Associations of PNPLA3 and LEP genetic polymorphisms with metabolic-associated fatty liver disease in Thai people living with human immunodeficiency virus
Kanuengnit Choochuay, Punna Kunhapan, Apichaya Puangpetch, Sissades Tongsima, Pornpen Srisawasdi,Abhasnee Sobhonslidsuk, Somnuek Sungkanuparph, Mohitosh Biswas, Chonlaphat Sukasem
Abstract BACKGROUND The prevalence of metabolic-associated fatty liver disease (MAFLD) is a growing public health issue in people living with human immunodeficiency virus (PLWH). However, the pathophysiology of MAFLD is still unknown, and the role of genetic variables is only now becoming evident.AIM To evaluate the associations of gene-polymorphism-related MAFLD in PLWH.METHODS The study employed transient elastography with a controlled attenuation parameter ≥ 248 dB/m to identify MAFLD in patients from a Super Tertiary Hospital in central Thailand. Candidate single-nucleotide polymorphisms (SNPs) were genotyped using TaqMan® MGB probe 5' nuclease assays for seven MAFLD-related genes. Statistical analyses included SNP frequency analysis, Fisher's Exact and Chi-square tests, odds ratio calculations, and multivariable logistic regression.RESULTS The G-allele carriers of PNPLA3 (rs738409) exhibited a two-fold rise in MAFLD, increasing by 2.5 times in MAFLD with human immunodeficiency virus infection. The clinical features and genetic patterns imply that LEP rs7799039 A-allele carriers had a nine times (P = 0.001) more significant chance of developing aberrant triglyceride among PLWH.CONCLUSION The current study shows an association between PNPLA3 rs738409 and LEP rs7799039 with MAFLD in PLWH.
Key Words: PNPLA3; LEP; Metabolic-associated fatty liver disease; People living with HIV; Thai
INTRODUCTION
Metabolic-associated fatty liver disease (MAFLD) related to systemic insulin resistance is defined as an accumulation of fat in the hepatocytes of more than 5%, consisting of steatosis, non-alcoholic steatohepatitis, fibrosis, and cirrhosis[1,2]. Nowadays, the pathogenesis of MAFLD remains unclear. Furthermore, the prevalence of MAFLD in people living with human immunodeficiency virus (PLWH) was reported to be 40%-55%, based on different MAFLD phenotype-proven techniques in multiple ethnicities[3,4]. Additionally, liver disease is the second leading cause of death in PLWH[5]. Since 2005, several studies have suggested that MAFLD is common in human immunodeficiency virus (HIV) patients, and its prevalence appears to be increasing. Therefore, HIV infection remains a contributing factor of MAFLD that directly activates insulin resistance in adipose tissue and generates mitochondrial toxicity and reactive oxygen species in hepatocytes, which worsens the MAFLD prognosis[6].
Genetic differences in the risk of MAFLD or NASH progression in the general population are well described[7]. One of the strongest and most consistent associations with the presence and progression of MAFLD in certain populations is associated with the single-nucleotide polymorphism (SNP) on thePNPLA3rs738409[8-12]. However, only limited studies exist regarding the role of rs738409 SNP on thePNPLA3gene among people living with HIV. A previous report analyzed the association betweenPNPLA3rs738409 polymorphism and the severity of liver disease, insulin resistance, and obesity in patients co-infected with HIV/hepatitis C virus, which was not associated with the duration of the HIV infection or antiretroviral therapy (ART), specific antiretroviral drugs, a history of opportunistic infection, the patient’s immune status, or the duration of the aminotransferase elevation[13,14]. Additionally, other studies indicate that genes involved in the lipid metabolism pathway, such asAPOC3, APOB, APOA5,andLIPC, are associated with MAFLD[15-22]. Moreover,GHRLandLEPalso exhibit an association with MAFLD pathogenesis[23-25]. In a previous study, it was shown that theAPOC3rs2854116 is associated with elevated serum levels of triglycerides, while this genotype did not affect the incidence of lipoatrophy after adjusting for gender and stavudine (d4T)-containing regimens in Thai people living with HIV[26]. Importantly,GHRLgene polymorphism was significantly correlated with insulin resistance, which is a hallmark of MAFLD and increased type 2 diabetes mellitus risk, particularly among Chinese people and in other populations[27-29]. However, the genome-wide associations replicated in people living with HIV and MAFLD remain inconclusive. A previous study showed both positive and negative associations between candidate SNP and MAFLD, making it difficult to determine the significance of these findings[7,30]. Hence, our study aimed to evaluate the association between several genes related to MAFLD in Thai people living with HIV.
