Assessing pathological features of breast cancer via the multimodal information of multiphoton and Raman imaging
Bing-Ran Gao(高冰然), Xi-Wen Chen(陈希文), Bao-Ping Zhang(张宝萍), Ivan A.Bratchenko,Jian-Xin Chen(陈建新), Shuang Wang(王爽), and Si-Yuan Xu(许思源),†
1Institute of Photonics and Photon-Technology,Northwest University,Xi’an 710127,China
2Key Laboratory of Opto-Electronic Science and Technology for Medicine of Ministry of Education,Fujian Provincial Key Laboratory of Photonics Technology,Fujian Normal University,Fuzhou 350007,China
3Laser and Biotechnical Systems Department,Samara National Research University,Samara 443086,Russia
Keywords: nonlinear multiphoton microscopic imaging,Raman microspectral imaging,breast cancer
1.Introduction
Nowadays, pathological analysis is mainly based on white-light microscopy, which provides low contrast and diagnostic specificity because of the reflection or attenuation of light.For improving specificity, tissue sections are usually subjected to hematoxylin and eosin(H&E)staining,allowing the visualization of nuclei through the application of hematoxylin,while eosin highlights the cytoplasm and extracellular matrix.[1]It enables the pathologist to easily differentiate between the nuclear and cytoplasmic parts of a cell due to the fact that nuclei are rendered blue or dark-purple, whereas the extracellular matrix is stained pink.This staining approach has withstood the test of time and, as of now, it has become the gold standard for disease diagnostic in clinical.[2]However,the major drawback of staining-based techniques is that the diagnostic results are not immediately available.Moreover,the inexperienced investigations may lead to false negative in clinical trials,requiring multiple repetitions before making final decisions.If the analysis should be performed intraoperatively, e.g., in the case of breast conservation surgery, frozen sections are commonly utilized for determining whether the removed tissue still contains tumor cells or the malignant tissue has been entirely removed.[3]Nevertheless,surgeons may face problems in grading tissue and accessing real-time diagnostic information.[4]Due to the rapid development of novel optical techniques,a significant progress has been achieved in the collection of histopathological information by advanced microscopy based on the unique tissue physical properties other than the reflection and refraction of white light.
For distinguishing molecular and morphological information in biomedical imaging, significant efforts have been made on the label-free and molecular specific characterization of cells and tissue samples.Exogenous optical contrast agents will not be needed to highlight cellular components,and the visualization of healthy and cancerous areas becomes possible via various imaging and spectroscopic tools.Among them, Raman imaging (RI), allowing one to achieve spectroscopic and graphic analytical data,has been greatly improved for clinical applications in recent years.[5,6]It combines Raman spectroscopy and digital imaging technology to simultaneously visualize the chemical composition and molecular structure of the material.[7]The advantages of Raman imaging in biological and medical research are high specificity,low sensitivity to water, and minimal requirement for sample preparation, as well as label-free molecular imaging nature.Moreover,the analytical value of Raman imaging is that each pixel in an image corresponds to a specific chemical data or species within a finite sampling volume.[5]Currently, Raman imaging microscopy systems often operate in a confocal pointscanning configuration,named confocal Raman microspectral imaging (CRMI), which increases the image resolution and spectral signal-to-noise ratio.However, the scan duration is usually extended to collect enough spectral information in a large area,which limits the utility of these systems for routine sample characterization.
In addition to the visualization of specific constitutional information by Raman imaging, structural differences in tissues or cells can be detected and evaluated using other imaging techniques.For example,multiphoton microscopy(MPM)is based on the nonlinear optical effects of the two-photon excited fluorescence(TPEF)and second harmonic generation(SHG),which enable one to perform experiments with a minimal invasion over long periods of time, thereby providing exquisite details of inherently dynamic biological processes within time scales from microseconds to days or weeks.[8]The particular interest of researchers in MPM relies on the high optical resolution for revealing morphological features and the absence of chemical contrast enabling differential diagnosis, which is beneficial for imaging of living and intact tissues.[9,10]MPM requires an ultrafast (typically femtosecond pulse duration) laser in order to achieve the extremely high photon density at the focal plane when exciting a twophoton absorption-based fluorescence signal.[11]As a result,excitable endogenous fluorescent species such as nicotinamide adenine dinucleotide(NAD(P)H),flavin,and elastin can be reliably highlighted via TPEF,thereby furnishing information on tissue microstructure,while SHG allows visualization of noncentrosymmetric molecules,e.g.,tissue collagen contents.[12]
Based on the diversified optical properties of tissues,many studies focus on particular aspects of the sample’s characteristics by using multimodal imaging information, which potentially provides a complementary view of the underlying pathology with optimized diagnostic significance.[13]Therefore,for unveiling pathological progressing features of breast cancer, MPM and CRMI techniques are both utilized in this work to address the histopathology characteristics of healthy(H),ductal carcinomain situ(DCIS),and invasive ductal carcinoma(IDC)tissues.In addition to the assessment of specific biomarkers,the combination of multimodal optical investigations offers new opportunities for exploiting optical modalities in breast-conserving surgery and other clinical directions.
