A crowd-sourced genomic project to assess hybrid content in a rare avian vagrant (Azure Tit Cyanistes cyanus (Pallas, 1770))
Mrtin Irestedt, Filip Th€orn, Per G.P.Erison, Hein vn Grouw, Yroslv A.Red'kin,Alexnder Hellquist, Frnk Johnsson, John A.A.Nylnder
aDepartment of Bioinformatics and Genetics, Swedish Museum of Natural History, Stockholm, Sweden
bDepartment of Zoology Stockholm University, Sweden
cBird Group, Department of Life Sciences, Natural History Museum, Tring, United Kingdom
dZoological Museum, Lomonosov Moscow State University, Moscow, Russia
eInstitute of Biological Problems of Cryolithozone SB RAS, Yakutsk, Russia
fDepartment of Women's and Children's Health, Uppsala University, Sweden
gDepartment of Ecology and Genetics, Uppsala University, Uppsala, Sweden
Keywords:Crowdsourcing Cyanistes cyanus Hybridization Museomics
ABSTRACTThe aim of this study was to correlate plumage variation with the amount of genomic hybrid content in hybrids between Azure Tits Cyanistes cyanus(Pallas,1770)and European Blue Tit Cyanistes caeruleus(Linnaeus,1758),by re-sequencing the genomes of museum specimens of non-hybrids and presumed hybrids with varying plumages.The project was funded by crowdsourcing and initiated when two presumed Azure Tits,observed by hundreds of Swedish birdwatchers, were rejected as hybrids based on minor plumage deviations assumed to indicate hybrid contents from the European Blue Tit.The results confirm that hybrids with intermediate plumages, so called Pleske’s Tits,are first generation hybrids(F1 hybrids).Individuals,whose plumages are similar to Azure Tits,but assessed as hybrids based on minor plumage deviations, are all backcrosses but vary in their degree of hybrid content.However, some individuals morphologically recognized as pure Azure Tits expressed similar degrees of hybrid content.The results indicate that:(1)hybrid content may be widespread in Azure Tits in the western part of its habitat distribution;(2)plumage deviation in backcrosses is not linearly correlated with the genetic degree of hybrid origin;and(3)all Azure Tits observed in Europe outside its natural distribution may have some degree of hybrid origin.We therefore suggest that it is very difficult to phenotypically single out hybrids beyond first generation backcrosses.We argue that decreased sequencing costs and improved analytical tools open the doors for museomic crowd-sourced projects that may not address outstanding biological questions but have a major interest for lay citizens such as birdwatchers.
1.Introduction
It is a well-known phenomenon that mating across species boundaries occasionally occurs and results in hybrid offspring.For a long time, hybrids were generally believed to be occasional vagaries with no or little biological significance.This view has now been challenged as it has been shown that gene flow between species can be important in several biological processes (Feder et al., 2005; Dasmahapatra et al., 2012; Lamichhaney et al.,2015;Meier et al.,2017;Marques et al.,2019).In birds,new cases of species that regularly hybridize are published at an increasing rate (e.g., Ottenburghs et al., 2015).There is also a trend in ornithology to upgrade taxa previously classified as subspecies to species level (Sangster, 2009).With a refined taxonomy, instances of introgression classified as interspecies hybridization are expected to increase as species boundaries are generally more permeable in closely related species(Roux et al.,2016).Taxonomic changes and the increased awareness that birds hybridize has led to that individuals that originally were accepted as pure taxa have been rejected,either as the documentations is too poor to assign them to species level in the upgraded taxonomy or because hybrid origin cannot be excluded.Hence, more information on species that hybridize regularly is needed.
