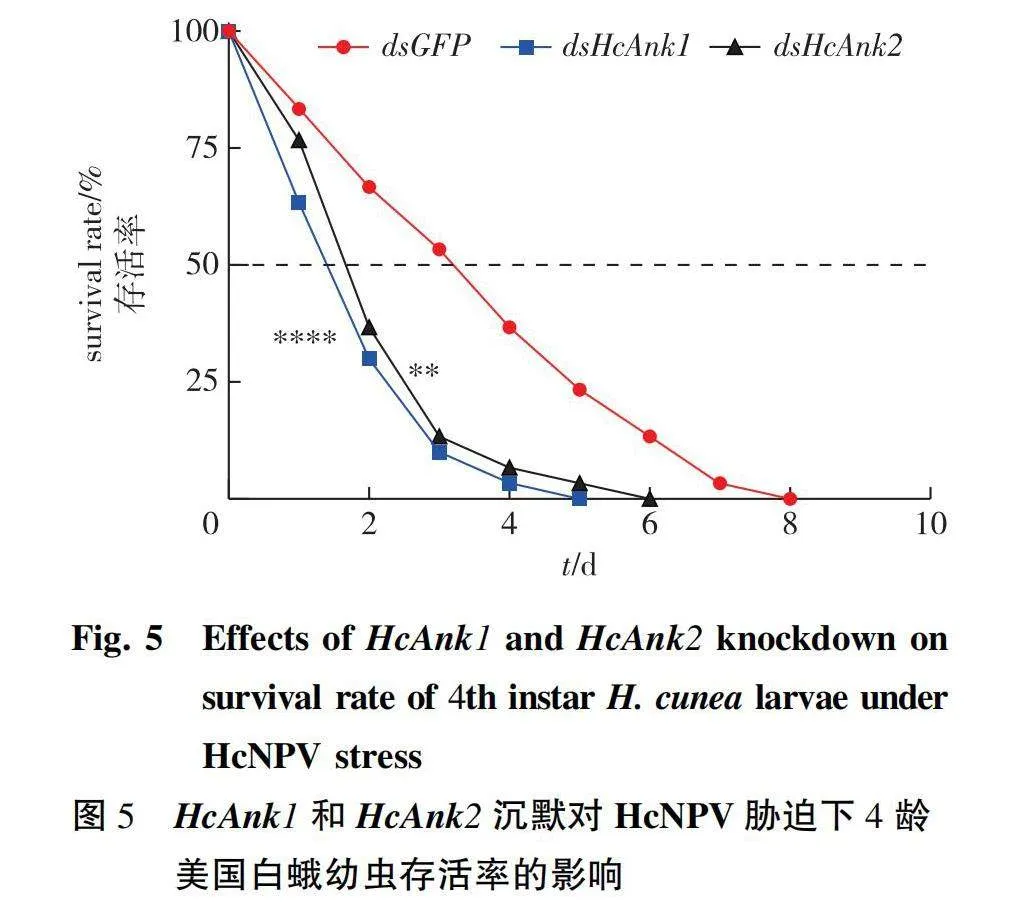
Excel 2019 was used to collect and analyze the data. For statistical analysis, GraphPad Prism 8.0 was used (GraphPad Software Inc., La Jolla, CA, USA). Log rank tests (Mantel-Cox log-rank test) were preformed to analyze trends in lifespan in the HcNPV treatments assays. The differences in expression levels at the different developmental stages were evaluated with one-way analysis of variance (ANOVA), followed by Tukey’s test, in SPSS 16.0 (Plt;0.05). Two-tailed Student’s t-test and One-way analysis of variance (ANOVA) were used to analyze the relative expression level assays to test any relationships between the different genotypes and HcNPV stress concentrations. Asterisk symbols reflect significant differences (*. Plt;0.05; **. Plt;0.01; ***. Plt;0.001; ****. Plt;0.000 1).
2 Results
2.1 cDNA cloning and bioinformatic analysis of Ankyrin genes in H. cunea
Two full-length cDNA of Ankyrins gene in H. cunea, HcAnk1 and HcAnk2 was identified from the transcriptome database of H. cunea, with open reading frame (ORF) of 1 392 and 1 866 bp encoding 463 and 621 amino acids. The molecular mass was predicted as 59.18 and 69.19 ku and the isoelectric point (pI) was predicted as 5.74 and 8.66 for HcAnk1 and HcAnk2, respectively. The 19 representative insect Ankyrin sequences were searched to improve the accuracy of phylogeny. Phylogenic analysis was carried out based on amino acid sequences of HcAnk1, HcAnk2 and other 19 lepidopteran Ankyrin proteins. HcAnk1 originated from the same evolutionary root as Trichoplusia ni (XP 026734686.1) with a bootstrap value of 92 (Fig. 1). However, HcAnk2 and 11 species of insects are clustered together species of insects are clustered together.