MATERlALS AND METHODS
Study subjects
We enrolled patients from a Super Tertiary Hospital in central Thailand and classified them into 4 groups: 83 PLWH and MAFLD (Group 1); 94 people living with HIV and without MAFLD (Group 2); 145 with NAFLD without HIV infection (Group 3), and 93 Chinese Dai genotyping data from the 1000 Genome Project phase (http://www.1000genomes.org) used to represent the Thai ethnicity (Group 4). The presence of MAFLD was confirmedviatransient elastography with a controlled attenuated parameter ≥ 248 dB/m, as prescribed by our colleague in a previous study[31]. The Infectious Disease Clinic enrolled PLWH who were on ART with full viral suppression and had no history of alcohol consumption in the trial. The key inclusion criteria were as follows: PLWH receiving ART with an undetectable HIV viral load for at least 6 months. Patients co-infected with hepatitis B or C virus, other known liver disorders such as cirrhosis or hepatocellular carcinoma, and critical liver disease were all excluded. The study protocol is shown in Figure 1.
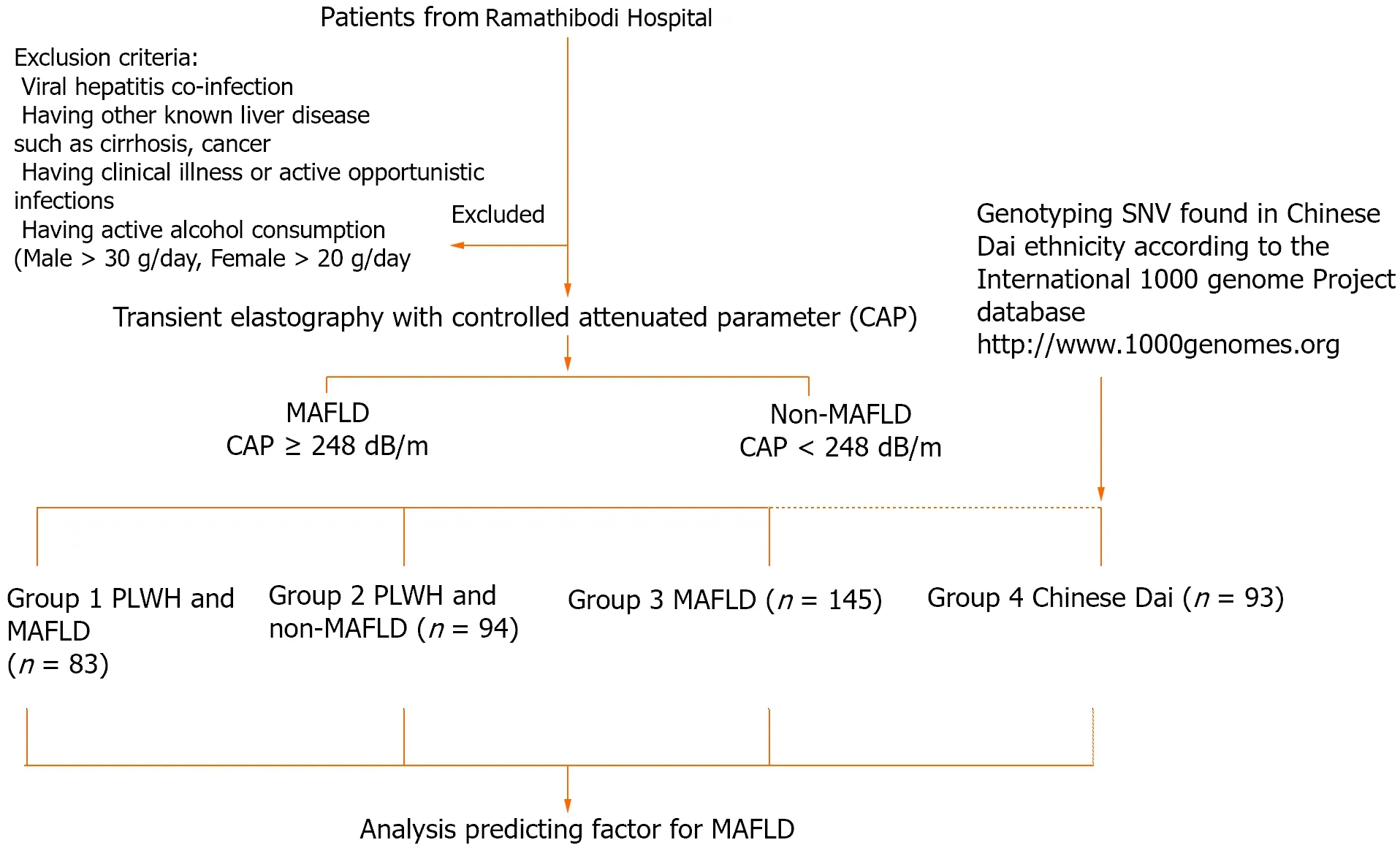
Figure 1 Protocol flowchart. MAFLD: Metabolic-associated fatty liver disease; PLWH: People living with HIV; SNV: Single nucleotide variant.
Genotyping analysis
The genotyping of seven genes related to MAFLD was performed using an allele-specific TaqMan®MGB probe 5’ nuclease assay with a real-time polymerase chain reaction (PCR) ViiA7™ system (Applied Biosystems, Life Technologies). The allele-specific