The traditional way to assess hybrid contents in birds has been morphological assessments (McCarthy, 2006).A recent example from Sweden is the re-evaluation of the Swedish records of Azure Tit Cyanistes cyanus (Pallas, 1770), where two out of four Swedish observations became rejected as hybrids based on minor plumage deviations (Corell et al., 2019).It is well known that Azure Tits and European Blue Tits Cyanistes caeruleus (Linnaeus, 1758) occasionally hybridize.In fact, hybrids with intermediate plumages,first generation hybrids(F1 hybrids),where originally described as a distinct species, the Pleske’s Tit Parus(Cyanistes)pleskii Cabanis,1877.A detailed description of the taxonomic history of Pleske’s Tit is presented in the supplementary material (Appendix A).However, the plumages of the two rejected Swedish individuals were not typical for what one would expect in F1 hybrids between Azure and European Blue Tits (Fig.1A).Overall, the plumages of these individuals were much more like those of “pure” Azure Tits.Minor plumage characters,such as the amount of white in the tail and on the wing covers,were nevertheless assessed to indicate a European Blue Tit hybrid content (Corell et al., 2019).Several publications have described the plumage variation in Azure Tits as well as plumage deviations that indicate hybrid origin (e.g., Lawicki, 2012).Yet, one may argue that minor plumage deviations,as in the two rejected individuals,may constitute natural plumage variation or mere plumage aberration.In addition,several genomic studies in other taxa have shown that there is no linear correlation between the degree of hybrid content and plumage deviation (Toews et al., 2016; Brelsford et al., 2017; Wang et al., 2020;Natola et al.,2022).One may thus also question if it is possible to exclude that individuals morphologically assessed as pure Azure Tits(such as the two Azure Tits retained on the Swedish official bird list), have a hybrid content.
Birdwatching is a recreational activity that attracts people worldwide and hence has both a social and an economic significance.In the United States, it is estimated that more than 46 million people birdwatch and spend 41 billion USD ($) on this hobby annually (Fish and Wildlife Service,2018).To keep track and to count the birds that one observes is common among birdwatchers.There is even a term,“twitcher”,used for birdwatchers that are prepared to travel long distances to observe a rare bird (Oddie, 1980).Several books (Kaufman, 2006; Obmascik, 2011;Gooddie,2013)and a movie(Frankel,2011)have been produced based on this“twitcher”phenomenon.As the dismissed Swedish Azure Tits had been observed by hundreds of birdwatchers (Fig.1B), we initiated a crowdsourcing project to explore the willingness of the Swedish bird“twitcher”community to co-finance a genomic project with the purpose to link plumage variation with hybrid content in the Azure Tit.While this would,from a scientific point of view,result in a better understanding of how plumage characters may reflect the degree of hybridization in Azure Tits, the impetus for the Swedish birdwatcher community was a potentially change in the verdict of the two dismissed Azure Tits.As no DNA was available for the two individuals determined as hybrids by the Swedish rarity committee,we sampled DNA from museum study skins of individuals with a similar plumage and compared them with samples from “pure” Azure and Blue Tits.In this paper, we present the results from the genetic investigation and also discuss how crowd sourcing may be useful to finance museomic projects that are of great interest to birdwatchers and other laymen, but that may not attract traditional research funding.
2.Materials and methods
2.1.Financing and sampling
This project was advertised in the magazine Roadrunner(No.1 2019)and followed by additional advertises in later issues as well as updates on the progress on Club300’s webpage (https://www.club300.se).When 50%of the estimated laboratory and sequencing cost had been covered,the project was initiated.In total, the genomes for 38 samples were resequenced (26 museum specimens and 12 fresh tissue samples); 9 European Blue Tits,12 Azure Tits,and 17 individuals that had been assessed as hybrids.The latter group varied in plumage morphology from being extremely similar to European Blue Tits to those similar to Azure Tits.The pure Azure Tits and hybrids similar to Azure Tits were re-examined based on the plumage criteria used by the Swedish rarity committee(Corell et al.,2019).Voucher numbers,sampling localities and affinities for the included samples are shown in Table 1.The genome of the Great Tit Parus major(Linnaeus,1758),was downloaded from GenBank(NCBI Parus major genome version 1.1, GCA_001522545.3) and used as reference.