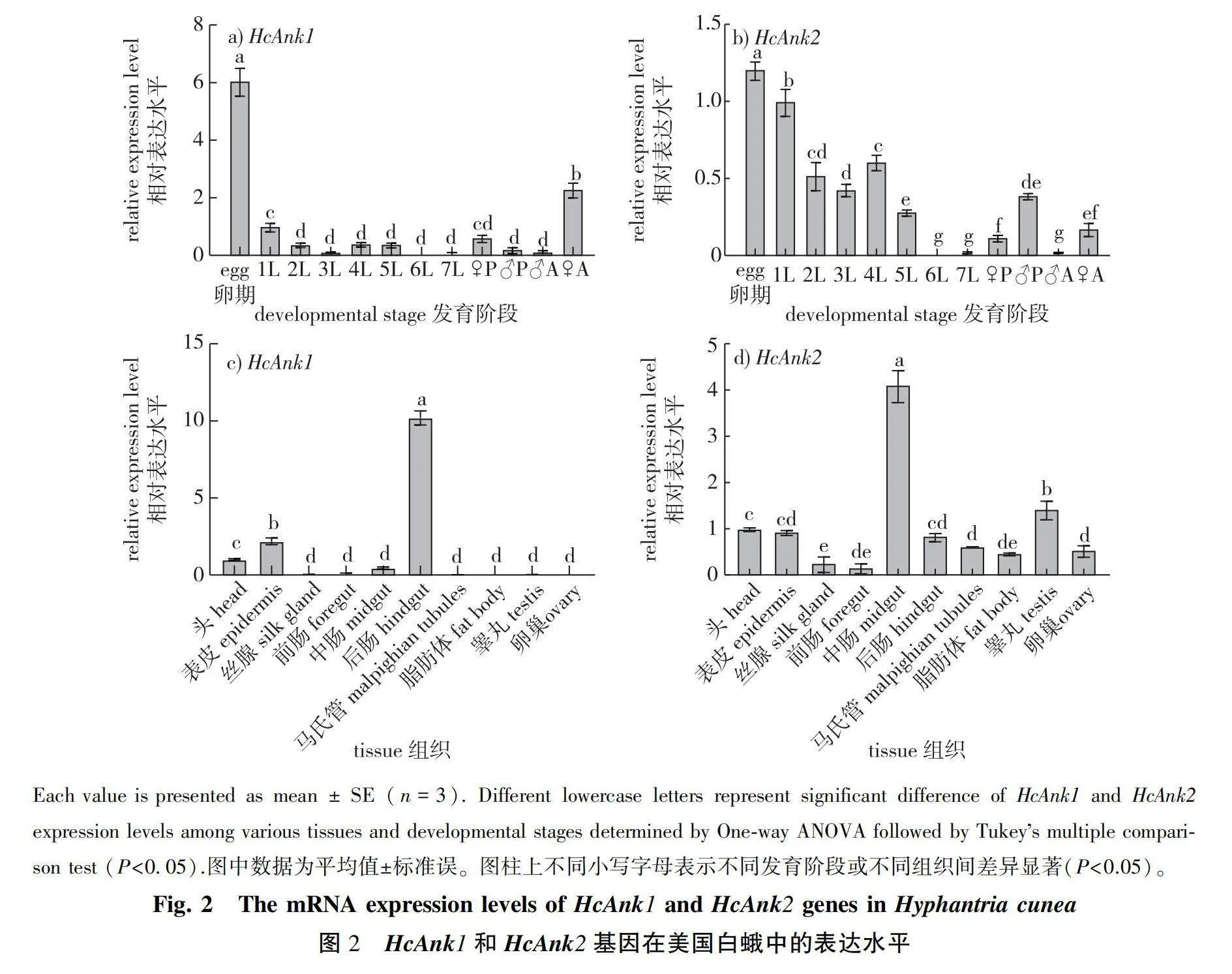
2.2 Specific expression of the HcAnks in different stages and tissues
To understand the physiological function of HcAnk1 and HcAnk2 genes, RT-qPCR was applied to investigate temporal and spatial expression patterns. The mRNA transcription levels of the HcAnk1 and HcAnk2 genes in the H. cunea eggs, seven different larval instars (1L to 7L), pupae, adults and ten tissues were analyzed, respectively. HcAnk1 gene was expressed at all the developmental stages of H. cunea, but it exhibited stage-specific patterns (Fig. 2a). The highest expression level was observed at egg stage with 6.05-fold of that in 1st instar larvae, while the lowest expression level was observed at 6th instar larvae with 0.004-fold of that in 1st instar larvae. HcAnk1 gene was downregulated in the adult males compared with that in the 1st larval stage (0.12-fold), but the HcAnk1 gene was upregulated in the adult females compared with the 1st larval stage (2.3-fold) (Fig.2a). However, the highest expression level of HcAnk2 was observed at egg stage (1.21-fold) while the lowest expression level was observed at 6th instar larvae (0.01-flod) compared with that in 1st instar larvae. The relative expression levels of 1st- to 3rd-instar larvae were increasingly downregulated, but those of 4th- to 6th-instar larvae were upregulated. At the adult stage, the expression level of HcAnk2 in female adults was 6.28-fold higher than that in male adults (Fig. 2b). It is speculated that HcAnk2 may play an important role in the reproduction of H. cunea females.
