染色体维持蛋白基因家族在头颈癌中的差异表达和预后价值


[摘要]目的探讨染色体维持蛋白(MCM)在头颈癌中的表达和预后治疗效果,旨在为头颈癌潜在的生物标志物以及治疗靶点提供更多的证据。方法使用Oncomine、GEPIA和Human Protein Atlas在线数据库分析MCM在头颈癌中的表达差异;运用GeneMANIA和STRING在线平台预测并构建MCM蛋白质相互作用网络;通过Metascape数据库对MCM进行功能和通路富集分析;通过Kaplan-Meier plotter分析平台研究MCM在头颈癌中的预后价值;使用Timer在线工具完成MCM与头颈癌中的免疫浸润细胞的相关性研究。结果头颈癌中MCM2/3/4/5/6/7/8/10的mRNA表达较正常头颈部组织显著增加(Plt;0.01),其中MCM2/3/7在转录和翻译阶段均过表达。MCM2-MCM7与DNA复制密切相关,并且发现细胞周期相关基因与MCM复合体密切相关。功能和通路富集分析表明其生物学功能主要与DNA复制和cGMP-PKG信号通路有关。MCM1/3/5/6/10的高表达与头颈癌患者总生存期的改善明显相关(Plt;0.05)。此外,人类乳头瘤病毒(HPV)感染状态对MCM基因家族的表达会造成一定影响,头颈鳞癌HPV阳性(HNSCC-HPV-pos)中MCM的表达与免疫浸润的丰度显著相关,尤其是B细胞、CD8+T细胞和CD4+T细胞。HNSCC-HPV-pos亚组的B细胞、CD8+T细胞和中性粒细胞免疫浸润明显增加(Plt;0.05),这与MCM的表达水平有关。结论MCM2/3/7的异常高表达提示其在头颈癌发生发展中起重要作用。MCM1/3/5/6/10可作为判断头颈癌患者生存率的候选预后标志物。
[关键词]MCM基因家族;头颈癌;预后;生物标志物;生物信息学
doi:10.3969/j.issn.1674-7593.2023.05.004
Exploring Gene Expression Disparities and Prognostic Significance of Mini-ChromosomeMaintenance Family in Head and Neck Cancer
Zheng Zhaoyang1,Wang Xianzeng2,Zhang Man3,Chen Shuangshuang1,Yang Ying3,Liu Hongchun3**
1The Second Clinical Medical College,Henan University of Traditional Chinese Medicine,Zhengzhou450002;2Department of Thoracic Surgery,Linzhou People's Hospital,Anyang456550;3Department of Medical Laboratory,the First Affiliated Hospital of Zhengzhou University,Zhengzhou450052
**Corresponding author:Liu Hongchun,email:xingyunerliu@163.com
[Abstract]ObjectiveTo explore the expression disparities and prognostic significance of mini-chromosome maintenance(MCM) in head and neck cancer(HNC),towards unveiling novel biomarkers and therapeutic targets.MethodsOncomine,GEPIA and Human Protein Atlas were used to analyze the expression patterns of MCM in HNC.Providing valuable insights into its differential expression in tumor tissues compared to normal tissues.Furthermore,to gain a comprehensive understanding of the functional implications and regulatory networks associated with MCM,we predicted and constructed a protein-protein interaction network using GeneMANIA and STRING.This network analysis allowed us to explore potential interactions and functional relationships of MCM with other proteins in HNC.Additionally,functional enrichment analysis and signaling pathway analysis of MCM were performed using Metascape.This analysis shed light on the biological processes and pathways in which MCM is involved,offering insights into its potential roles in HNC pathogenesis.To assess the prognostic value of MCM in HNC,we employed Kaplan-Meier plotter,enabling us to analyze survival data and evaluate the correlation between MCM expression levels and patient outcomes.Lastly,we investigated the correlation between MCM expression and immune infiltrating cells in HNC using the TIMER database.This analysis provided valuable information regarding the potential interactions between MCM and the tumor immune microenvironment in HNC.ResultsThere was a significant increase in the expression levels of MCM2,MCM3,MCM4,MCM5,MCM6,MCM7,MCM8,and MCM10 in HNC samples(Plt;0.01).Specifically,MCM2,MCM3,and MCM7 exhibited both transcriptional and translational upregulation.The MCM2-7 complex,known for its involvement in DNA replication,demonstrated a close association with cell cycle-related genes.Functionally,MCM was primarily implicated in DNA replication processes and the cGMP-PKG signaling pathway.Notably,higher expression levels of MCM1,MCM3,MCM5,MCM6,and MCM10 were significantly associated with improved overall survival in HNC patients(Plt;0.05).Furthermore,the expression of MCM was influenced by human papilloma virus(HPV) infection status.Specifically,MCM expression in HPV-positive head and neck squamous cell carcinoma(HNSCC-HPV-pos) correlated with the abundance of immune invasion,particularly B cells,CD8+ T cells,and CD4+ T cells.In the HNSCC-HPV-pos subgroup,there was a significant increase in immune infiltration of B cells,CD8+ T cells,and neutrophils(Plt;0.05),which was linked to the expression levels of MCM.These findings suggest that MCM plays a crucial role in HNC,with dysregulated expression patterns observed in tumor samples.The upregulation of MCM genes,particularly MCM2,MCM3,and MCM7,highlights their potential as biomarkers for HNC prognosis.Additionally,the impact of HPV infection on MCM expression and its association with immune infiltration further emphasizes the complex interplay between MCM and the tumor microenvironment in HNC.ConclusionThe aberrantly elevated expression of MCM2,MCM3,and MCM7 strongly suggests their pivotal involvement as oncogenes in HNC pathogenesis.Moreover,the study identifies MCM1,MCM3,MCM5,MCM6,and MCM10 as potential candidate prognostic biomarkers for HNC.