2.2.Laboratory procedures
Genomic DNA was extracted from fresh samples (n ¼ 12) with a KingFisher Duo magnetic particle processor (ThermoFisher Scientific)using the KingFisher Cell and Tissue DNA Kit.Library preparation,using Illumina TruSeq DNA Library Preparation Kit, and sequencing on Illumina Nowaseq(S4 2 150 bp)was performed by SciLifeLab,Stockholm.Genomic DNA from footpad samples (n ¼ 26) was extracted with the QIAamp DNA Micro Kit (Qiagen) following the procedures described in Irestedt et al.(2022).For library preparation of DNA from footpads we followed the protocol of Meyer and Kircher(2010).Library preparation included blunt-end repair,adapter ligation,and adapter fill-in,followed by four independent index PCRs.For more detailed descriptions of laboratory procedures from degraded historical DNA samples see Irestedt et al.(2022).The finished libraries were pooled at equal ratio with other museum samples and run on approximately 1.25 lanes on the Illumina NovaSeq(S4 2 100 bp)platform.All raw reads generated for this study have been deposited at the European Nucleotide Archive (ENA), accession number PRJEB64125.
2.3.Bioinformatic analyses
Sequence data for all samples were mapped against the reference genome using the nextflow pipeline nf-core/eager v2.4.3 (Di Tommaso et al., 2017; Yates et al., 2021).This pipeline includes sequence quality and cleaning steps using AdapterRemoval (Schubert et al., 2016) and FastP(Chen et al.,2018),mapping with bwa mem(Li et al.,2009),and PCR duplicate removal with MarkDuplicates (Picard Toolkit, 2019).In addition,we also used the pipeline to generate genotype likelihood files with ANGSD (Korneliussen et al., 2014).The resulting bam files from nf-core/eager where filtered for coverage, and principal component analyses of inferred allele frequencies, as well as estimating admixture proportions where performed using PCAngsd v1.10 (Meisner and Albrechtsen, 2018).The two latter steps were done using the nextflow pipeline nf-GL_popstructure (https://github.com/FilipThorn/nf-GL_pop structure).As the Z chromosome is hemizygous in females and may show different introgression patterns than autosomes,which potentially may influence the results, the analyses were conducted both with the Z-chromosome included and excluded, respectively.Sequences mapped to the mitochondrial reference contig (P.major, Genbank accession NC_040875.1) where extracted for each sample from the genotype files generated by nf-core/eager, then aligned together with the reference using MAFFT v7.453 (Katoh and Standley, 2013).A phylogenetic tree where estimated from the multiple sequence alignment using RaxML-NG v.1.1.0(Kozlov et al.,2019)using the GTR þ G substitution model,and rooted with P.major as outgroup.To assess the sex of the sequenced individuals, mapping with bwa mem was done against genomic sequences from the Zebra Finch(Taeniopygia guttata Vieillot,1817),using the W and Z chromosomes (Genbank accessions NC_045028.1 and NC_044241.2,respectively).The ratio of mapping frequency against the two chromosomes was calculated and compared to the expectancy that females having both W and Z, while males having two Z chromosomes.These calculations was performed using the script determine-sex.R provided in https://github.com/Ahinsu/Chicken-sex-determination.
3.Results
3.1.Crowdsourcing
The total laboratory and sequencing costs for this project was ca 112,000 SEK (~11,000 EUR).The funding from Club300 members and other contributors reached 83,000 SEK,which means that nearly 75%of this project has been covered by crowdsourcing.
3.2.Mapping and SNP results
We obtained sequence data from all samples in the study.The mean genome coverage when mapped against the Great Tit chromosomes ranged between 6 and 27 for the fresh samples and between 2 and 16 for toe pad samples (see Table 1 for individual coverage).For calculating admixture proportions and generating PCA plots, the bam files were filtered to only retain sequences that mapped to reference contigs having a minumum of 3 coverage for all samples.Further
filtering in ANGSD setting minimum quality score to 20, minimum mapping score to 20,and removing allele frequencies with less than 0.05,yielded a total number of genome-wide SNPs of 16,123,565.