The highest expression of HcAnk1 gene was found in the hindgut (10.21-fold) compared with that in the head (Fig. 2c). The expression of HcAnk1 in the midgut was 0.23-fold of that in the head (Fig. 2b). The expressions of HcAnk1 and HcAnk2 genes in silk gland, foregut and fat body were low among all tissues. However, the highest expression of HcAnk2 gene was found in the midgut (4.11-fold) compared with that in the head (Fig. 2d). The expression levels in epidermis, hindgut, Malpighian tubules were close to those in head (Fig. 2d).
2.3 Effects of HcNPV infection on gene expression
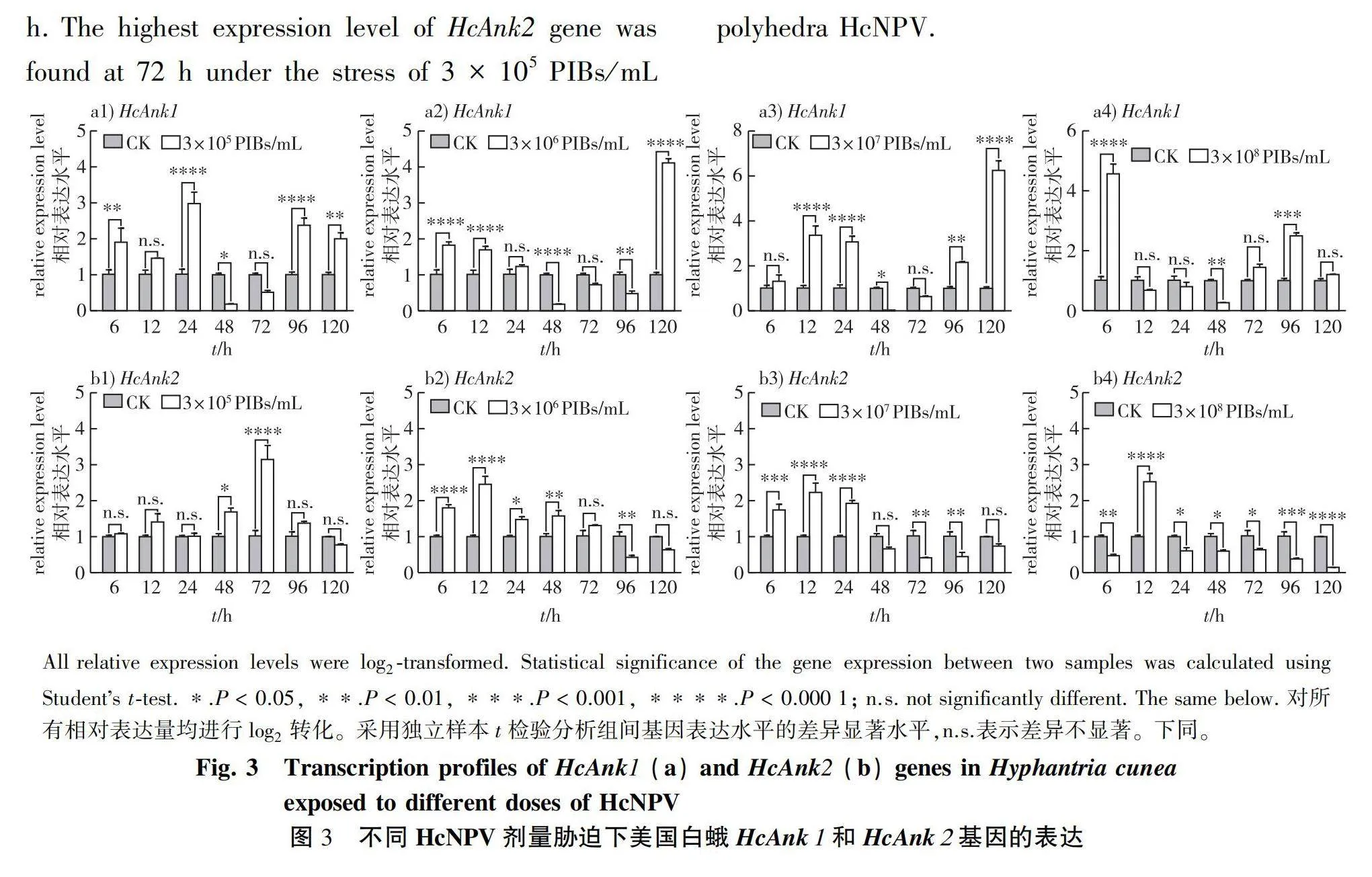
After exposure to four different concentrations of HcNPV,HcAnk1 expression in H. cucea was upregulated. With the extension of exposure to HcNPV, the HcAnk1 expression levels were significantly increased, except at 48 and 72 h. Under 3 × 108 PIBs/mL polyhedra HcNPV stress, the HcAnk1 expressions at 6, 72, 96, 120 h were upregulated by 4.57-, 1.45-, 2.50- and 1.21-fold of that in the control (Fig. 3a). The expression of HcAnk1 gene was the highest in H. cucea larvae treated by the 3 × 107PIBs/mL polyhedra HcNPV for 120 h, the four HcAnk1 genes were induced by 8-fold of that in the control.