2.Material and methods
2.1.Tissue sample preparation
Breast tissue samples including healthy tissue (n= 6),DCIS (n=12), and IDC (n=12) were obtained during the breast-conserving surgery at the Department of Breast Surgery of the First Affiliated Hospital of Xi’an Jiaotong University of China.All biopsy procedures such as sample collection, tissue section preparation, and microscopic analysis were compliant with the current laws of the People’s Republic of China.The tissue biopsies were prepared and sliced into five sections.Among them,the first and second sections with 10µm thicknesses were placed on a glass substrate for multiphoton microscopic imaging.The third slice had a thickness of 5 µm and was exposed to H&E staining for pathological evaluation.Finally,the fourth and fifth slices(both 15µm in thickness)were put onto a gold-plated substrate for Raman imaging(RI).Prior to MPM and RI studies,white light microscopy images of all prepared sections were acquired to identify the parallel similar regions of the samples at the same scale.Investigated by one clinician in the hospital using an H&E stained sample,the tissue types and their pathological statuses were identified for further MPM and CRMI experiments.Large-area multiphoton imaging(5000µm×4000µm,8000µm×6000µm,and 1400 µm×2000 µm) was implemented as a guide to identifying regions of interest for each sample, and then specific small regions (900 µm×700 µm, 600 µm×500 µm, and 700µm×1000µm)were selected for Raman imaging.
2.2.Multiphoton microscopic imaging
A multiphoton microscopic imaging system used in the study consisted of a laser scanning microscope (LSM Model 880,Zeiss)and a mode-locked titanium sapphire femtosecond laser(Chameleon Ultra,Coherent).Nonlinear optical imaging of the tissue was performed with the excitation light at a wavelength of 810 nm, and the signal was acquired by a 20× objective(plan-apochromat,NA=0.8,Zeiss).The laser emitted by the mode-locked titanium sapphire femtosecond laser was reduced to 5 mW–10 mW by the action of an acousto-optic modulator, which enabled one to decrease the risk of laserinduced damage in the tissue.The excited backscattered signals were collected simultaneously by two independent channels, namely, SHG and TPEF.The first channel filtered the 395 nm–415 nm spectral bandwidth signals and the second one received the 428 nm–695 nm signals.
2.3.Raman microspectral imaging and data analysis
A confocal Raman microspectral imaging system(Alpha 500R, WITec GmbH) was applied for spectral and imaging investigations.The Raman spectra of the tissues were excited using a 532 nm semiconductor continuous laser,and collected with a 100× microscope objective (NA=1.25, EC Epiplanneofocal, Zeiss).The spectra were then acquired by a spectrometer(UHTS300 model,WITec GmbH)with a 600 mm-1grating and a deep-depletion, back-illuminated CCD camera(Du401A-BR-DD-352,Andor Technology)cooled to-60◦C.The hyperspectral Raman datasets were collected by moving the sample in the focus plane on a piezo scanning stage(P-524 K081, PI GmbH).Before Raman imaging, the whitelight microscopic images of the tissue samples were obtained with a 20×microscope objective(NA=0.4,Epiplan-neofular,Zeiss).For each breast tissue, the Raman images were acquired at a rate of 1 s per pixel within the areas selected according to the H&E-stained section and MPM imaging results.The scanning areas for healthy, DCIS,and IDC tissues were 2900 µm×700 µm, 600 µm×500 µm, and 700 µm×1000µm,respectively.