[Key words]Mini-chromosome maintenance protein;Head and neck carcinoma;Prognosis;Molecular biomarkers;Bioinformatics
头颈癌(Head and neck cancer,HNC)是全球第六大最常见的恶性肿瘤,具有预后差、存活率低等特点[1]。头颈癌的危险因素包括吸烟、饮酒和人类乳头瘤病毒(Human papilloma virus,HPV)感染。近期研究人员发现,HPV相关的头颈癌的发病率正在增加[2]。目前头颈癌的治疗方案包括手术、放疗和化疗[3]。尽管已经在检测和治疗方面取得了进步,但头颈癌恶性程度高,进展快,确诊人群多处于中晚期[4],因此,有必要寻找有效的肿瘤生物标志物和合适的靶点来预测患者的预后,提高生存率。
DNA复制是肿瘤发生和发展研究的核心内容之一[5]。染色体维持蛋白(Mini-chromosome maintenance,MCM)家族是DNA正常复制所必需的。近年来,MCM已被证实在多种癌症的发病机制中起着关键作用[6-7]。然而,目前尚未有研究表明它们在头颈癌中是否发挥同样重要的作用。
本研究首次利用公共数据库对头颈癌中MCM基因家族的表达水平、功能富集,基因或蛋白分子潜在的相互作用、基因预后价值以及免疫浸润状态进行了全面探索,以期为研究头颈癌的分子机制提供新见解,并为开发头颈癌新型治疗靶点提供更多理论依据。
1对象与方法
1.1Oncomine、GEPIA和Human Protein Atlas
Oncomine(https://www.oncomine.org)是一个癌症微阵列数据库和综合数据挖掘平台,提供癌症和正常样本中基因的表达信息[8]。GEPIA数据库(http://gepia.cancer-pku.cn)整合了来自癌症基因组图谱(The cancer genome atlas,TCGA)和基因类型组织表达(Genotype-tissue expression,GTEx)项目的9 736个肿瘤和8 587个正常样本的基因表达数据[9]。Human Protein Atlas平台(https://www.proteinatlas.org)包含了近20种最常见癌症的免疫组织化学的表达数据[10]。本研究利用以上数据库分析了头颈癌组织中和相应的邻近正常对照组织中MCM家族成员的mRNA以及蛋白表达水平。
1.2GeneMANIA和STRING