Exposed to HcNPV concentrations of 3 × 106 PIBs, 3 × 107 and 3 × 108 PIBs/mL polyhedra, the HcAnk2 mRNA level in H. cucea was remarkably decreased compared with that of the control group with the extension of time (Fig. 3b). Under the stress of HcNPV with the different concentrations of 3 × 105, 3 × 106, 3 × 107 and 3 × 108 PIBs/mL polyhedra for 12 h, the HcAnk2 expressions were upregulated by 1.41-, 2.46-, 2.23- and 2.53-fold of that in the control. Under the stress of 3 × 106 PIBs/mL polyhedra HcNPV, the HcAnk2 expressions were upregulated at 6, 12, 24, 120 h and mainly downregulated at 48-96 h. The highest expression level of HcAnk2 gene was found at 72 h under the stress of 3 × 105 PIBs/mL polyhedra HcNPV.
2.4 Effects of HcAnkys silencing on H. cunea performance
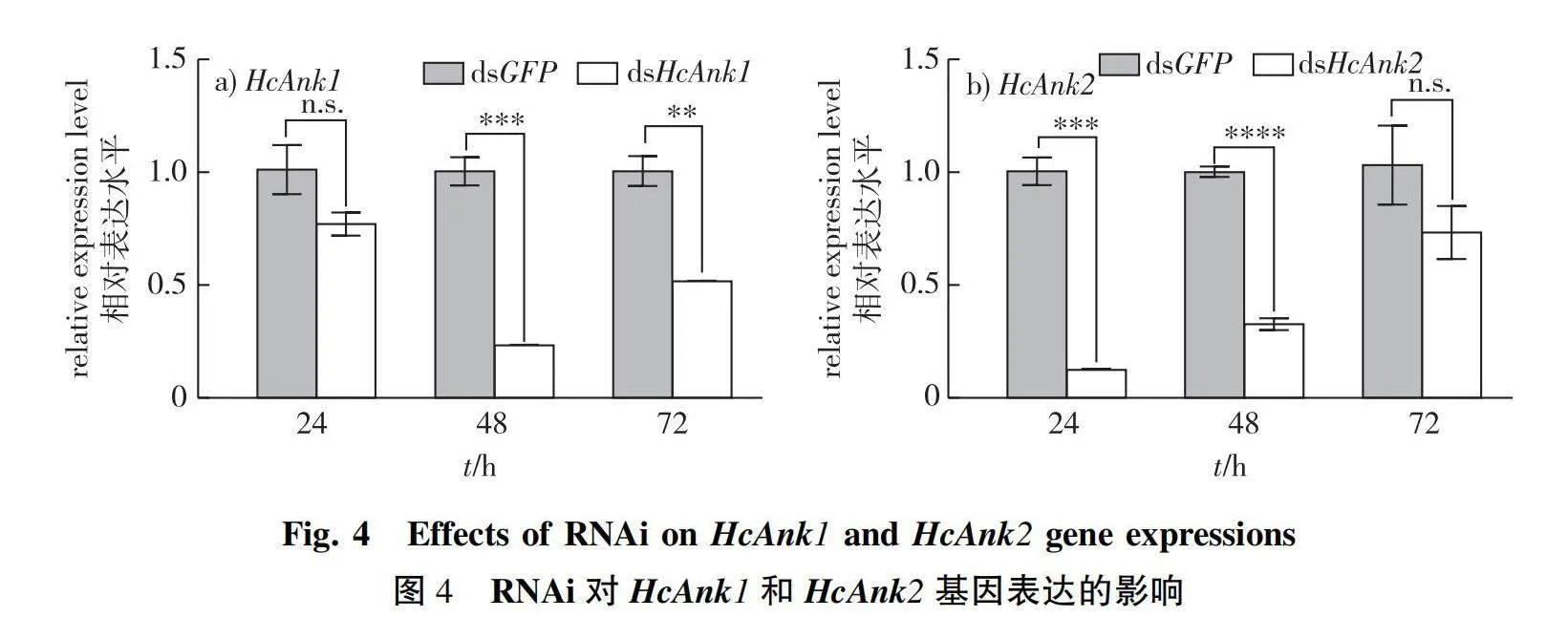
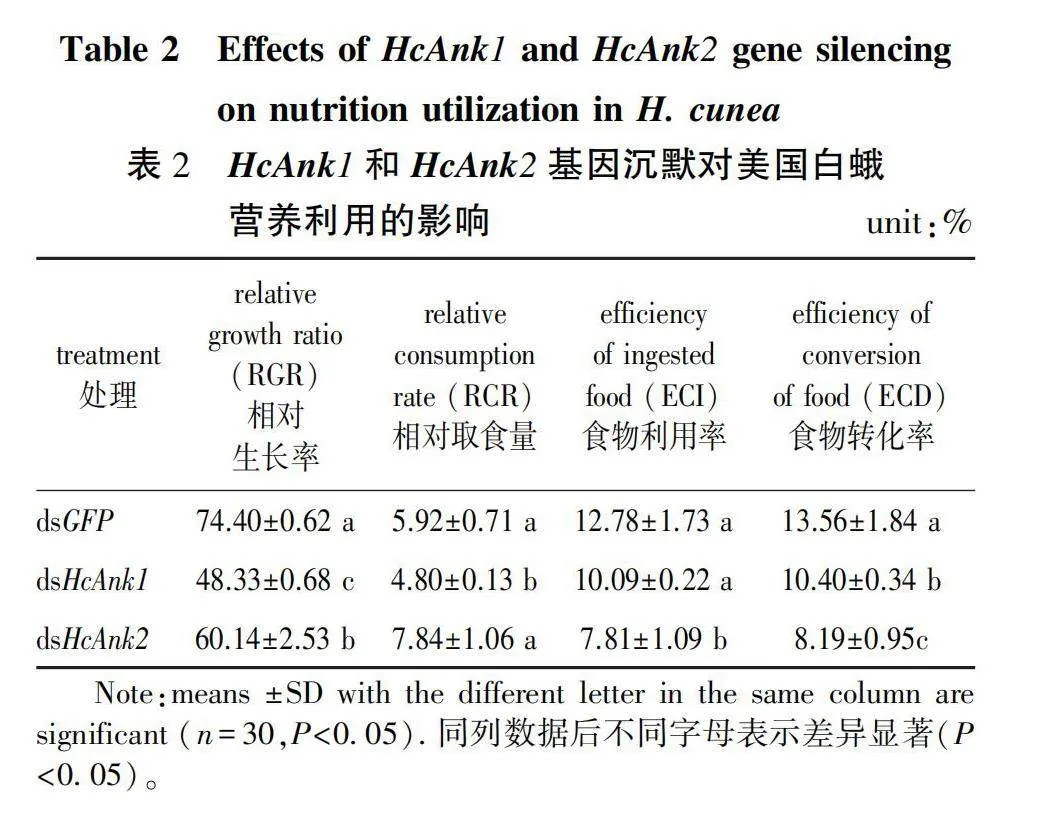
To investigate the functions of HcAnk1, HcAnk2, RNAi technology was used to silence the genes in the 4th instar H. cunea larvae. The transcript levels of HcAnk1 and HcAnk2 were downregulated by 22.97% (P gt; 0.05) and 87.59% (Plt;0.001) at 24 h, respectively. The transcript levels of HcAnk1 and HcAnk2 were significantly downregulated by 76.70% and 67.27% at 48 h, respectively. And