TaqMan®MGB probe 5’ nuclease chain reaction assay was performed with primers ofPNPLA3(rs738409);APOC3(rs2854116);APOA5(rs662799);APOB(rs10495712);LIPC(rs1800588);LEP(rs7799039); andGHRL(rs27647). Each 6 μL of the PCR mixture contained 2 μL of genomic DNA in a concentration of 5 ng/μL, 2.5 μL of the TaqMan®Genotyping Master mix, 0.25 μL of allele-specific TaqMan®MGB probe and a sequence-specific primer kit, and 1.25 μL of DNase-free water. The thermal cycler program started with 10 min at 95°C, followed by 50 cycles of 15 s at 92°C and 90 s at 60°C. The allelic discrimination plot was analyzed using ViiA7™ software (Applied Biosystems, Life Technologies). Allele 1 was labeled with VIC®dye fluorescence, and allele 2 was labeled with FAM®dye fluorescence.
Statistical analysis
The frequencies of all SNPs were checked for Hardy-Weinberg equilibrium using the R statistic, version 3.6.1, from the R Foundation for Statistical Computing. Fisher’s Exact and Chi-square tests were used to determine the statistical difference between the minor alleles between MAFLD patients and control patients using SPSS version 22.0 for Windows, SPSS Inc., Chicago, IL, United States. The association of the candidate genes’ polymorphisms with MAFLD was assessed by calculating the odds ratios and the corresponding 95% confidence intervals. Backward stepwise multivariable logistic regression analysis was used to assess whether one or more genetic factors predicted MAFLD. APvalue of less than 0.05 was considered significant.