All the acquired spectral data were pre-processed by using the NWU-spectral-analysis(NWUSA)toolbox,which integrated all analysis functions of our home-made software Northwest University spectra analysis(NWUSA)[14]and Raman spectral imaging toolbox(NWU-RSIT).[15]The acquired spectroscopic datasets were firstly preprocessed following the steps of cosmic ray removal, fingerprint regions selection(600 cm-1–1800 cm-1and 2800 cm-1–3000 cm-1),spectral background subtraction by a 9th-order polynomial fitting,and 5th-order Savitzky–Golay smoothing.K-mean cluster analysis (KCA) of the data was afterward performed to provide a detailed biochemical composition and morphological structure information within the scanned regions.[15]It is noteworthy that KCA as an unsupervised multivariate analysis method allows one to represent the maps of cluster members as separated color-coded images that can be used to gather the relevant micromorphological information.In addition, the biochemical composition of various clusters can be established by calculating their average spectra to reflect the variation within the characteristic Raman peaks from the specimen.[16]
3.Results
3.1.The analysis of healthy breast tissue
The H&E-stained image of a healthy breast tissue is illustrated in Fig.1(a),where the black squares represent the MPM imaging area (Fig.1(b)), and the red frames indicate the Raman imaging area(Fig.1(c)).Endogenous fluorophores inside breast tissue such as NAD(P)H and flavin adenine dinucleotide(FAD),were prone to producing strong TPEF signals.Meanwhile, collagen fibers in the extracellular matrix were more likely to be detected in SHG signals.[17]Based on this fact,the complete ductal and lobular morphology of the normal breast tissue and the epithelial cells inside the ducts and the basement membrane can be observed via both the MPM and Raman imaging techniques.Figure 2 depicts the MPM and Raman images of healthy breast tissues, in which Fig.2(a) indicates the selected imaging area(the yellow box),and Fig.2(b)displays the MPM imaging of healthy breast tissues.Figures 2(c)and 2(d)show the SHG and TPEF images in green and red colors,respectively,and the reconstructed Raman image is given in Fig.2(e).

Fig.1.The microscopic images of healthy breast tissue.The H&Estained image of a healthy breast tissue is illustrated in panel(a),in which the black squares represent the MPM imaging area(b),and the red frames indicate the Raman imaging area(c).
According to Fig.2(b), the parenchyma of the breast mainly consisted of the ducts,plenty of connective tissues and adipose tissues.Each duct was composed of an inner layer of epithelial cells,an outer layer of myoepithelial cells,and a basement membrane wrapped around the outer layer.Collagen in the extracellular matrix can be identified by the SHG signal(green)in Fig.2(c),while cells and elastic fibers are visualized by the TPEF signal(red)in Fig.2(d).In Fig.2(d),the mammary ducts consist of an epithelial cell layer and a myoepithelial cell layer.Meanwhile,the black circular structures in the middle of the cells were the non-fluorescent nuclei.The breast ducts were surrounded by the connective tissues consisting of numerous collagen bundles.The collagen fibers were closely and orderly distributed in the normal breast tissue,and the basement membrane was intact, which is consistent with the H&E-stained pictures in Fig.1(a).
To further characterize the composition of healthy breast tissue, Raman imaging was performed within the same region highlighted in the yellow frame in Fig.2(a).The obtained spectral dataset of the scanned region was analyzed by the KCA method, which enabled one to automatically classify similar spectral features and reconstruct the pseudo-color images.The mean spectra in each sub-cluster depicted the constitutional information in different regions, exhibiting the spatial distribution of various biochemical components across the scanned area.[17,20]Figures 2(e)–2(i) show the KCA images of the ductal structures of healthy breast tissue, whereas Fig.2(e) displays a combined image to describe the overall morphology of the sampling region.Figures 2(f)–2(h)depict the individual sub-clusters whose average spectra are given in Fig.2(i).