the silencing effect of dsHcAnk1 and dsHcAnk2 was still detected at 72 h (Fig. 4a and 4b), and the best silencing effect of the HcAnk1 and HcAnk2 genes was found at 24 h and 48 h, respectively, suggesting that HcAnk1 and HcAnk2 were successfully silenced by RNAi (Fig. 4a and 4b). HcAnks silencing affects nutritional utilization of H. cunea (Table 2). The RGR, ECI and ECD of H. cunea larvae silenced by HcAnk1 and HcAnk2 were lower than that of the control H. cunea larvae silenced by GFP. However, the RCR of H. cunea larvae injected with dsHcAnk2 was higher than that of the H. cunea larvae injected with dsGFP (P gt; 0.05), and the RCR of H. cunea larvae injected with dsHcAnk1 was still lower than that of the control group injected with dsGFP (Table 2).
After microinjection of dsRNA, the H. cunea larvae were infected with HcNPV (3×108 PIBs/mL polyhedra). The larval survival percentage was recorded until the eighth day. The results showed that the survival percentage of the larvae microinjected with dsHcAnk1, dsHcAnk2 was lower than that of larvae microinjected with dsGFP at every observation time point (Fig. 5), which indicated that HcAnk1 and HcAnk2 gene silencing decreased the resistance of H. cunea to HcNPV.