GeneMANIA(http://www.genemania.org)是用于构建具有相似功能的基因,查询基因之间交互式功能关系的网络[11]。STRING(https://string-db.org/)是一个资源丰富的数据库,其目标是收集、评价和整合蛋白质-蛋白质相互作用网络的所有公共资源,并用潜在的计算功能来预测补充这些资源[12]。本研究构建了MCM家族基因及其潜在相互作用分子的蛋白-蛋白间交互作用(Protein-protein interaction,PPI)网络,以表明MCM之间及其相互作用基因之间的功能关联。
1.3Metascape
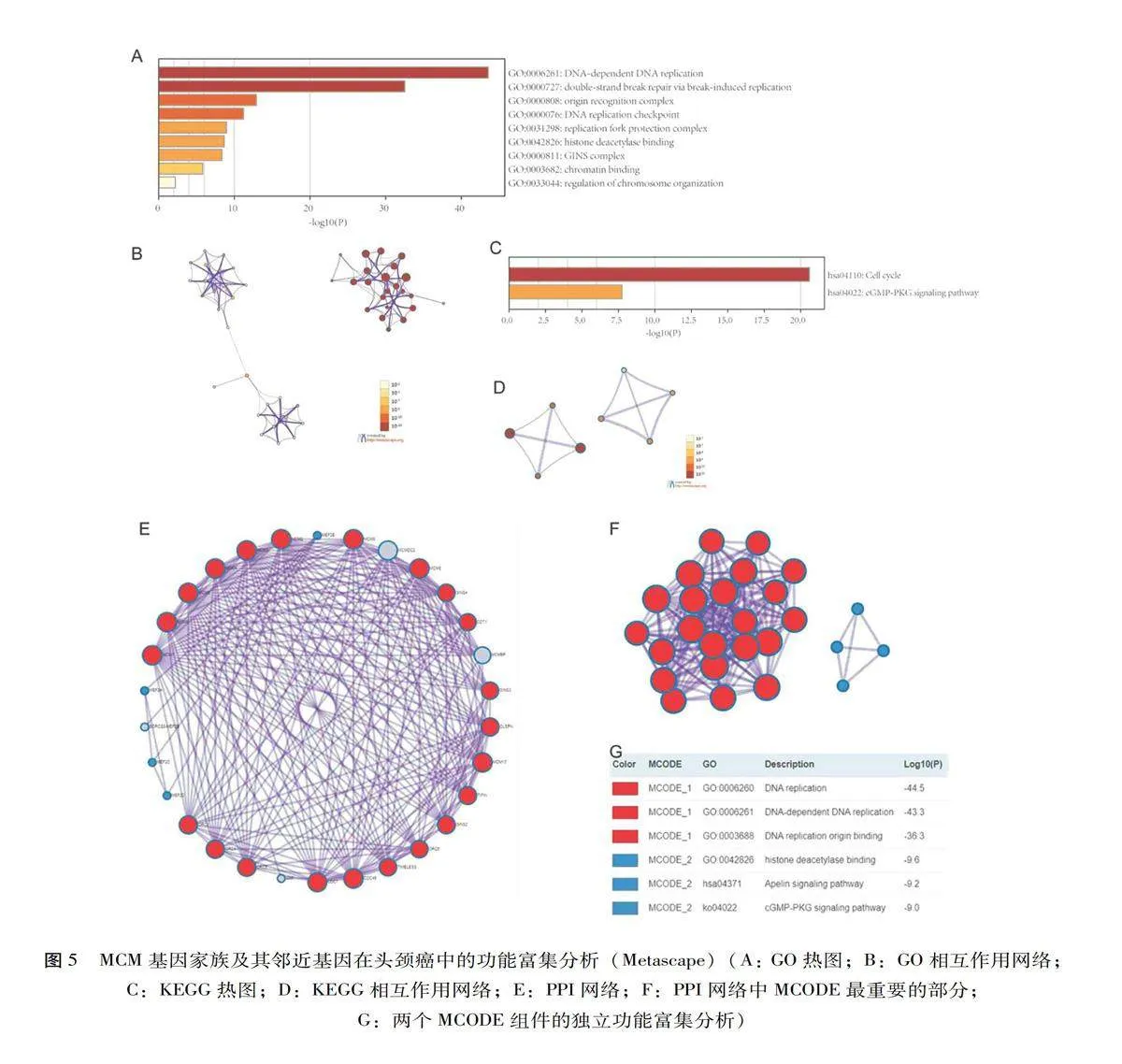
Metascape(http://metascape.org)是一种用于基因注释和功能富集分析的工具[13]。基于Metascape在线工具进行生物学过程、细胞成分和分子功能的基因富集分析(Gene ontology,GO),以及通路富集分析(Kyoto encylopaedia of genes and genomes,KEGG)。本研究利用Metascape在线工具得到GO和KEGG富集热图以及它们的作用网络,并应用分子复合体检测(Molecular complex detection,MCODE)的算法来识别连接紧密的网络成分。
1.4Kaplan-Meier Plotter
Kaplan-Meier plotter(http://kmplot.com/)是一个在线工具,可以在18 674个肿瘤样本中探索54 675个基因的预后作用[14]。在本研究中,根据基因表达水平的中位数将头颈癌患者分为高表达组和低表达组,并通过该工具评估了MCM家族成员与患者总生存期(Overall survival,OS)和无复发生存期(Relapse-free survival,RFS)的相关性。
1.5Timer
Timer(https://cistrome.shinyapps.io/timer/)是一款可靠、直观的软件,用于系统分析各种免疫细胞的浸润情况及其临床影响[15]。通过“基因模块”探讨MCM与头颈鳞癌HPV阳性患者(Head and neck squamous cell carcinoma-Human papilloma virus-positive,HNSCC-HPV-pos)中免疫细胞浸润的丰度之间的相关性。利用“生存模块”的生存曲线图分析了免疫细胞浸润和MCM表达的生存差异。
1.6统计学方法