Three grouped sub-clusters of healthy breast tissue were mainly manifested by the spectral bands at 754 cm-1(symmetric breathing of tryptophan),[18]1004 cm-1(phenylalanine, CH3rocking coupled with C–C stretching of carotenoids),[19]1155 cm-1(C–C and C–N stretching;carotenoids),[19]1265 cm-1(υ(CN),δ(NH) amide III,α-helix, collagen (protein assignment)),[20]1307 cm-1(CH3/CH2twisting or bending mode of collagen/lipid),[21]1450 cm-1(δ(CH2)/δ(CH3), lipids)),[22]1520 cm-1(–C=C– carotenoids),[19]1579 cm-1(pyrimidine ring, nucleic acids),[19]1658 cm-1(amide I (C=O stretching mode of proteins,α-helix conformation)/C=C lipid stretching),[23]2860 cm-1(CH2symmetric stretch of lipids),2900 cm-1(CH stretching of lipids and proteins),and 2930 cm-1(CH stretching of lipids and proteins).[19]It revealed that the blue cluster had a lower overall intensity than the other two clusters with heterogeneous spectral signals, while, other three clusters showed spectral differences in the 1400 cm-1–1800 cm-1and 2800 cm-1–3000 cm-1ranges.Referring to the H&E and MPM imaging results, it could be concluded that red clusters are secretions in the ducts,whereas an intense lipid signal from the green cluster(2860 cm-1)as a distinct feature means that the cluster can be assigned to a ductal stroma.These observations are consistent with the SHG images in Fig.2(c).In turn.the blue region was a tissue slit in the epithelial tissue of the duct,which exhibits a low signal-to-noise ratio spectral signal.
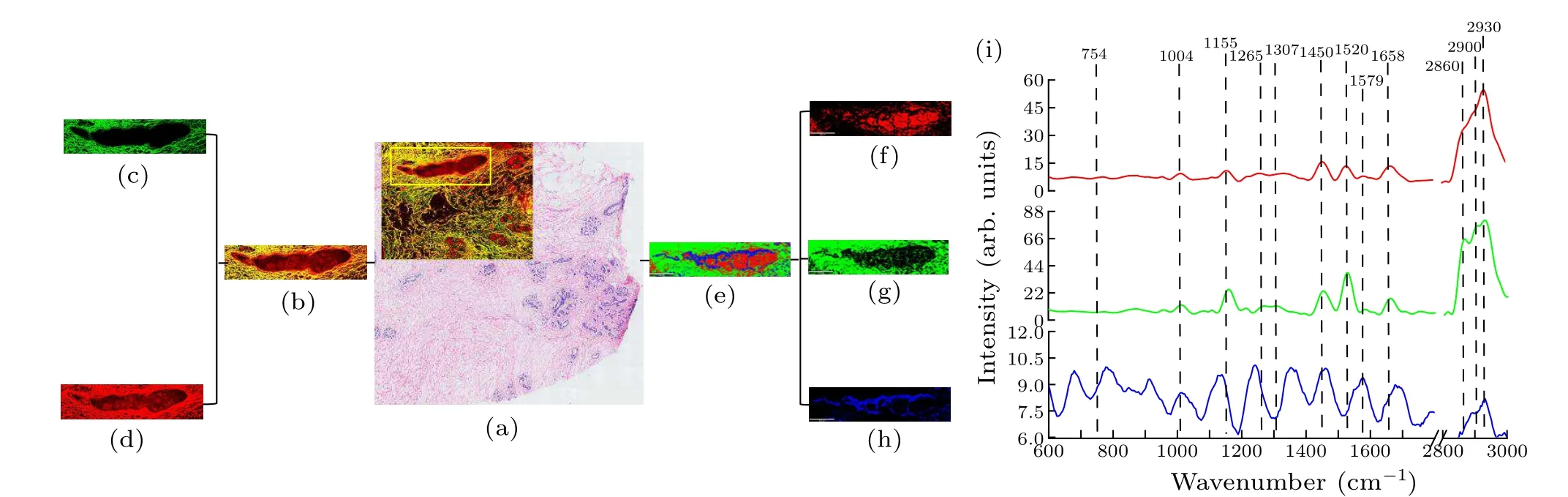
Fig.2.MPM and Raman images of healthy breast tissue.Subfigure(a)indicates the selected imaging area(the yellow box),and(b)displays the combined MPM results of healthy breast tissues.Subfigures(c)and(d)show the SHG and TPEF images(in green and red colors,respectively).The reconstructed Raman image by the KCA algorithm is given in panel(e),which displays a combined image to describe the overall morphology of the sampling region.Subfigures(f)–(h)depict the individual sub-clusters whose average spectra are given in panel(i).Scale bars of subfigures(e)–(h): 600µm.