3.3.Mitochondrial phylogeny and molecular sexing
In the mitochondrial phylogeny(Appendix B:Fig.S1)all individuals included in the study group in two well-supported clades.The first mitochondrial clade includes all individuals morphologically and genetically assessed as pure European Blue Tits and those that are backcrosses with European Blue Tits (individuals estimated to have a genome-wide content of European Blue Tit that is well over 50%).The second mitochondrial clade is formed by“pure”Azure Tits,hybrids that are backcross with Azure Tit(in general less than ca 25%European Blue Tit)and all individuals that are estimated to be F1 hybrids(estimated as first generation hybrids as they have a genome-wide content of close to 50% from the European Blue Tit and the Azure Tit, respectively).The determination of the sex show that 10 females and 27 males (and one individual whose sex is undetermined)were included in this study.This bias of more males is present in the Azure Tits,the European Blue Tits as well as in individuals with more or less hybrid content.Several of the hybrids are also females and of individuals assessed as F1 hybrids one individual out of eight is a female.
3.4.Population structure and levels of hybrid contents
The admixture analyses (genotype likelihoods) and the principal component analysis(PCA)based on genome-wide SNP data show similar patterns between themselves as well as when the Z-chromosome was excluded and deleted, respectively.The result from the analyses where the Z-chromosome was excluded is found in the supplementary material(Appendix B: Figs.S2 and S3), while the results from the analyses with the Z-chromosome included is given below.In the principal component analysis(Fig.2),the PCA 1 axis explains 35.57%of the genetic variation and is the axis that primarily separates the European Blue Tits from Azure Tits.Along this axis,all European Blue Tits group tightly together.In the PC2 axis that explains 2.89%of the genetic variation,one European Blue Tit individual (NRM20126772) is well separated from the other European Blue Tits.However,this individual belongs to a separate subspecies and has been collected geographically distant (Crete in Greece) from other European Blue Tits in this study.In the PCA 1 axis,one of the most eastern European Blue Tits included in the study (UWBM 83447) is placed slightly towards the Azure Tits, which may indicate a minor hybrid content.In the admixture analysis none of the European Blue Tits shows signs of hybrid content.In contrast to the European Blue Tits,theAzure Tits have a greater spread along the PCA 1 axis, where samples collected in the eastern part of their distribution are most divergent from the European Blue Tit, while Azure Tits collected at more westerly locations are positioned closer towards the European Blue Tits along the PCA 1-axis.In the admixture analysis(Fig.3)three of the most westerly collected Azure Tits (assessed as pure Azure Tits on plumage) are suggested to have European Blue Tit hybrid content.Of the 17 samples that beforehand had been assessed as hybrids(labeled as such in the museum collections), 14 are assessed as hybrids to various degree in the admixture analysis,while two were assessed as European Blue Tits and one as a pure Azure Tit.The latter is assessed as a pure Azure Tit when applying the plumage criteria used by the Swedish rarity committee.The plumages of the two individuals genetically assessed as pure European Blue Tits(BMMH 1901.5.4.300 and BMNH 1901.5.4.317 in the admixture plot)are overall very similar to plumages of European Blue Tits, but slightly paler.It is likely that these two individuals have been misclassified as hybrids based on some pigmentation disorder.A majority of the hybrids in the admixture analyses show close to 50%genetic contents from Azure Tit and Blue Tit, respectively.These individuals are assessed as F1 hybrids, and they all exhibit a typical “Pleske’s Tit” plumage (Fig.4).It is notable that all these individuals have average genome content of Blue Tit that are slightly higher than 50%(between 52%and 59%).All other individuals assessed to be hybrids vary in their degree of hybrid contents which make it difficult to evaluate their backcrossing history.However,two individuals (BMNH 1901.5.4.236) and (BMNH 1901.5.4.349) are most likely first generation backcrosses (BC1) with Blue Tit and Azure Tit, respectively.

Fig.2.Principal component analysis (PCA) of 16M SNPs to infer population genetic structure among the samples.The PC1 explains 35% of the total variation among the SNPs and is the axis that mainly separates Azure Tits, European Blue Tits and hybrids from each other.Individuals assessed as F1 hybrids are those between the dashed lines, while estimated F2 backcrosses are marked with a star.(For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In summary, our analysis (admixture) showed that individuals that morphologically scored as hybrids could also be pure Blue Tits or Azure Tits based on genomic data.In addition, the analysis showed that the percentage of Blue Tit genome vary widely in individuals score as hybrids(from 16%to 86%).Finally,some individuals scored as pure Azure Tits had a low percentage on Blue Tit genome (7–14%), while the opposite pattern was not found.