RESULTS
Characteristics of people living with HIV and with MAFLD (Group 1) and of non-MAFLD patients (Group 2) and MAFLD patients (Group 3)
When comparing the people living with HIV and with and without MAFLD, we enrolled a higher proportion of males with a higher BMI. The levels of fasting glucose, HbA1C, triglyceride, aspartate transaminase (AST), alanine transaminase (ALT), and gamma-glutamyl transferase were significantly higher in the MAFLD group compared to the non-MAFLD group in the metabolic profiles and liver function tests. When comparing the HIV treatment regimens, the proportion of non-nucleoside reverse transcriptase inhibitor, nucleoside reverse transcriptase inhibitor (NRTI), protease inhibitor (PI)-based, or alternative regimens did not differ between the two groups (P= 0.573), but comorbidities of dyslipidemia, hypertension, and diabetes mellitus were higher in people living with HIV and MAFLD (P =0.002;P= 0.001 andP= 0.005, respectively) (Table 1).

Table 1 Baseline characteristics of metabolic-associated fatty liver disease patients and controls
Distribution of SNPs in people living with HIV and MAFLD (Group 1) or without MAFLD (Group 2), with MAFLD (Group 3), and Chinese Dai (Group 4)
All the SNP genotyping experiments were successful. Throughout the entire study, the genotype frequencies of each SNP did not deviate from Hardy-Weinberg equilibrium (P> 0.05). Table 2 shows the genotype distribution and minor allele frequency of the investigated SNPs in people living with HIV with or without MAFLD, MAFLD patients, and Chinese Dai. All potential SNP genotype distributions (PNPLA3rs738409, APOC3rs2854116, APOA5rs662799, APOBrs10495712,LIPCrs1800588, LEPrs7799039, andGHRLrs27647) in people living with HIV with MAFLD were similar to those seen in patients living with HIV without MAFLD. In comparison to Chinese Dai, patients with MAFLD had a higher frequency of thePNPLA3G-allele (P= 0.035). The frequencies of the other SNPs were not significant in persons living with HIV and those with or without MAFLD.