3 Discussion
Ankyrin proteins are known to be involved into a multitude of functions, such as cell-cycle regulation, transcriptional regulation, cytoskeleton interactions, signal transduction, development, intracellular trafficking, sex differentiation, and they can also act as toxins[22-24]. Some researchers have confirmed that Ankyrin proteins play a major role in insect-pathogen interactions and the evolution of infections[25-26]. The expression patterns of Ankyrin vary among the developmental stages and species[5,8]. The expression patterns of the two Ank genes (Dank1 and Dank2) are different in Drosophila melanogaster, the Dank1 was evenly expressed throughout the entire life cycle of the D. melanogaster and also expressed in Drosophila S2 tissue culture cells; and the expression of Dank2 is restricted to the nervous system in the Drosophila embryo[5]. Previous studies have shown that D. melanogaster Ankyrins play important roles in cell cycle regulation and early stages of development[27-28]. The transcription level of human Ank2 peaked in early and middle embryonic development and reached a plateau at later developmental stages, while the transcriptional level of mouse Ank2 peaked in late embryonic development[29-32]. In this study, the two Ankyrins in H. cunea were highly expressed at the egg stage, but lowly at the stage of post-embryonic development. Because the egg stage is a critical phase of embryonic development, the expressions of these genes may imply their involvement of cell-cycle regulation, transcriptional regulation, development and sex differentiation[22-33]. Our present results showed that expressions of HcAnks were high in head and epidermis, whereas HcAnk1 has a higher transcription in the hindgut, and HcAnk2 has a higher expression level in midgut and testis. Ankyrin is a structural platform for protein-protein interactions, the differential expression of Anks in different tissues may play an important role in the process of cell development regulated by multiple signal pathways[34-35].
It was shown that Ank proteins of certain pathogenic intracellular bacteria or polydnaviral are secreted into the host cytoplasm and interact with host factors, involving in the immune response of insects[36-38]. In polydanvirus, Ankyrin plays an important inhibitory and interference effects in the signal transmission response to the host insect’s immune system[13,15-16, 39]. Salvia et al.[40] reported that Toxoneuron nigriceps Bracovirus Ankyrin 1(TnBVANK1), induces apoptosis by interacting with Alg-2 interacting protein X (Alix), suggesting a role of TnBVANK1 in the suppression of host immune response observed after parasitisation by T. nigriceps. Gueguen et al.[16] reported Ichnoviral Vankyrins differentially impede Toll-NF-κB-dependent hematopoietic and immune signaling in a heterologous system in vivo Drosophila, and can block NF-κB signaling in fruit fly larvae. Researches of polydnaviral ankyrin proteins in Drosophila revealed that immune-suppressive viruses may block both cellular and humoral immunity in insects to win the biological ‘arms race’. However, there is limited information on the response of Ankyrins to the stress of insect viruses. Ankyrin in the host insect did not take a positive response to the virus stress. The previous studies, researchers mainly discussed the changes of ankyrin during the parasitism of the host Drosophila. Interestingly, we found that the expression of mRNA of Ankyrins was induced by the stress of different concentrations of HcNPV in this study. In addition, the expression patterns of HcAnk1 and HcAnk2 were totally different in response to the stress of HcNPV. Under the stress of different concentrations from 3×105 to 3×108 PIBs/mL polyhedra, HcAnk1 expression was upregulated at 120 h, which was 1.21-6.25 fold higher than that in the control. However, the HcAnk2 expression was mainly inhibited at 120 h, which was only 0.15-0.77 fold of that in the control. These results indicate that the Anks gene plays an important role in the immune response of H. cunea to HcNPV stress. The role of Ank gene in response to biotic stress is conserved, and similar results are also found in plants[41]. Further functional verification confirmed knockdown of HcAnk1 and HcAnk2 in larvae caused 1.93-fold and 1.86-fold higher mortality than that of the control larvae microinjected with dsRNA of GFP under HcNPV stress, respectively. The results indicated that HcAnks are the key genes in the response to HcNPV infection. It is speculated that the decreased expression of Ank results in weakened interaction with the ANK repeat domain and the inability to normally activate downstream defense signaling components, which in turn leads to the increased sensitivity of the larvae to HcNPV. However, the molecular mechanism of ankyrins in response to HcNPV stress needs to be further verified. In the present study, we have functionally characterized HcAnk1 and HcAnk2 gene in H. cunea to better understand their roles in response to HcNPV stress. The reveal of its molecular mechanism contributes to evaluate its potential as a target gene to be applied in controlling H. cunea through RNAi approaches.
Author Contributions
M. P.and L. S. designed the research and wrote the manuscript. H. W., J.Z and L. Y. performed the experiments and analyzed the data. H.W. and C. C. revised the manuscript. All authors listed have approved the manuscript for publication.
Funding
This work was supported by grants from
Competing ofInterest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
参考文献(reference):
[1]BENNETT P B.Anchors aweigh!:ion channels,cytoskeletal proteins,and cellular excitability[J].Circ Res,2000,86(4):367-368.DOI: 10.1161/01.res.86.4.367.
[2]HRYNIEWICZ-JANKOWSKA A,CZOGALLA A,BOK E,et al.Ankyrins,multifunctional proteins involved in many cellular pathways[J].Folia Histochem Cytobiol,2002,40(3):239-249.
[3]BOULEY M,TIAN M Z,PAISLEY K,et al.The L1-type cell adhesion molecule neuroglian influences the stability of neural ankyrin in the Drosophila embryo but not its axonal localization[J].J Neurosci,2000,20(12):4515-4523.DOI: 10.1523/JNEUROSCI.20-12-04515.2000.