4.Discussion
4.1.No linear correlation between plumage deviation and hybrid content

Fig.3.Admixture plot(K¼2)showing the ancestry components of European Blue Tit(blue)and Azure Tit(white)in the investigated the individuals.Samples with red labels are individuals marked as hybrids in museum collections that showed no sign of hybrid origin in the present analyses.Admixture proportions are given at the top.(For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
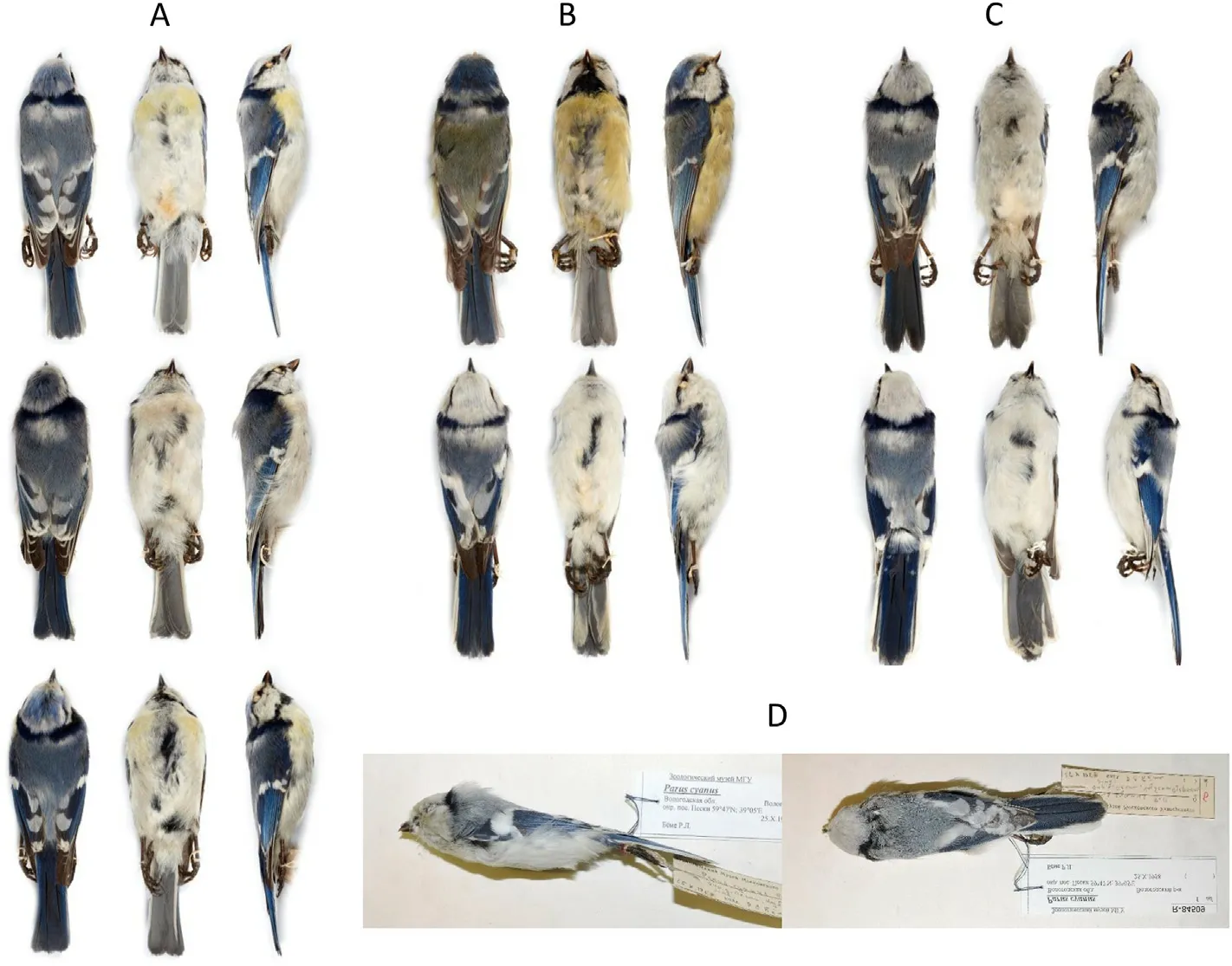
Fig.4.Examples of hybrids between Azure Tits and Blue Tits with various degree of hybrid content.(A)Birds assessed as F1 hybrids (NHMUK 1901.5.4.304,NHMUK 1901.5.4.308 and NHMUK 1901.5.4.309),(B) birds estimated as first generation backcrosses(BC1 hybrids)with European Blue Tit above(NHMUK 1901.5.4.236) and Azure Tit below (NHMUK 1901.5.4.349), (C) morphologically assessed hybrids with smaller amount of European Blue Tit hybrid contents (NHMUK 1901.5.4.310 and NHMUK 1965.M.15606), and (D) individual morphologically assessed as pure Azure Tit but genetically expressing European Blue Tit hybrid contents (ZMMU R84509).(For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Several species are currently recognized within the Blue Tit complex,but only the European Blue Tit has a distribution that borders to and partly overlaps the distribution of the Azure Tit(del Hoyo et al.,2007).One population of Azure Tits in south-central Asia is sometimes considered as a separate species (Yellow-breasted Tit C.flavipectus (Severtsov,1873)).The combined range of the European Blue Tit and the Azure Tit extends across the entire Palearctic, with the former species occupying mainly Europe,and the latter Asia(Martin,1991;del Hoyo et al.,2007).Hybrids between the two species are reported from North-West Europe since early 20th century (Pleske, 1912).The hybrids vary in their plumages from appearing almost as pure Azure Tits to almost as pure Blue Tits (Dementiev and Gladkov, 1954), suggesting that first generation (F1) hybrids are able to backcross with the parental species.Phylogenetic studies of the genus Cyanistes suggest that the Azure Tit is nested within the Blue Tit complex, as sister to the European Blue Tit(Kvist et al.,2005;del Hoyo et al.,2007;Illera et al.,2011).