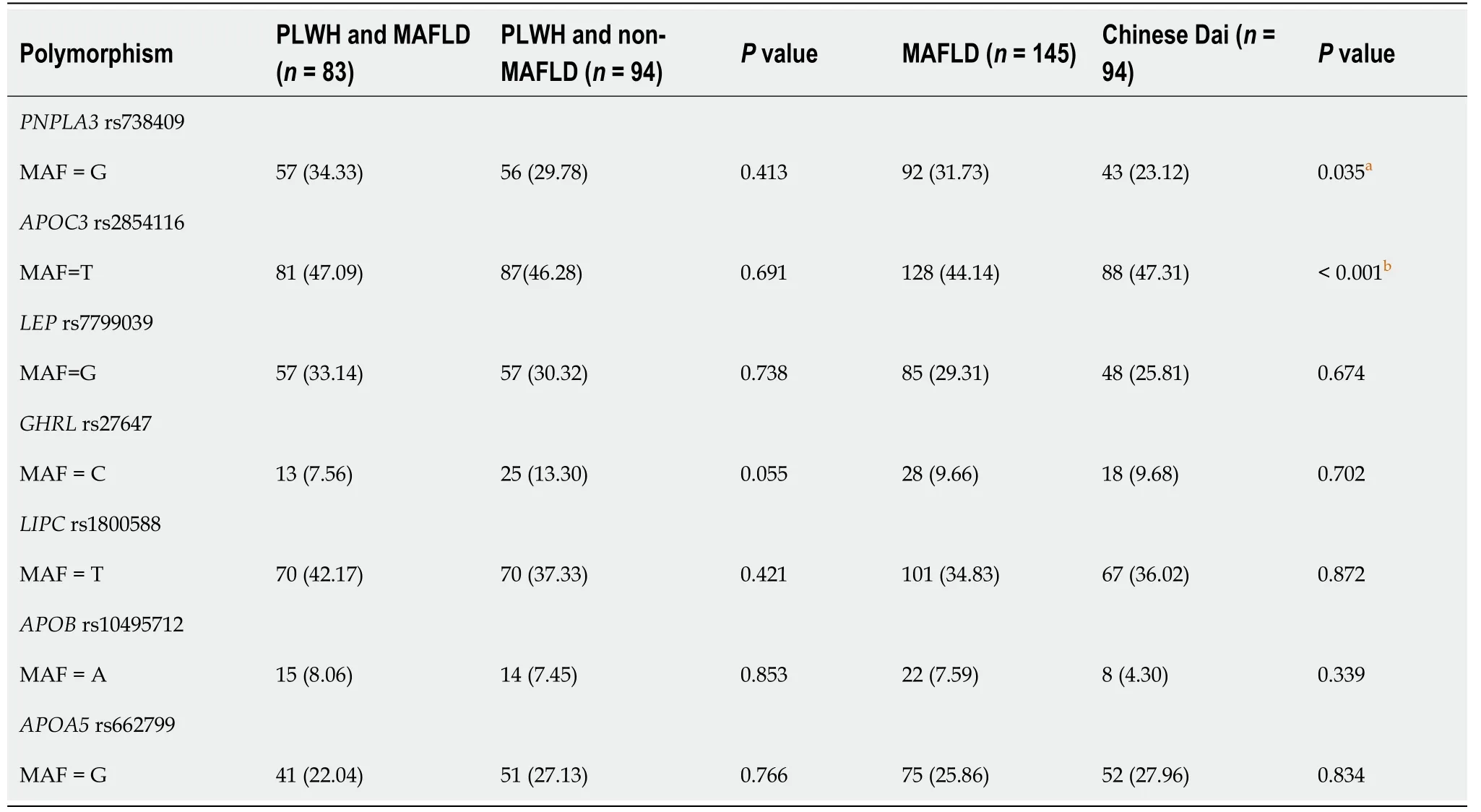
Table 2 Genotype distributions and Minor allele frequency of candidates single-nucleotide polymorphisms

Table 3 Genotype and allele frequencies of the single-nucleotide polymorphisms in the people living with human immunodeficiency virus and metabolic-associated fatty liver disease compared with people living with human immunodeficiency virus and non-metabolicassociated fatty liver disease group

Table 4 Genotype and allele frequencies of the single-nucleotide polymorphisms in the metabolic-associated fatty liver disease compared with Chinese Dai
Association between PNPLA3 and other candidate SNPs with MAFLD
The data given in Tables 3 and 4 show the well-establishedPNPLA3rs738409 gene, which is found on chromosome 22 and has a function related to lipid droplet formation in the hepatocytes; the G-carrier patients had an approximately twofold higher risk of developing MAFLD when compared to MAFLD with Chinese Dai (P= 0.012). Importantly, people living with HIV and MAFLD exhibited a 2.5-fold increased risk (P= 0.002) when compared to Chinese Dai (Group 4). In addition,GHRL(Ghrelin) rs27647 is a promising susceptibility gene for insulin regulation; the C-allele carrier has exhibited a protective effect in MAFLD in people living with HIV. The odds of having MAFLD is 53% lower if the people living with HIV are C-carriers ofGHRLrs27647 than if they are not C-carriers (P= 0.047). However, there was no statistically significant association withGHRLin the other groups. Furthermore,APOC3rs2854116 C-allele carriers were also statistically significant in the MAFLD group, exhibiting a six-fold higher risk in the dominant model with Chinese Dai as the comparison (P< 0.001).