基于Oncomine和GEPIA数据库,头颈癌组织和正常头颈组织之间的MCM基因家族的差异表达采用t检验进行比较分析;使用Kaplan-Meier生存分析法探究MCM基因家族的表达水平与头颈癌患者预后的关系,采用对数秩检验比较不同表达水平的生存差异从而得出P值;MCM基因家族分子之间的相关性分析采用Spearman等级相关性检验。在上述分析中,Plt;0.05为差异有统计学意义。
2结果
2.1MCM基因家族在头颈癌中的转录和翻译水平研究
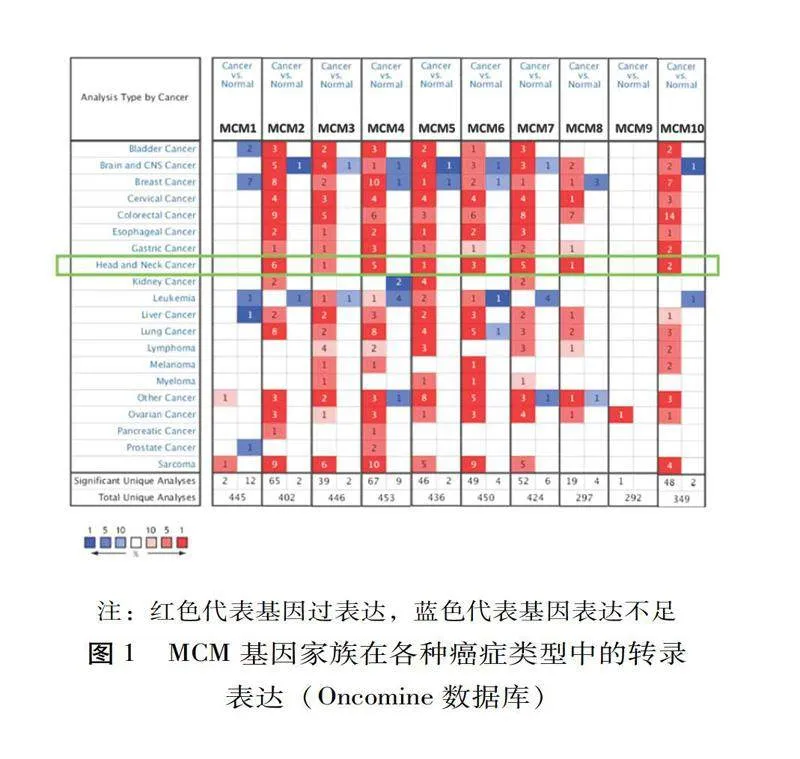
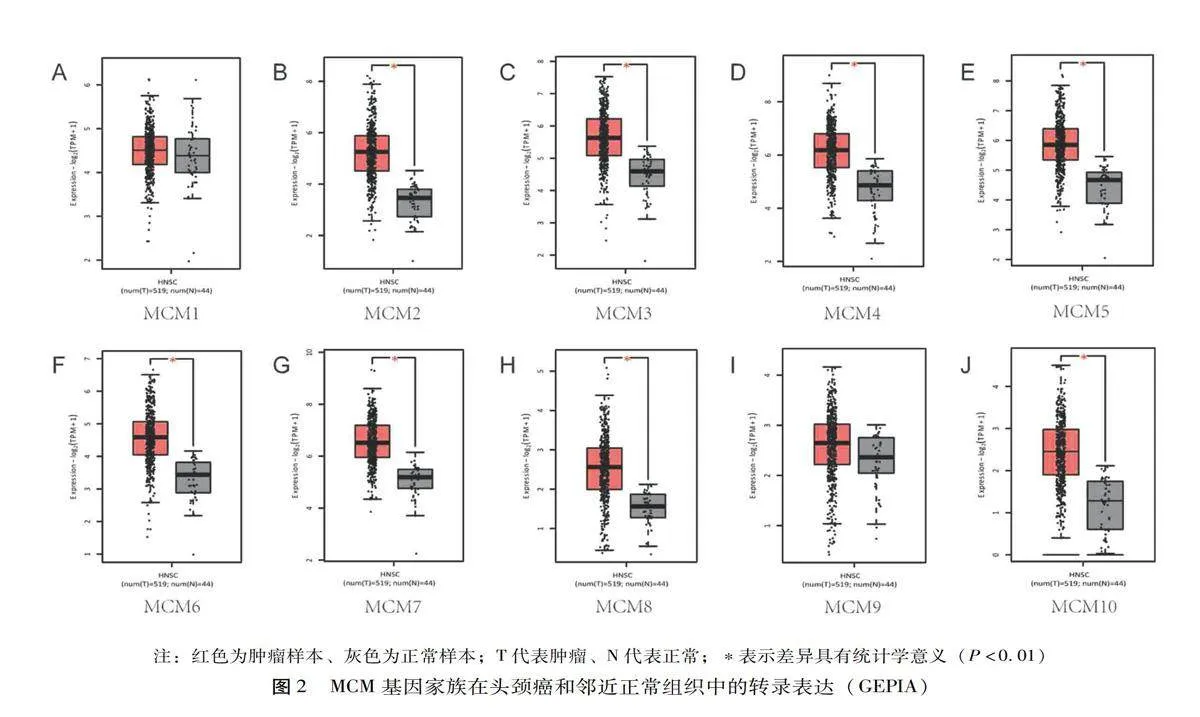
利用Oncomine数据库检测20种癌症中10种MCM的mRNA表达,并与正常组织进行比较,见图1。MCM2/3/4/5/6/7/8/10的mRNA在多组肿瘤组织中的表达明显增高。通过GEPIA比较了GTEx和TCGA数据集中头颈鳞癌(Head and neck squamous cell carcinoma,HNSCC)患者和正常样本MCM转录水平的差异。如图2所示,与正常样本相比,肿瘤组织样本中MCM2/3/4/5/6/7/8/10(图2 B-H,J)的mRNA表达显著上调,这一结果与Oncomine数据库的结果一致。
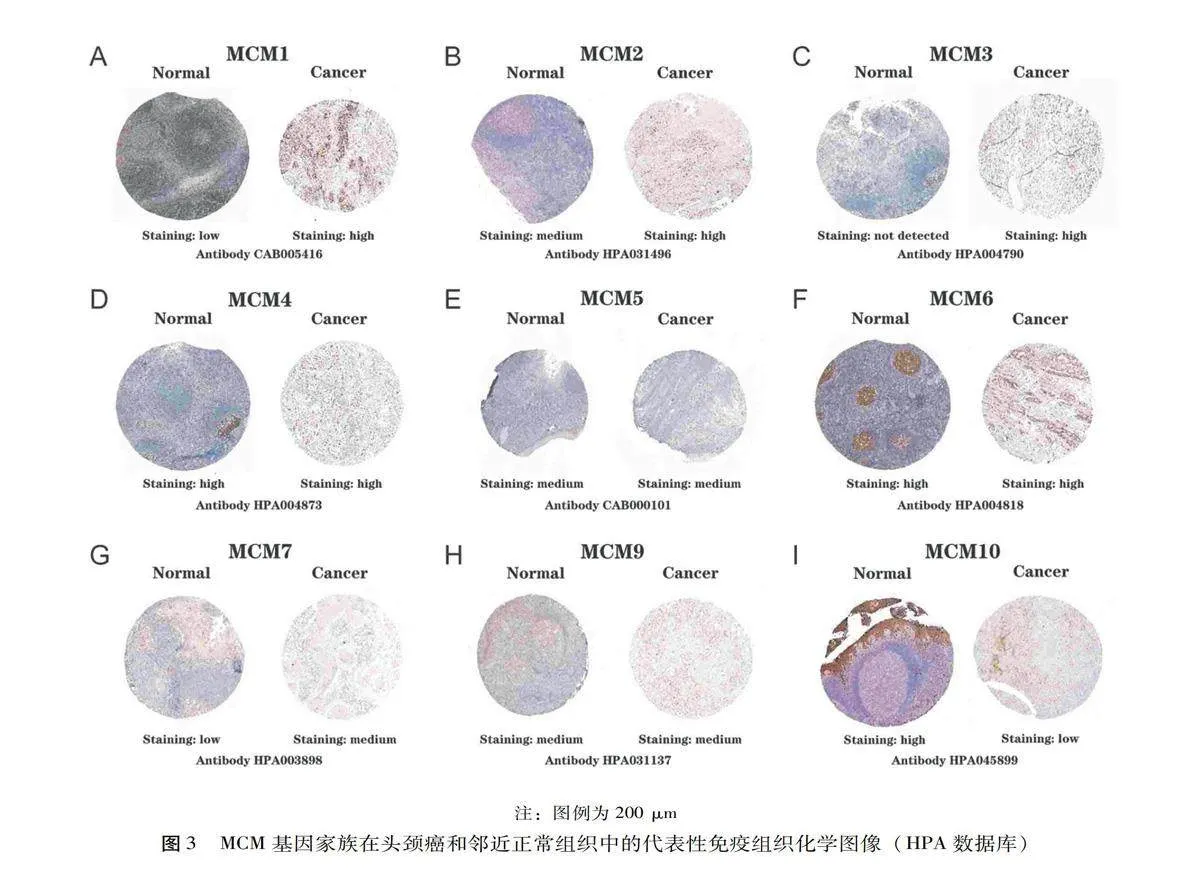
检测头颈癌中MCM的mRNA表达模式后,运用Human Protein Atlas数据库探索MCM在头颈癌中的蛋白表达。如图3所示,MCM1/7在正常组织中低表达,而在头颈癌组织中高表达。MCM2-MCM3蛋白在正常组织中低表达或不表达,而在肿瘤组织中高表达。该数据库丢失了MCM8数据。结果表明,在头颈癌患者中,MCM2/3/7在转录和翻译中都过度表达。
2.2MCM基因家族在头颈癌中的基因和蛋白水平的交互作用网络
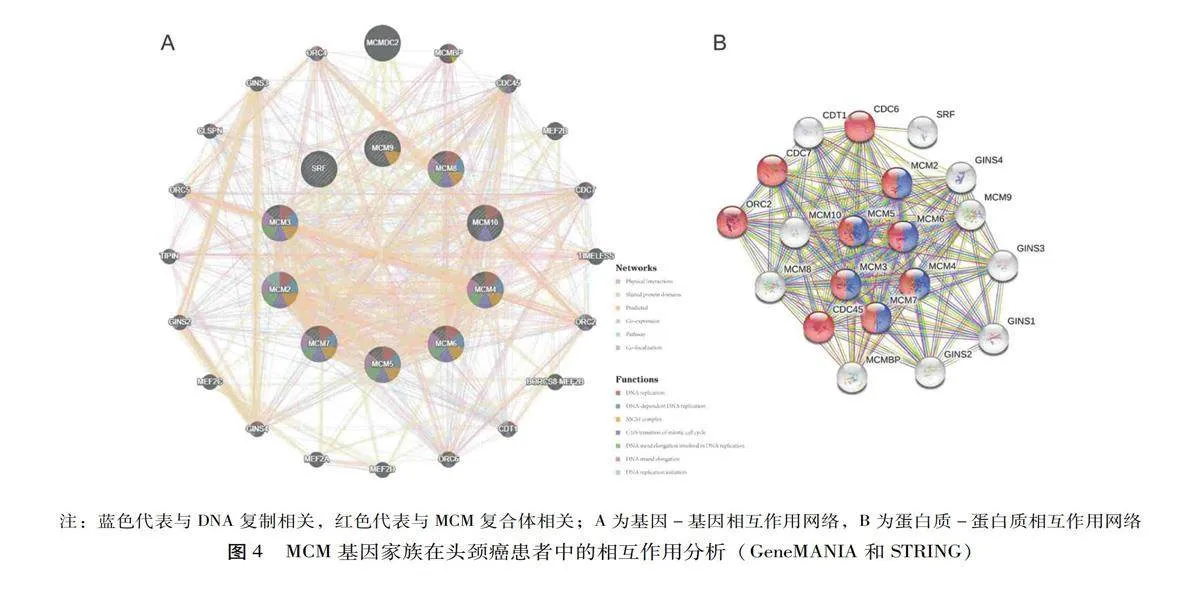
利用GeneMANIA对MCM1-MCM10及其邻近基因进行基因水平相关分析,这些基因涉及MCMDC2、ORC4、GINS3、CLSPN、ORC5、TIPIN、GINS2、MEF2C、GINS4、MEF2A、MEF2D、ORC6、CDT1、BORCS8-MEF2B、ORC2、TIMELESS、CDC7、MEF2B、CDC45和MCMBP,见图4A。结果揭示了物理相互作用、共享蛋白结构域、预测、共表达和相同通路之间的关系。