The results of this study confirm that the species boundary between the European Blue Tit and the Azure Tit is semipermeable, as we have been able to confirm backcrosses in both directions(Figs.2 and 3).The Admixture analysis identified a hybrid content in several individuals morphologically assessed as pure Azure Tits but not in any of the European Blue Tits (Fig.3).A similar pattern is shown in the population structure analysis (PCA, Fig.2) where the Azure Tits are spread more widely along the axis that separates the two species, whereas the European Blue Tits group more closely together.This may indicate that gene flow between the two species is predominantly in the direction from the European Blue Tit to the Azure Tit,a so-called asymmetric hybridization.It is notable that all individuals assessed as F1 hybrids are estimated to have average genome content of Blue Tit that are higher than 50%(between 53%and 59%)in the admixture plot.This indicate that European Blue Tits hybridize with Azure Tit populations that themselves possess small levels of Blue Tit hybrid contents.
All individuals assessed as F1 hybrids and backcrosses with Azure Tits have mitogenomes belonging to the Azure Tit branch in the mitogenomic tree,while only backcrosses with Blue Tits have mitogenomes belonging to the Blue Tit branch (Appendix B: Fig.S1).As the mitochondrion is maternally inherited this suggest that Blue Tit males hybridize with female Azure Tits more frequently than vice versa.Given the limited sample size of this study and the biased sampling towards individuals with plumages similar to pure Azure Tits,our results are only tentative.To determine that hybridization in this system is asymmetric or sex biased will require more data.Given that hybrids are rare outside the region where the two species overlap and that the hybrid zone is narrow(Martin,1991),there is no doubt that there are reproductive barriers that prevent extensive gene flow and total fusion of the two species.Although it is out of scope to study this herein, such barriers could either be pre-zygotic assortative mating (Jiang et al., 2013), where individuals prefer to mate with individuals of the same species,and/or post-zygotic genetic incompatibility making hybrids less fit (Maheshwari and Barbash, 2011).According to Haldane's rule (Haldane, 1922), genetic incompatibilities are expected to be more frequent and severe in the heterogametic sex (females in birds).As our sampling is limited and biased in male/female ratio it is impossible to make firm conclusions whether pre- or post-zygotic barriers are most important in upholding barriers to free gene flow between these two species.However, we consider it most likely that assortative mating is the main barrier that prevent free mixing of these two species based on the observation that one of the F1 hybrids is a female.