Association between candidate SNPs and the lipid profile, liver function, and glucose metabolisms
We performed a subgroup analysis of people living with HIV in terms of their metabolic profiles and compared the genotypes of candidate genes. As shown in Table 5, the mean or median values of the lipid profile [triglyceride, total cholesterol, LDL]; liver function (AST, ALT); and glucose metabolisms (HbA1C and fasting plasma glucose) were higher in the people living with HIV and MAFLD than in the control group (people living with HIV and non-MAFLD). The association between the genotypes of theAPOA5rs662799 SNP and serum lipid parameters in the control group is presented in Table 5 and Supplementary Table 1. Serum total cholesterol levels in control patients differed between the AA and AG/GG genotypes (P< 0.05). TheAPOA5rs662799 G allele carriers had a lower proportion of total cholesterol levels in the normal range (< 200 mg/dL) than the A allele non-carriers and indicated the protective effect ofAPOA5rs662799 in an abnormal range of total cholesterol (> 200 mg/dL); these results showed statistical significance (P= 0.045). Furthermore,LEPrs7799039 AG and AA carriers exhibited a significant nine-fold higher risk in an abnormal range of triglyceride (> 150 mg/dL) when compared with non-carriers (P= 0.001). Unfortunately, none of the individual SNPs were associated with LDL. Moreover, in men,APOC3rs2854116 TT alleles also showed a protective effect on the highdensity lipoprotein (HDL) profile (Table 5 and Supplementary Table 1). Furthermore, AST is known to be a reliable surrogate marker for outcome measures in MAFLD. Table 6 shows thatPNPLA3rs738409 G-carrier patients have an approximately 2.5 times higher chance of AST abnormality (> 34 U/L) when compared with non-carriers (statistically significant atP= 0.010).

Table 6 Association between genetic polymorphism and metabolic traits
Association between the genetic profile, clinical factors, and MAFLD
A stepwise multiple logistic regression was performed to investigate the relationship between the genetics profiles, clinical factors, and MAFLD. Sixteen variables, including gender, AST, ALT, total cholesterol, triglycerides, HDL, LDL, fasting plasma glucose, HbA1C, and the genetic profiles ofPNPLA3rs738409, APOC3rs2854116, APOA5rs662799, APOBrs10495712, LIPCrs1800588, LEPrs7799039,andGHRLrs27647 were entered into the original equation. The results showed that seven variables, namely, AST, total cholesterol triglycerides, LDL, fasting plasma glucose,APOBrs10495712, andAPOA5rs662799, were significantly associated with MAFLD (Table 7).

Table 7 Logistic regression analysis of factors associated with metabolic-associated fatty liver disease (people living with human immunodeficiency virus and metabolic-associated fatty liver disease vs people living with human immunodeficiency virus and nonmetabolic-associated fatty liver disease)
DlSCUSSlON
Risk factors for MAFLD in people living with HIV (PLWH) include the normal factors seen in the general population, such as components of metabolic syndrome (obesity, diabetes, hypertension, dyslipidemia, a sedentary lifestyle, and excessive dietary intake)[1]. Hepatic steatosis and mitochondrial oxidative stress are pivotal to MAFLD pathogenesis. In PLWH with MAFLD, HIV-specific factors such as lipodystrophy, ART, and HIV infection itself are strongly linked to the development of MAFLD[31,32]. However, our study did not find NRTI-based and PI-based regimens to be predictive factors for MAFLD.
The strongest and most consistent associations with the presence and progression of MAFLD in the studied populations are related to the SNP on thePNPLA3rs738409, which was discovered by the first GWAS in 2003[8]. Our study demonstrated the significance ofPNPLA3rs738409 in MAFLD when compared to the general population, indicating the impact of genetic factors. Moreover, we evaluated the effect of both HIV infection and genetic factors by conducting a comparison between people living with HIV and Chinese Dai, finding that it increased the chance of the development of MAFLD between 2 and 2.5 times when compared to the genetic factor alone. Moreover, our results agree with previous studies that demonstrated the significant association withPNPLA3rs738409 and biopsy-proven fibrosis or steatosis among HIV/hepatitis C virus or HBV co-infected patients, HIV-mono infection, and the group with no viral infection[33-35].
Insulin resistance has been characterized as the crucial pathophysiological factor in MAFLD. The advanced reports found that insulin resistance is associated with the reduction of circulating ghrelin level[21,36,37]. Interestingly, our study has shown that the G/A genotype and G/G genotype ofGHRLrs27647 were associated with a 53% decreased risk of MAFLD in people living with HIV when compared with non-MAFLD patients. Moreover, a previous study observed higher levels of ghrelin in patients with hypertriglyceridemia, as well as a positive correlation between ghrelin and trigly-ceride levels in patients with hypertriglyceridemia[38,39]. Unfortunately, our study failed to detect the association between SNP and triglyceride levels in people living with HIV.