此外,对MCM基因家族与邻近基因在蛋白表达水平上的相互作用进行了STRING分析。其中MCM2-MCM7与DNA复制密切相关,并且发现CDC6、CDC7、CDC45和ORC2等细胞周期相关基因与MCM复合体密切相关,见图4B。
2.3MCM基因家族及其邻近基因的功能和通路富集分析
利用Metascape对MCM基因家族及其邻近基因进行了GO和KEGG分析,MCM家族成员及其邻近基因在DNA复制、断裂诱导的双链断裂修复、DNA复制检查点等生物学过程上表现出较强的丰富性,见表1、图5 A-B。MCM基因家族分子功能主要是DNA复制起始结合、组蛋白脱乙酰酶结合和染色质结合,DNA复制预起始复合体、MCM复合体、起源识别复合体等是细胞成分富集的主要部分。表1、图5 C-D显示了MCM家族成员及其邻近基因的前3条KEGG通路,包括细胞周期、DNA复制和cGMP-PKG信号通路。
在基因列表中确定的蛋白质-蛋白质相互作用网络和MCODE组分如图5 E~G所示,结果表明其生物学功能主要与DNA复制和cGMP-PKG信号通路有关。
2.4MCM基因家族在头颈癌中基因表达的预后价值
利用Kaplan-Meier plotter探究MCM mRNA表达在头颈癌中的预后价值。结果如图6所示,MCM1(HR=0.73,95%CI:0.56~0.95,P=0.020)、MCM3(HR=0.660,95%CI:0.49~0.89,P=0.006)、MCM5(HR=0.68,95%CI:0.50~0.93,P=0.015)、MCM6(HR=0.74,95%CI:0.55~1.00,P=0.047)和 MCM10(HR=0.64,95%CI:0.46~0.88,P=0.007)与较长的总生存期相关。然而,MCM mRNA的表达与头颈癌患者的无复发生存期无明显相关性。
2.5MCM基因家族在头颈癌中的免疫浸润
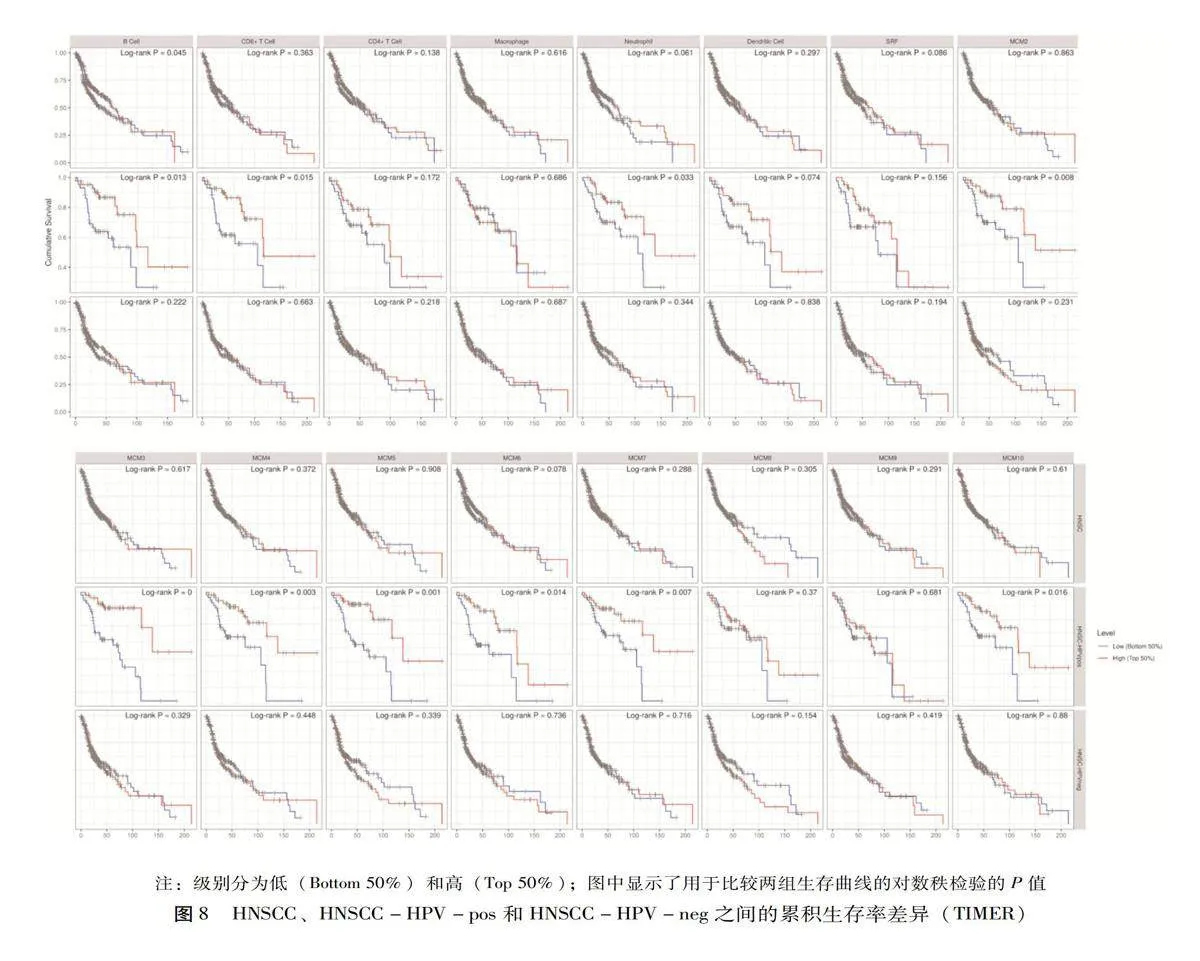
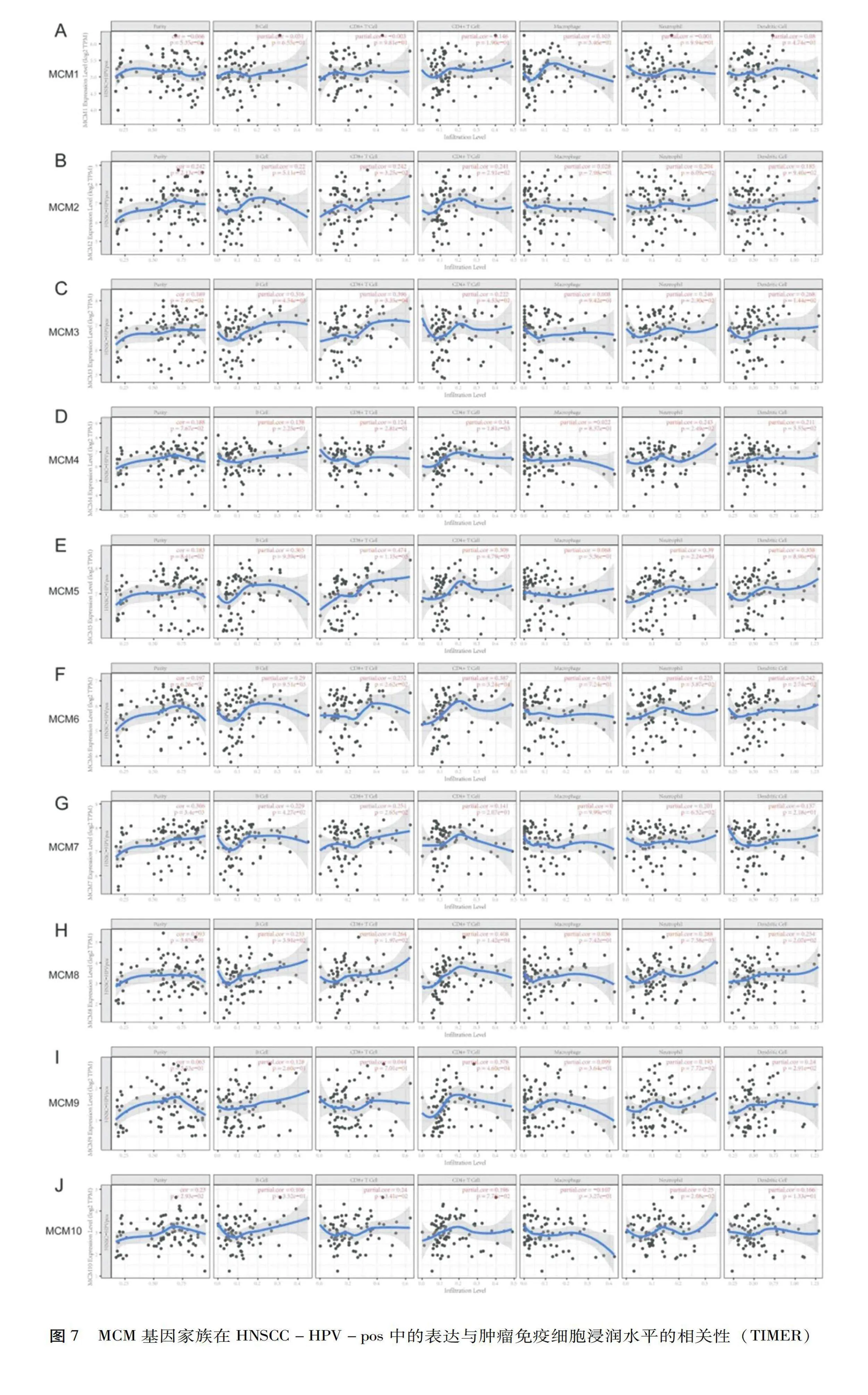
使用Timer在线工具分析了MCM表达与免疫浸润丰度的相关性、临床转归与免疫浸润丰度以及MCM表达的相关性。随着HPV感染状态的不同,MCM表达与免疫细胞浸润的相关性也会不同。如图7所示,除巨噬细胞外,HNSCC-HPV-pos中MCM的表达与免疫浸润的丰度显著相关,尤其是B细胞、CD8+T细胞和CD4+T细胞。