Fig.3.The univariate Raman images of healthy breast tissue were reconstructed by the integration of specific Raman peaks at 754 cm-1(tryptophan), 1004 cm-1 (phenylalanine), 1155 cm-1 (carotenoid), 1265 cm-1 (collagen), 1307 cm-1 (collagen), 1520 cm-1 (carotenoid),1579 cm-1 (nucleic acid), 1658 cm-1 (lipid), and 2930 cm-1 (lipid).Image (j) shows the combined image of nucleic acid (red), carotenoids(green),and collagen(blue);image(k)shows the combined image of carotenoids(green),nucleic acids(red),and lipids(blue);image(l)shows the combined image of carotenoid(green)and collagen(red).Scale bars of subfigures(a)–(l): 600µm.
To understand the origin of biochemical components spatially distributed in the healthy tissue, the univariate Raman images were reconstructed and presented in Fig.3.These images displayed the presence of tryptophan (754 cm-1),nucleic acid (1579 cm-1), carotenoid (1155 cm-1and 1520 cm-1), collagen (1265 cm-1and 1307 cm-1), phenylalanine(1004 cm-1), and lipid(1658 cm-1and 2930 cm-1)within the scanned region.It could be observed that phenylalanine, carotenoids, and collagen components were mainly distributed in the stroma.Nucleic acids were primarily detected in the epithelial tissue of the breast ducts, and a small amount was located in the stroma.In contrast, lipids uni-
formly spread in the nucleic acids and were primarily distributed in the epithelial tissue of the breast ducts, whereas their small fraction was found in the stroma and inside the ducts.Figure 3(j) represents the spatially merged images of nucleic acids, carotenoids, and collagen, while, Fig.3(k) depicts the merged images of carotenoids, nucleic acids, and lipids.Combined distribution characteristics of nucleic acids and collagen in healthy breast ducts are shown in Fig.3(l).The morphological characteristics of the breast ducts in these superimposed images were also consistent with the MPM results, which means a finger-spectral based component interpretation and validation from Raman imaging.
3.2.The analysis of ductal carcinoma in situ breast tissue
The H&E-stained image of the ductal carcinomain situ(DCIS) tissue was illustrated in Fig.4(a), where the black squares represented the MPM imaging area (Fig.4(b)), and the red frames indicated the Raman imaging region(Fig.4(c)).According to the H&E-stained image, different from the normal breast ducts,the DCIS tumor cells filled the lumens of the breast ducts, making them significantly larger.Figure 5 displays the MPM and Raman imaging results of the DCIS breast tissues.Figure 5(a) indicates the area selected for scanning(the yellow box), and Fig.5(b) displays the MPM images of DCIS breast tissues.Figure 5(c) depicts the SHG image in green color,and Fig.5(d)shows the TPEF image in red color.The reconstructed Raman image is given in Fig.5(e).

Fig.4.The microscopic images of ductal carcinoma in situ (DCIS) tissue.The H&E-stained image of a DCIS tissue is illustrated in panel(a),in which(b)the black squares represent the MPM imaging area,and(c)the red frames indicate the Raman imaging area.
According to Fig.5(b), unlike the normal breast tissue,there was a significant increase of malignant epithelial cells in the DCIS breast tissue, which filled almost entirely the lumens of the ducts without breaking through the duct basement membrane.Meanwhile, the collagen bundle in Fig.5(c) was not significantly changed, and it surrounded the breast ducts as in the case of the normal tissue.Although the basement membrane was well defined and intact,it was significantly dilated, and the surrounding collagen layer became thinner as the duct expanded.The phenomenon of small and uniform nuclei as part of the ducts can be observed in Fig.5(d).These morphological changes are consistent with the structural features observed in the corresponding H&E-stained images in Fig.4(a).