TheAPOC3gene plays a crucial role in the circulation and clearance of very-low-density lipoprotein, HDL, and chylomicron remnants[40,41]. The polymorphism in the promotor region of theAPOC3rs2854116 (-455T>C) gene has been extensively studied and has been found to be related with insulin resistance at the transcriptional level. Consequently, the overexpression ofAPOC3, which functions to inhibit lipoprotein lipase and the cellular uptake of triglyceride-rich lipoprotein particles, may result in hypertriglyceridemia, as has been confirmed byin vivoand clinical studies[23,24,42-44]. In this study, we showed thatAPOC3rs2854116 C-allele carrier patients have a six-fold higher risk of developing MAFLD in a dominant model. Our findings are also consistent with previous reports that theAPOC3 rs2854116genetic variant leads to increased plasma concentrations of apolipoprotein C3, resulting in hepatic insulin resistance and MAFLD in multiethnic populations[23,45,46].
Our results show a similar trend to those of a previous report, which demonstrated a positive correlation between AST levels and the accumulation of intrahepatic triglyceride[47]. Interestingly, our results indicate a robust association inLEPrs7799039 with the lipid profile, especially with triglyceride levels. According to a subgroup analysis of patients infected with HIV, a patient who is a carrier of the A-allele (AG and AA) has a nine-times-higher risk of exhibiting abnormal triglyceride levels (> 150 mg/dL). Further information suggests thatLEPrs7799039, located on chromosome 7, encodes 167 amino acid peptide variants with a molecular weight of 16 ku, which may subsequently affect the biological functions ofLEP[48]. In recent years,LEPhas been found to regulate the energy balance in coordination with the regulation of the glucose and lipid metabolisms. Thus, it plays a vital role in the development of MAFLD. This finding aligns with that of previous reports that evaluated the association betweenLEPrs7799039 and diabetes mellitus, metabolic syndrome, MAFLD, and cardiovascular disease[49,50]. Our findings should be interpreted while bearing in mind several potential limitations. First, the small sample size of each group may have limited the study’s ability to detect a significant relationship. Second, the patients included in this study were exclusively Thai, so our findings may not apply to patients of other ethnic origins. Further, long-term studies are still needed to confirm these findings in other ethnicities. Although the results of the available research are satisfactory, they have not been proven in randomized control trials. Further studies of genetic predispositions for MAFLD with the absence or presence with MAFLD will certainly provide a better understanding of the molecular mechanisms of MAFLD.
CONCLUSlON
The prevalence of MAFLD in people living with HIV is increasing, representing a public health concern. The existing evidence suggests that AST, fasting plasma glucose, triglyceride, total cholesterol, LDL, and the genetic factorsPNPLA3rs738409 andLEPrs7799039 indicate genetic susceptibility for PLWH, leading to improvements in the treatment of MAFLD.
ARTlCLE HlGHLlGHTS
Research background
Metabolic-associated fatty liver disease (MAFLD), which is characterized by hepatocyte fat accumulation, poses substantial health risks; it affects a significant number of people globally, especially those living with obesity, diabetes, dyslipidemia, hypertension, and metabolic syndrome. Despite its prevalence, the precise mechanisms underlying MAFLD, which involve factors including viral hepatitis, human immunodeficiency virus (HIV), antiretroviral treatment, and genetics, remain unclear.
Research motivation
MAFLD is prevalent among individuals with HIV, with rates ranging from 40% to 55%; it is influenced by both antiretroviral medications and specific genetic variants. Notably, thePNPLA3rs738409 variant, a genetic factor, plays a significant role in the development of MAFLD.
Research objectives
The present investigation sought to assess the correlation between gene polymorphisms and MAFLD in individuals living with HIV.