为了直观地显示生存差异,绘制了免疫浸润细胞和基因的生存曲线图,分析了HNSCC、HNSCC-HPV-pos和头颈鳞癌HPV阴性(Head and neck squamous cell carcinoma-Human papilloma virus-negative,HNSCC-HPV-neg)之间的累积存活率差异,见图8,结果发现HNSCC-HPV-pos亚组的B细胞、CD8+T细胞和中性粒细胞免疫浸润明显增加(Plt;0.05),这与MCM的表达水平有关。
3讨论
MCM1也称为血清反应因子(Serum response factor,SRF),参与细胞的生物学功能及多种疾病的发病机制,并参与辅因子募集到基因启动子的过程,在转录激活中发挥重要作用[16-17]。本文经数据研究发现,虽然MCM1在头颈癌与正常组织的mRNA表达量差异无统计学意义,在预后价值上差异也无统计学意义,但在蛋白层面上检测到MCM1在头颈癌组织中呈高表达,提示MCM1的蛋白表达水平可能对头颈癌的发展与预后具有一定的参考价值。
MCM2是MCM六聚物复合体的关键蛋白,在G1/S期的DNA复制阶段起着重要作用。Kaczkowski等报道了MCM2是一种泛癌生物标志物,这表明其作为多种肿瘤的生物标志物和治疗靶点的潜力[18]。MCM3已被发现与多种类型的人类癌变有关,其过表达使其成为检测肿瘤细胞的理想生物标志物[19]。MCM4中高度保守的序列对于形成稳定的MCM2-MCM7六聚体是必不可少的[20]。先前的研究表明,MCM5在头颈癌中的高表达与肿瘤分期和临床病理参数密切相关[21]。已经证实MCM6基因敲除延迟了S/G2细胞周期进程[22]。MCM7在DNA复制和细胞周期的起始过程中起关键作用,保证了DNA复制在1个周期中只发生1次[23]。本研究发现,与正常组相比,头颈癌患者的MCM2-MCM7显著高表达。在蛋白层面上,MCM2/3/7的高表达仍具有统计学意义,基因与蛋白层面同时高表达,暗示其在头颈癌发生发展中的关键作用。
到目前为止,关于MCM8、MCM9和MCM10在头颈癌中的研究很少。MCM8的沉默诱导了G0/G1期停滞和凋亡,阻止DNA复制过程,并抑制细胞增殖和自我更新[24]。MCM9参与DNA复制和同源重组,延长了复制应激后细胞周期的重新进入,这可能意味着MCM9具有抑制肿瘤的功能[25]。MCM10可以通过增强CDC45-MCM2-7-GINS复合物的组装来促进染色体复制,同时也是染色体不稳定的抑制剂[26]。本研究表明,MCM8和MCM10仅在头颈癌的mRNA水平过表达。生存分析表明,MCM1/3/5/6/10的高表达与头颈癌患者的良好预后相关,提示其可能成为头颈癌患者评估预后的标志物。