As with the characterization of healthy breast tissue,Raman imaging was performed within the same region of the DCIS tissue (highlighted with the yellow frame in Fig.5(a))to understand its biological composition.Figures 5(e)–5(i)display the KCA images of ductal structures of DCIS breast tissue,wherein Fig.5(e)shows a combined image to describe the overall morphology of the sampling region, Figs.5(f)–5(i) depict the separated sub-clusters, and the corresponding average spectra of each sub-cluster are plotted in Fig.5(j).The four sub-clusters of DCIS breast tissue mainly exhibited the following spectral bands: 754 cm-1, 1004 cm-1,1155 cm-1,1265 cm-1,1307 cm-1,1450 cm-1,1520 cm-1,1579 cm-1, 1658 cm-1, 2860 cm-1, and 2930 cm-1.These bands are highly related to biochemical components such as nucleic acids, lipids, and proteins in breast tissue.Comparing the average spectra of each subcluster, it was found that the overall spectral intensities of the red and green subclusters were higher than those of the yellow and blue clusters.The intensity of the red cluster exceeded that of the green cluster,meaning that the former had a higher content of biological components than the latter,which might be caused by the large aggregation of cancer cells.Based on that,the red cluster was identified as the cancer cell aggregation area inside the duct,whereas the green cluster was referred to as a duct basement membrane.The yellow and blue clusters with the lower spectral signal-to-noise ratios were suspected to be impurities and tissue cracks.These KCA results were consistent with the corresponding H&E-stained and MPM imaging data.
The univariate Raman images of DCIS breast tissue are shown in Fig.6,where the spatial distribution of different biochemical components can be observed.Among them,tryptophan(754 cm-1),[18]nucleic acid(1579 cm-1),[19]carotenoid(1155 cm-1and 1520 cm-1),[17]collagen (1265 cm-1and 1307 cm-1),[20]phenylalanine (1004 cm-1),[19]and lipid(1658 cm-1and 2930 cm-1)[19]were detected throughout the scanned region.These biological components were present inside the ducts of DCIS breast tissue as well as the interstitium.Although their distributions varied slightly, the brighter areas indicated the higher amounts of the components.Figure 6(j)represents the spatially merged images of nucleic acids and collagen.Figure 6(k)displays the merged images of collagen,nucleic acids, and lipids.Finally, Fig.6(l) depicts the combined distribution characteristics of collagen, carotenoid, and lipids in DCIS breast ducts.A visual inspection of these univariate imaging maps and compositional overlay images revealed that nucleic acids, lipids, and carotenoids were more abundant inside the ducts,while collagen was predominant in the basement membrane of the ducts.Therefore, the MPM images were in good agreement with Raman imaging from the constitutional perspectives.
3.3.The analysis of invasive ductal carcinoma breast tissue
The H&E-stained image of IDC breast tissue is illustrated in Fig.7(a),where the black squares represent the MPM imaging area (Fig.7(b)), and the red frames indicate the Raman imaging area (Fig.7(c)).According to the H&E-stained image,in contrast to DCIS breast tissue,tumor cells in IDC penetrated the basement membrane and infiltrated into the surrounding tissue.Figure 8 depicts the MPM and Raman imaging results of IDC breast tissues.Figure 8(a)indicates the selected imaging area(the yellow box).Figure 8(b)displays the MPM images of IDC breast tissues.The SHG image is shown in Fig.8(c)in green color,the TPEF image is given in Fig.8(d)in red color, and the reconstructed Raman image is available in Fig.8(e).
According to Fig.8(c), due to the infiltration and encroachment of the tumor, the basement membrane was destroyed or even disappeared,and the ductal structure could no longer be observed.At the same time,the collagen fibers were elongated and their arrangement became more disorganized.Inspecting from the TPEF image in Fig.8(d), the tumor cells were no longer confined to the ducts as in ductal carcinomain situ,but broken through the basement membrane and infiltrated into the surrounding stroma.These tumor cells varied in shape and showed a high degree of pleomorphism.The results are consistent with the H&E staining image in Fig.7(a).
Figures 8(e)–8(j)display the KCA images of ductal structures in IDC breast tissue, in which Fig.8(e) illustrates a combined image to describe the overall morphology of the sampling region.The individual sub-clusters are present in Figs.8(f)–8(i),and the corresponding average spectra of each sub-cluster are plotted in Fig.8(j).The four sub-clusters of IDC breast tissue mainly exhibit the following spectral bands:754 cm-1, 1004 cm-1, 1155 cm-1, 1265 cm-1, 1307 cm-1,1450 cm-1,1520 cm-1,1579 cm-1,1658 cm-1,2860 cm-1,2900 cm-1,and 2930 cm-1.These bands are highly related to biochemical components such as nucleic acids,lipids,and proteins in breast tissue.Comparing the average spectra of each sub-cluster,it was found that the overall spectral intensities of the red and green subclusters were higher than those of the yellow and blue clusters.Meanwhile,the spectral features of the red and yellow clusters were very similar with a little intensity variations.However,the spectra of green clusters showed significant differences from red and yellow clusters at 2900 cm-1,indicating that green clusters were richer in lipids.Based on that, the red and yellow clusters were assigned to cancer cell aggregation areas in the stroma.The yellow cluster was referred to cancer cells aggregating at the edge of the tissue,and the green cluster was associated with the tissue stroma.The blue cluster exhibited a cluttered spectral feature,and the relevant region was assumed to be the substrate or impurity.