Research methods
We employed transient elastography and set a threshold for the controlled attenuated parameter at ≥ 248 dB/m for the identification of MAFLD. All participants underwent genotyping for candidate single-nucleotide polymorphisms.
Research results
Individuals carrying the G-allele ofPNPLA3(rs738409) demonstrated a two-fold increased risk of developing MAFLD; this risk rose to 2.5 times in cases of MAFLD with HIV infection. The clinical characteristics and genetic profiles suggested that carriers of the A-allele ofLEPrs7799039 had a nine-fold higher likelihood of developing abnormal triglyceride levels among individuals living with HIV.
Research conclusions
The present research reveals a connection betweenPNPLA3rs738409 andLEPrs7799039 and MAFLD in individuals with HIV.
Research perspectives
Genetic factors play a crucial role in the pathophysiology of MAFLD. In upcoming research, targeting thePNPLA3gene in clinical trials may emerge as a promising direction for precision medicine in the treatment of MAFLD.
ACKNOWLEDGEMENTS
We appreciate the study patients' participation and the support of the attending staffs and all staffs in the Divisions of Personalized medicine and Personalized Medicine, research nurses in the Divisions of Infectious Diseases and Gastroenterology and Hepatology, and the Department of Radiology.
FOOTNOTES
Author contributions:Choochauy K performed the majority of experiments and wrote the manuscript; Sukasem C conceptualized, validated and designed the study and corrected the manuscript; Kunhapan P, Puangpetch A, and Tongsima S involved in analytical tools; Srisawasdi P participated to the collection of the human material and clinical data; Sobhonslidsuk A and Sungkanuparph S served as scientific advisors and participated in the collection of human materials; Biswas M critically reviewed the manuscript to improve overall clarity and quality; Sukasem C was the guarantor.
Supported bythe Faculty of Medicine, Ramathibodi Hospital, Mahidol University.
lnstitutional review board statement:The study was approved by the ethics committee of the Faculty of Medicine, Ramathibodi Hospital, Mahidol University (Bangkok, Thailand) (COA. MURA2019/645). All study procedures were conducted in accordance with the 1964 Helsinki Declaration.
lnformed consent statement:In this investigation, genomic material was isolated from residual specimens of the study subjects, demonstrating minimal risk to patients and participant consent was not necessary.
Conflict-of-interest statement:No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
Data sharing statement:No additional data are available.
STROBE statement:The authors have read the STROBE Statement - checklist of items, and the manuscript was prepared and revised according to the STROBE Statement - checklist of items.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin:Thailand
ORClD number:Kanuengnit Choochuay 0000-0002-1871-1631; Punna Kunhapan 0000-0001-9301-4206; Apichaya Puangpetch 0000-0002-5558-2752; Sissades Tongsima 0000-0002-1491-1839; Pornpen Srisawasdi 0000-0001-5718-2130; Abhasnee Sobhonslidsuk 0000-0002-3730-5532; Somnuek Sungkanuparph 0000-0002-1055-2963; Mohitosh Biswas 0000-0003-1432-7701; Chonlaphat Sukasem 0000-0003-0033-5321.
S-Editor:Liu JH
L-Editor:A
P-Editor:Cai YX
 World Journal of Hepatology2024年3期
World Journal of Hepatology2024年3期
- World Journal of Hepatology的其它文章
- Update in lean metabolic dysfunction-associated steatotic liver disease
- Retrospective study of the incidence, risk factors, treatment outcomes of bacterial infections at uncommon sites in cirrhotic patients
- Palliative long-term abdominal drains vs large volume paracenteses for the management of refractory ascites in end-stage liver disease
- Comprehensive prognostic and immune analysis of sterol Oacyltransferase 1 in patients with hepatocellular carcinoma
- Prediction model for hepatitis B e antigen seroconversion in chronic hepatitis B with peginterferon-alfa treated based on a responseguided therapy strategy
- lnfluence of nonalcoholic fatty liver disease on response to antiviral treatment in patients with chronic hepatitis B: A meta-analysis