近年来,大量研究表明,肿瘤浸润性免疫细胞(Tumor infiltrating immune cells,TICs)的系统评价对临床预后的预测和免疫治疗的发展具有重要意义。本研究首次探讨了头颈癌中肿瘤浸润性免疫细胞与MCM的关系以及HNSCC、HNSCC-HPV-pos和HNSCC-HPV-neg之间累积生存期的差异。观察到MCM的表达水平与免疫细胞浸润密切相关,并且这些免疫细胞浸润对头颈鳞癌的预后具有显著影响,提示MCM的表达在头颈鳞癌免疫浸润中的重要作用,以及作为评估头颈鳞癌患者预后标志物的可能性,这将有助于临床进一步了解头颈癌患者的免疫状态,从而寻求有效的免疫治疗方法。
作为复制许可因子,MCM基因家族对DNA复制启动和细胞增殖至关重要[6,27],进而在肿瘤的发生发展中起着关键作用。MCM2-MCM7是由6种高度保守的蛋白质形成的一个与DNA结合的六聚物复合体,在DNA合成过程中促进细胞增殖和解旋酶活性[28]。据报道,MCM8和MCM9作为六聚体ATP酶/解旋酶复合物,可能参与同源重组修复,说明MCM8-MCM9复合体在解决复制应激问题中具有重要意义[29]。MCM10可以通过招募一些转录因子来促进细胞周期的持续复制[30]。本研究利用GeneMANIA和STRING构建了MCM和20个邻近基因的分子作用网络,对该分子互作网络进行功能和通路富集分析,结果表明这些基因主要富集在DNA复制和cGMP-PKG信号通路中,本研究进一步揭示了MCM基因家族及其相关信号通路的复杂性,为深入研究MCM基因家族影响头颈癌患者预后的内在机制以及开展针对MCM基因家族的靶向治疗方案提供了线索。
综上所述,该研究系统地分析了MCM基因家族在头颈癌中的表达水平、预后价值和免疫状态,为阐明头颈癌的发病机制提供了新的思路。但研究还存在一定的局限性:仅从在线数据库中获得相关数据,缺少体外或体内实验来验证数据分析结果。其次,对MCM基因家族潜在的机制没有进行更深入的探索。本研究后续将通过一系列实验,验证MCM基因家族在头颈癌中的表达,并结合临床信息验证其预后价值,寻找有效的免疫治疗方案。
参考文献
[1]Sung H,Ferlay J,Siegel RL,et al.Global cancer statistics 2020:GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J].CA Cancer J Clin,2021,71(3):209-249.