The univariate Raman images of IDC breast tissue are shown in Fig.9, displaying the spatial distribution patterns of tryptophan (754 cm-1), nucleic acid (1579 cm-1),carotenoid(1155 cm-1and 1520 cm-1),collagen(1265 cm-1and 1307 cm-1), phenylalanine (1004 cm-1), and lipid(1658 cm-1and 2930 cm-1) across the scanned region.A visual inspection of these images revealed that each component was distributed in the IDC tissue, but phenylalanine and carotenoids were predominant in the upper right corner of the scan area, whereas lipid components were abundant and evenly distributed throughout the tissue.Figure 9(j) represents the spatially merged images of nucleic acids,carotenoid,and lipids.Figure 9(k) displays the merged images of collagen,carotenoid,and lipids.Finally,Fig.9(l)depicts the combined distribution characteristics of collagen and nucleic acids in IDC breast ducts.
4.Discussion
In this study, MPM and CRMI techniques were combined for in-depth investigation of the progression of breast ductal carcinoma and thoroughex vivoanalysis of the pathological features of healthy, ductal carcinomain situ, and invasive ductal carcinoma breast tissue.The collagen in the stroma of the breast tissue and the collagen fibril component in the basement membrane of the breast ducts had a non-centrosymmetric structure that generated the SHG signals.In addition, adipocytes in breast tissue, as well as epithelial and tumor cells in ducts and blood vessels, produced strong autofluorescence signals through TPEF.According to Figs.2(c), 5(c), and 8(c), there were a significant change in the distribution characteristics of collagen with tumor progression.In the healthy breast ducts(Fig.2(c)),the collagen fibers were arranged in an orderly manner outside the lumens.At that moment,cells were distributed along the basement membrane of the duct with secretions inside the lumens of the duct(Fig.2(d)).As shown in Fig.5(c),once the epithelial cells became cancerous and started to proliferate, the duct morphology changed considerably, the lumen expanded, and the collagen fibers were transformed from curved to straight due to stretching.However, the collagen fibers in the stroma were still arranged around the ducts as in healthy tissue, and the basement membrane was thin but clear and intact.Based on the TPEF results (Fig.5(d)), tumor cells were also found to be aggregated inside the ducts.As the extent of carcinoma continued to progress,the basement membrane was destroyed,and the cancer cells had infiltrated into the tissue stroma(Fig.8(b)).Meanwhile,the collagen bundles in the IDC tissue changed in both morphology and arrangement,from the original orderly distribution to a disorderly arrangement, and the ductal structure disappeared (Fig.8(c)).This transformation in ductal morphology because of collagen fibers acted as the skeletal structures of the extracellular matrix.Moreover,with the infiltration of tumor cells,collagen fibers were locally degraded under the action of matrix-degrading enzymes, which disturbed its arrangement structure and provided the migration space for tumor infiltration.[24,25]
To further gather the comprehensive biochemical composition information on breast cancer tissues, Raman spectroscopy was applied to the same area from both the spectral and image perspectives,and the pathological features of breast ductal carcinoma were then analyzed at different stages of proliferation.The results of the KCA analysis were consistent with those of MPM images, where the lumens and external stroma of healthy breast ducts could be clearly distinguished.The external interstitial region of the duct was found to be rich in lipid components (Fig.2(g)), and was also very similar to the morphological structure reflected in the SHG signal(Fig.2(c)).As the cancer continued to evolve within the duct,the latter was dilated(Fig.5(g)),which was consistent with the SHG results(Fig.5(c)).Also,cancer cells aggregating inside the duct were identified (Fig.5(f)), which coincided with the TPEF images(Fig.5(b)).In IDC tissue(Fig.8(e)), the KCA analysis showed the same disorganized mesenchymal structure destroyed by cancer cells and the disappearance of the ductal basement membrane was revealed via multiphoton microscopic imaging.In addition, a comparison of the average spectra acquired at the interior of healthy ducts(Fig.2(f))and the interior of ducts with ductal carcinomain situ(Fig.5(f))enabled one to conclude that the Raman peaks of tryptophan(754 cm-1) and nucleic acid (1579 cm-1) were attributed to the red cluster of DCIS tissue, indicating that the content of nucleic acids and proteins gradually increased as the cancer cells continued to proliferate inside the ducts.This was due to the loss of tumor suppressors or activation of oncogenes during cancer development,resulting in the uncontrolled proliferation of cancer cells and the synthesis of large amounts of proteins.During this process,DNA in daughter cells continuously replicated,leading to an increase in nucleic acid content.[26,27]While the ductal structure could not be observed in the KCA pseudo-color image(Fig.8(e)),the high-intensity spectral features within the range of 2800 cm-1–3000 cm-1indicated that the tissue was rich in fatty components.Since the interstitium of breast tissue was mainly composed of fat and connective tissue,the spectral features further revealed that the cancer cells were broken through the ductal basement membrane and simultaneously infiltrated into the interstitium.