[4]HORTSCH M,PAISLEY K L,TIAN M Z,et al.The axonal localization of large Drosophila Ankyrin2 protein isoforms is essential for neuronal functionality[J].Mol Cell Neurosci,2002,20(1):43-55.DOI: 10.1006/mcne.2002.1113.
[5]RUBTSOV A M,LOPINA O D.Ankyrins[J].FEBS Lett,2000,482(1/2):1-5.DOI: 10.1016/s0014-5793(00)01924-4.
[6]BENNETT V,BAINES A J.Spectrin and Ankyrin-based pathways:metazoan inventions for integrating cells into tissues[J].Physiol Rev,2001,81(3):1353-1392.DOI: 10.1152/physrev.2001.81.3.1353.
[7]CAI X J,ZHANG Y H.Molecular evolution of the ankyrin gene family[J].Mol Biol Evol,2006,23(3):550-558.DOI: 10.1093/molbev/msj056.
[8]OTSUKA A J,FRANCO R,YANG B,et al.An ankyrin-related gene (unc-44) is necessary for proper axonal guidance in Caenorhabditis elegans[J].J Cell Biol,1995,129(4):1081-1092.DOI: 10.1083/jcb.129.4.1081.
[9]DUBREUIL R R,YU J.Ankyrin and beta-spectrin accumulate independently of alpha-spectrin in Drosophila[J].Proc Natl Acad Sci USA,1994,91(22):10285-10289.DOI: 10.1073/pnas.91.22.10285.
[10]PIELAGE J,FETTER R D,DAVIS G W.A postsynaptic spectrin scaffold defines active zone size,spacing,and efficacy at the Drosophila neuromuscular junction[J].J Cell Biol,2006,175(3):491-503.DOI: 10.1083/jcb.200607036.
[11]SPURRIER J,SHUKLA A K,BUCKLEY T,et al.Expression of a fragment of Ankyrin2 disrupts the structure of the axon initial segment and causes axonal degeneration in Drosophila[J].Mol Neurobiol,2019,56(8):5689-5700.DOI: 10.1007/s12035-019-1477-6.
[12]SILVERMAN N,MANIATIS T.NF-kappaB signaling pathways in mammalian and insect innate immunity[J].Genes Dev,2001,15(18):2321-2342.DOI: 10.1101/gad.909001.
[13]THOETKIATTIKUL H,BECK M H,STRAND M R.Inhibitor kappaB-like proteins from a polydnavirus inhibit NF-kappaB activation and suppress the insect immune response[J].Proc Natl Acad Sci USA,2005,102(32):11426-11431.DOI: 10.1073/pnas.0505240102.
[14]FALABELLA P,VARRICCHIO P,PROVOST B,et al.Characte-rization of the IkappaB-like gene family in polydnaviruses associated with wasps belonging to different Braconid subfamilies[J].J Gen Virol,2007,88(Pt 1):92-104.DOI: 10.1099/vir.0.82306-0.
[15]BITRA K,SUDERMAN R J,STRAND M R.Polydnavirus Ank proteins bind NF-κB homodimers and inhibit processing of Relish[J].PLoS Pathog,2012,8(5):e1002722.DOI: 10.1371/journal.ppat.1002722.
[16]GUEGUEN G,KALAMARZ M E,RAMROOP J,et al.Polydnaviral ankyrin proteins aid parasitic wasp survival by coordinate and selective inhibition of hematopoietic and immune NF-kappa B signaling in insect hosts[J].PLoS Pathog,2013,9(8):e1003580.DOI: 10.1371/journal.ppat.1003580.
[17]CHEN Y R,ZHONG S L,FEI Z J,et al.Transcriptome responses of the host Trichoplusia ni to infection by the baculovirus Autographa californica multiple nucleopolyhedrovirus[J].J Virol,2014,88(23):13781-13797.DOI: 10.1128/JVI.02243-14.
[18]SUN L L,LIU P,SUN S H,et al.Transcriptomic analysis of interactions between Hyphantria cunea larvae and nucleopolyhedro virus[J].Pest Manag Sci,2019,75(4):1024-1033.DOI: 10.1002/ps.5212.
[19]TAMURA K,STECHER G,PETERSON D,et al.MEGA6:molecular evolutionary genetics analysis version 6.0[J].Mol Biol Evol,2013,30(12):2725-2729.DOI: 10.1093/molbev/mst197.
[20]PFAFFL M W,HORGAN G W,DEMPFLE L.Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR[J].Nucleic Acids Res,2002,30(9):e36.DOI: 10.1093/nar/30.9.e36.
[21]SUN L L,WANG Z Y,WU H Q,et al.Role of ocular albinism type 1 (OA1) GPCR in Asian Gypsy moth development and transcriptional expression of heat-shock protein genes[J].Pestic Biochem Physiol,2016,126:35-41.DOI: 10.1016/j.pestbp.2015.07.004.