F1 hybrids of Azure Tit and Blue Tit are characterized by a suite of intermediate plumage characters.Compared to the Azure Tit,the typical characters for hybrids include darker cap, paler greyish upperparts,less white in the tail, reduced white in the tertials and upper wing coverts,and underparts with traces of a dark collar and or yellowish wash(Harrap and Quinn,1991).Such intermediate plumage characters are typical for the so called Pleske’s Tit(Fig.4A)and all individuals genetically assessed as F1 hybrids in this study possess them.Individuals genetically assessed as first generation backcrosses (BC1) are overall superficially similar to pure Blue Tits or pure Azure Tits,respectively(Fig.4B).Individuals with less European Blue Tit hybrid content become even more similar to pure Azure Tits, by having more white in primary coverts and tail (Fig.4C).These results support the relevance of the plumage clues that are used to assess the hybrid content in Azure Tits (Lawicki, 2012; Corell et al.,2019),as the degree of hybrid content is reflected to a certain extent in the plumage.However, recombination during hybridization and backcrossing will create mosaic genomes in backcross hybrids that vary individually.In several avian species pairs genome-wide association mapping against plumage characters suggests that relatively few genes are involved in the expression of plumage phenotypes, but also that hybrid phenotypes are not directly correlated with overall hybrid content(Toews et al., 2016; Wang et al., 2020).A good example of this is the extensive genomic study of the hybrid zone of Red-breasted and Red-napped Sapsuckers, where there is considerable genetic overlap between morphological hybrids and morphological pure individuals(Natola et al.,2022).In backcrossed hybrids with Azure Tit appearance,one can thus expect that the phenotypes are correlated with alleles of genes involved in plumage expression that have been inherited into their mosaic genomes, rather than to their average hybrid content.It is thus notable that some individuals that morphologically are identified as pure Azure Tits (Fig.4D) express similar levels of European Blue Tit hybrid contents as individuals morphologically assessed as hybrids.Overall,the results support that plumage deviation in backcrosses is not linear correlated with the genetic degree of hybrid origin, that minor hybrid content is widespread in Azure Tits(scored as“pure”based on plumage)in the eastern part of its distribution area,and that all Azure Tits observed in Europe outside its natural distribution may have some degree of hybrid content.
Although a non-scientific problem the question remains how much hybrid content is required for rejecting a specimen as a hybrid.Based on the results from this study and other studies of hybridizing bird complexes (Toews et al., 2016; Wang et al., 2020; Natola et al., 2022) it is obvious that it is difficult to estimate hybrid levels in backcrosses and to separate backcrosses from pure individuals phenotypically.A pragmatic cut-off would be to define F1 and morphologically clearly deviating individuals(potentially first generation backcrosses and offspring between backcrosses and F1 hybrids)as hybrids,and accept minor morphological deviations found in some (but not all) backcrosses as good members of their species.With this argumentation, the two presumed Azure Tits observed in Sweden should be classified as Azure Tits.
4.2.Museomics and crowdsourcing
Increased awareness that hybridization plays an important role in many biological processes, such as trait transfer (Dasmahapatra et al.,2012; Hedrick, 2013; Lamichhaney et al., 2015), adaptive radiations(Seehausen,2004),and the origin of new species(Mavares and Linares,2008; Abbott et al., 2013; Schumer et al., 2014) have made studies of hybridization a hot topic for research.In birds,several genomic studies of hybrid zones have resulted in publications with novel and interesting findings (Poelstra et al., 2014; Toews et al., 2016), which makes the Azure and Blue Tit complex an attractive study system.However,in-depth studies of hybridization require good source material(e.g.,DNA from many individuals), which unfortunately is currently lacking from the Azure and Blue Tit hybrid zone.That makes it less attractive to invest research time and funding in this particular study-system.Yet, linking morphological variation with hybrid content in the Azure and Blue Tit is of significant interest to birdwatchers,as shown by the willingness of the community to support this project financially.The massive bank of biological diversity stored in natural history collections (Duckworth et al.,1993)have become available for genomic studies through the advent of high-throughput sequencing (Bi et al., 2013).Today, museomics is a powerful tool in phylogenomics (McCormack et al., 2016; Knapp et al.,2019; Tsai et al., 2020), biogeography, conservation genetics (Dussex et al., 2018) and for the study of population fluctuations through time(Murray et al., 2017).Here we provide an example of how these resources can be utilized also for projects that has a narrow scientific scope but interest lay citizens.