Based on different optical principles, CRMI and nonlinear optical imaging (SHG and TPEF) techniques can reconstruct the histomorphological and structural features of different cancerous stages of breast ducts.In particular,multiphoton microscopic imaging methods underlying the concept of nonlinear optics allow one to obtain high-resolution microstructural images of tissues, while CRMI enables one to further analyze the changes in tissue morphology and composition of breast cancer tissues at different stages from spectroscopic and imaging perspectives.Therefore,combined MPM and Raman imaging approach is efficient for monitoring the tumor evolution at the micro-level,being also a powerful imaging tool for gathering information on microenvironmental changes during breast tumor development.
5.Conclusion
A multifaceted pathological study of different stages of breast ductal carcinoma development was implemented by combining Raman imaging and MPM techniques.Special attention was paid to a thorough analysis of the multimodal microscopic imaging results of healthy breast ductal tissue,DCIS,and IDC tissue.MPM-based imaging(TPEF and SHG)provided high-resolution images of the fine structure of breast tissue.In turn, Raman spectroscopy enabled one to not only accurately identify the structural and spectral features at various stages of ductal carcinoma from a spectral perspective,but also visualize the distribution of components in the tissue through univariate imaging.This evolutionary process was described in terms of both cells and extracellular stroma as follows.As the cancer cells continued to proliferate,the acquired MPM images showed that the cells first arranged around the basement membrane of the duct, then proliferated to fill the lumens of the duct,and were finally broken through the basement membrane to infiltrate into the stroma.At once, while the Raman imaging failed to visualize the cell distribution with high resolution,it still allowed one to distinguish between the cell aggregation area and the interstitial area.During tumor development, the ductal basement membrane in the stroma first changed from thick to thin and then vanished throughout as the cancer cells infiltrated into the stroma.This phenomenon was shown to be consistent for both the MPM and CRMI data.At the same time,CRMI not only explained spectroscopically the gradual increase of nucleic acid and protein components inside the ducts as cancer cells proliferated, but also displayed the distribution of each biological component in the breast tissue via univariate imaging.Thus,the combination of MPM and CRMI presented non-labeled structural and constitutional information with higher specificities and sensitivities.Without special sample preparations,it provided new insights into the pathological study and diagnosis of tissue malignant,especially if it needs to be performed intraoperatively with bulk tissue or excised frozen sections.The observations in this work also ensured technical support for the development of multimodal optical imaging techniques for advanced precise histopathological analysis.
Acknowledgments
Project supported by the National Natural Science Foundation of China (Grant No.61911530695) and the Key Research and Development Project of Shaanxi Province of China(Grant No.2023-YBSF-671).
——记4808工厂威海修船厂
- Chinese Physics B的其它文章
- The application of quantum coherence as a resource
- Special breathing structures induced by bright solitons collision in a binary dipolar Bose–Einstein condensates
- Effect of short-term plasticity on working memory
- Directional-to-random transition of cell cluster migration
- Effect of mono-/divalent metal ions on the conductivity characteristics of DNA solutions transferring through a microfluidic channel
- Off-diagonal approach to the exact solution of quantum integrable systems