[2]Johnson DE,Burtness B,Leemans CR,et al.Head and neck squamous cell carcinoma[J].Nat Rev Dis Primers,2020,6(1):92.
[3]Cramer JD,Burtness B,Le QT,et al.The changing therapeutic landscape of head and neck cancer[J].Nat Rev Clin Oncol,2019,16(11):669-683.
[4]孙树洋,张志愿.基于肿瘤临床前模型建立头颈癌新型药物基因组学的必要性及展望[J].山东大学学报(医学版),2021,59(9):57-63.
[5]Mertz TM,Harcy V,Roberts SA.Risks at the DNA replication fork:effects upon carcinogenesis and tumor heterogeneity[J].Genes(Basel),2017,8(1).doi:10.3390/genes8010046
[6]Wang YF,Chen HR,Zhang JL,et al.MCM family in gastrointestinal cancer and other malignancies:From functional characterization to clinical implication[J].Biochim Biophys Acta Rev Cancer,2020,1874(2):188415.
[7]Cao T,Yi SJ,Wang LX,et al.Identification of the DNA replication regulator MCM complex expression and prognostic significance in hepatic carcinoma[J].Biomed Res Int,2020,2020:3574261.
[8]Rhodes DR,Yu J,Shanker K,et al.ONCOMINE:a cancer microarray database and integrated data-mining platform[J].Neoplasia,2004,6(1):1-6.
[9]Tang ZF,Li CW,Kang BX,et al.GEPIA:a web server for cancer and normal gene expression profiling and interactive analyses[J].Nucleic Acids Res,2017,45(W1):W98-W102.
[10]Uhlen M,Oksvold P,Fagerberg L,et al.Towards a knowledge-based human protein atlas[J].Nat Biotechnol,2010,28(12):1248-1250.
[11]Warde-Farley D,Donaldson SL,Comes O,et al.The GeneMANIA prediction server:biological network integration for gene prioritization and predicting gene function[J].Nucleic Acids Res,2010,38(Web Server issue):W214-220.
[12]Szklarczyk D,Gable AL,Lyon D,et al.STRING v11:protein-protein association networks with increased coverage,supporting functional discovery in genome-wide experimental datasets[J].Nucleic Acids Res,2019,47(D1):D607-D613.
[13]Zhou YY,Zhou B,Pache L,et al.Metascape provides a biologist-oriented resource for the analysis of systems-level datasets[J].Nat Commun,2019,10(1):1523.
[14]Nagy ,Munkácsy G,Gyrffy B.Pancancer survival analysis of cancer hallmark genes[J].Sci Rep,2021,11(1):6047.
[15]Li TW,Fan JY,Wang BB,et al.TIMER:a web server for comprehensive analysis of tumor-infiltrating immune cells[J].Cancer Res,2017,77(21):e108-e110.
[16]Onuh JO,Qiu H.Serum response factor-cofactor interactions and their implications in disease[J].FEBS J,2021,288(10):3120-3134.
[17]Azam H,Pierro L,Reina M,et al.Emerging role for the Serum Response Factor(SRF) as a potential therapeutic target in cancer[J].Expert Opin Ther Targets,2022,26(2):155-169.
[18]Kaczkowski B,Tanaka Y,Kawaji H,et al.Transcriptome Analysis of Recurrently Deregulated Genes across Multiple Cancers Identifies New Pan-Cancer Biomarkers[J].Cancer Res,2016,76(2):216-226.
[19]Lkkegaard S,Elias D,Alves CL,et al.MCM3 upregulation confers endocrine resistance in breast cancer and is a predictive marker of diminished tamoxifen benefit[J].NPJ Breast Cancer,2021,7(1):2.
[20]魏敏,王丽娜,白雪飞,等.MCM4基因在子宫内膜癌中表达和临床意义的生物信息学分析[J].中国免疫学杂志,2022,38(11):1362-1365,1372.
[21]Gong B,Ma M,Yang X,et al.MCM5 promotes tumour proliferation and correlates with the progression and prognosis of renal cell carcinoma[J].Int Urol Nephrol,2019,51(9):1517-1526.
[22]Liu ZK,Li J,Chen J,et al.MCM family in HCC:MCM6 indicates adverse tumor features and poor outcomes and promotes S/G2 cell cycle progression[J].BMC Cancer,2018,18(1):200.
[23]Chuang TD,Luo X,Panda H,et al.miR-93/106b and their host gene,MCM7,are differentially expressed in leiomyomas and functionally target F3 and IL-8[J].Mol Endocrinol,2012,26(6):1028-1042.
[24]Wang XL,Zhang L,Song YF,et al.MCM8 is regulated by EGFR signaling and promotes the growth of glioma stem cells through its interaction with DNA-replication-initiating factors[J].Oncogene,2021,40(27):4615-4624.
[25]Guo T,Zheng Y,Li G Y,et al.Novel pathogenic mutations in minichromosome maintenance complex component 9(MCM9) responsible for premature ovarian insufficiency[J].Fertil Steril,2020,113(4):845-852.
[26]Paulson CN,John K,Baxley RM,et al.The anti-parasitic agent suramin and several of its analogues are inhibitors of the DNA binding protein Mcm10[J].Open Biol,2019,9(8):190117.
[27]Yu S,Wang GQ,Shi Y,et al.MCMs in cancer:prognostic potential and mechanisms[J].Anal Cell Pathol(Amst),2020,2020:3750294.
[28]Ishimi Y.Regulation of MCM2-7 function[J].Genes Genet Syst,2018,93(4):125-133.
[29]Li J,Yu DQ,Liu L,et al.Structural study of the N-terminal domain of human MCM8/9 complex[J].Structure,2021,29(10):1171-1181(e4).
[30]Murayama T,Takeuchi Y,Yamawaki K,et al.MCM10 compensates for Myc-induced DNA replication stress in breast cancer stem-like cells[J].Cancer Sci,2021,112(3):1209-1224.
(2022-12-29收稿)