To birdwatchers,the audience that is the target of the present study,it is arguably most interesting to: 1) improve species identification knowledge in difficult complexes;and 2)to have tools to assess degree of hybrid contents phenotypically in complexes where hybridization occurs.Studies like the present one,which links plumage variation with genomic hybrid content, are thus potential targets for forthcoming crowedsourced projects based on museum samples.Several studies have shown that mitochondrial data do not always mirror the species tree well due to processes such as incomplete linage sorting or mitochondrial introgression(Holder et al., 2001; Buckley et al., 2006; Andersen et al.,2021) and that only specific regions of the nuclear genome may have differentiated in recently diverged “species” (Poelstra et al., 2014;Penalba et al., 2022).Thorough genomic examinations of species boundaries in complexes where other cues (e.g., morphology, biogeography and vocalization) do not co-agree may thus be other potential project suitable for genomic avian crowed-sourced projects based on museum samples, not least as funding for pure phylogenetic and taxonomic research is limited.Decreased sequencing costs and improved pipelines for analyzing genomic data makes projects like this neither particularly expensive nor analytically complex.
Funding
This project was crowd sourced and mainly financed by interested birdwatchers.MI also acknowledge financial support from the Swedish research council(2019-03900) and Riksmusei v€anner.
Authors’contributions
MI and JN designed the study, JN and FT analyzed the data, MI drafted the manuscript with input from all co-authors.All authors read and approved the final manuscript.
Ethics statement
The work was conducted on old museum specimens and tissue samples already preserved at Natural History Museums.
Declaration of competing interest
The authors declare no conflict of interests.
Acknowledgements
This study was only possible thanks to the generous financial support from keen birdwatchers and they are all acknowledged.A special thank goes to S€oren Sahlin whose generous contribution was critical for the project.We are also grateful to Club300’s former and current president Yvonne Blomb€ack and Martin Alexandersson and the editorial staff at magazine Roadrunner (Magnus Corell and Hans Bister) for helping us with advertising and practicalities with this crowd-sourcing project.We are grateful to the following Natural history Museums that generously have contributed samples to this study: the Natural History Museum,Tring (Hein van Grouw and Mark Adams); the Burke Museum, Seattle(Sharon Birks); Swedish Museum of Natural History, Stockholm (Ulf Johansson) and Zoological Museum of Moscow University (Pavel Tomkovich).The authors acknowledge support from the National Genomics Infrastructure in Stockholm funded by Science for Life Laboratory, the Knut and Alice Wallenberg Foundation and the Swedish Research Council, and SNIC/Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure.MI acknowledge financial support from the Swedish research council(2019-03900)and Riksmusei v€anner.
Appendix.Supplementary data
Supplementary data to this article can be found online at https://do i.org/10.1016/j.avrs.2023.100130.
- Avian Research的其它文章
- Selecting the best: Interspecific and age-related diet differences among sympatric steppe passerines
- Morphology and morphometry of two hybridizing buntings at their hybrid zone in northern Iran reveal intermediate and transgressive morphotypes
- Quiet in the nest: The nest environment attenuates song in a grassland songbird
- Characteristics of cross transmission of gut fungal pathogens between wintering Hooded Cranes and sympatric Domestic Geese
- Fecal DNA metabarcoding reveals the dietary composition of wintering Red-crowned Cranes (Grus japonensis)
- Short-term night lighting disrupts lipid and glucose metabolism in Zebra Finches: Implication for urban stopover birds