[22]BORK P.Hundreds of Ankyrin-like repeats in functionally diverse proteins:mobile modules that cross phyla horizontally?[J].Proteins,1993,17(4):363-374.DOI: 10.1002/prot.340170405.
[23]SEDGWICK S G,SMERDON S J.The ankyrin repeat:a diversity of interactions on a common structural framework[J].Trends Biochem Sci,1999,24(8):311-316.DOI: 10.1016/s0968-0004(99)01426-7.
[24]MOSAVI L K,CAMMETT T J,DESROSIERS D C,et al.The ankyrin repeat as molecular architecture for protein recognition[J].Protein Sci,2004,13(6):1435-1448.DOI: 10.1110/ps.03554604.
[25]HABYARIMANA F,AL-KHODOR S,KALIA A,et al.Role for the Ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages[J].Environ Microbiol,2008,10(6):1460-1474.DOI: 10.1111/j.1462-2920.2007.01560.x.
[26]PAN X X,LÜHRMANN A,SATOH A,et al.Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors[J].Science,2008,320(5883):1651-1654.DOI: 10.1126/science.1158160.
[27]AXTON J M,SHAMANSKI F L,YOUNG L M,et al.The inhibitor of DNA replication encoded by the Drosophila gene plutonium is a small,ankyrin repeat protein[J].EMBO J,1994,13(2):462-470.DOI: 10.1002/j.1460-2075.1994.tb06281.x.
[28]ELFRING L K,AXTON J M,FENGER D D,et al.Drosophila PLUTONIUM protein is a specialized cell cycle regulator required at the onset of embryogenesis[J].Mol Biol Cell,1997,8(4):583-593.DOI: 10.1091/mbc.8.4.583.
[29]ASP M,GIACOMELLO S,LARSSON L,et al.A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart[J].Cell,2019,179(7):1647-1660.e19.DOI: 10.1016/j.cell.2019.11.025.
[30]KANG H J,KAWASAWA Y I,CHENG F,et al.Spatio-temporal transcriptome of the human brain[J].Nature,2011,478(7370):483-489.DOI: 10.1038/nature10523.
[31]FERTUZINHOS S,LI M F,KAWASAWA Y I,et al.Laminar and temporal expression dynamics of coding and noncoding RNAs in the mouse neocortex[J].Cell Rep,2014,6(5):938-950.DOI: 10.1016/j.celrep.2014.01.036.
[32]KAWANO S,BABA M,FUKUSHIMA H,et al.Autism-associated ANK2 regulates embryonic neurodevelopment[J].Biochem Biophys Res Commun,2022,605:45-50.DOI: 10.1016/j.bbrc.2022.03.058.
[33]ISLAM Z,NAGAMPALLI R S K,FATIMA M T,et al.New paradigm in ankyrin repeats:beyond protein-protein interaction module[J].Int J Biol Macromol,2018,109:1164-1173.DOI: 10.1016/j.ijbiomac.2017.11.101.
[34]BENNETT V,GILLIGAN D M.The spectrin-based membrane skeleton and micron-scale organization of the plasma membrane[J].Annu Rev Cell Biol,1993,9:27-66.DOI: 10.1146/annurev.cb.09.110193.000331.
[35]KREIS T,VALE R.Guidebook to the extracellular matrix,anchor,and adhesion proteins[M] Oxford, New York:Sambrook and Tooze Publication at Oxford University Press,1999:340-392.
[36]PARK J,KIM K J,CHOI K S,et al.Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins[J].Cell Microbiol,2004,6(8):743-751.DOI: 10.1111/j.1462-5822.2004.00400.x.
[37]LIN M Q,DEN DULK-RAS A,HOOYKAAS P J J,et al.Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection[J].Cell Microbiol,2007,9(11):2644-2657.DOI: 10.1111/j.1462-5822.2007.00985.x.
[38]RIKIHISA Y,LIN M Q.Anaplasma phagocytophilum and Ehrlichia chaffeensis type IV secretion and Ank proteins[J].Curr Opin Microbiol,2010,13(1):59-66.DOI: 10.1016/j.mib.2009.12.008.
[39]STRAND M R.Polydnavirus gene products that interact with the host immune system[M]//Parasitoid Viruses.Amsterdam:Elsevier,2012:149-161.DOI: 10.1016/b978-0-12-384858-1.00012-6.
[40]SALVIA R,GROSSI G,AMORESANO A,et al.The multifunctional polydnavirus TnBVANK1 protein:impact on host apoptotic pathway[J].Sci Rep,2017,7(1):11775.DOI: 10.1038/s41598-017-11939-x.
[41]LOPEZ-ORTIZ C,PEA-GARCIA Y,NATARAJAN P,et al.The ankyrin repeat gene family in Capsicum spp:genome-wide survey,characterization and gene expression profile[J].Sci Rep,2020,10(1):4044.DOI: 10.1038/s41598-020-61057-4.
(责任编辑 吴祝华)

