Qualitative and quantitative detection of aldehydes in DNA with 2-amino benzamidoxime derivative
N Li,Qin Zhou,Kun Li,Ting Jing,Xio-Qi Yu,b,*
a Laboratory of Green Chemistry and Technology (Ministry of Education),College of Chemistry,Sichuan University,Chengdu 610064,China
b Department of Chemistry,Xihua University,Chengdu 610039,China
Keywords:Aldehydes in DNA 2-Amino benzamidoxime derivative Intramolecular ring closure Fluorescent adducts Quantitative analysis
ABSTRACT An aldehyde-reactive probe based on 2-amino benzamidoxime (ABAO) framework was introduced,which can selectively label aldehydes in DNA through intramolecular ring closure under mild aqueous solutions.We screened ABAO derivatives that can undergo a cyclization with the formylated nucleobases to generate a fluorescence nucleoside,and of these derivatives 5–methoxy-ABAO (PMA) emerged as the optimal choice.PMA can sensitively and selectively react with 5fU,5fC and AP to form fluorogenic dihydroquinazoline derivatives,which also can quantify DNA damages induced by γ-irradiation.PMA-initiated labeling strategy provides great convenience for qualitative and quantitative detection of aldehydes in DNA.
Natural nucleobase modifications in genomic DNA have a significant and profound influence on the field of epigenetics [1],and aldehydes are highly important groups of these modifications due to their chemical reactivity [2].The formylated nucleobases include 5-formyluracil (5fU) [3],5-formylcytosine (5fC) [4] and abasic sites (AP) [5].5fU and 5fC are natural modified nucleobases produced by the oxidation of thymine and cytosine,respectively.Also,Fenton-type reagents,reactive oxygen species (ROS)[5] has been revealed to yield mutagenic 5fU in genomic DNA.5fC can be obtained by oxidation of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine(5hmC) [6],which can be deformylated to cytosine (C) and removed its amino group by AID/APOBEC enzyme to obtain 5fU [7].In addition,it can also remove pyrimidine bases through the action of thymine-DNA-glycosylase (TDG) to produce AP,and AP undergoes a base excision repair (BER) process to cytosine.In a sense,AP can be considered as a ‘nucleoside modification’.It is the most common lesion in DNA,which originates from spontaneous hydrolysis of theN-glycosidic bond and the shedding of base [8–10].5fC and AP are transient intermediates in the active DNA demethylation of 5mC by ten-eleven translocation (TET)family enzymes,with mounting evidence to suggest that they play a vital role in epigenetic functions [9,11].Moreover,5fU can be also removed from the genome by the BER glycosylase SMUG1 to generate AP,and then become unsubstituted thymine (T) again.In summary,there is a certain connection between aldehydes in DNA,which may be related to the epigenetic modification of organisms.Nucleic acid epigenetics mediates a variety of biological processes,including embryonic development,gene regulation [12],genome imprinting [13],cell differentiation [14] and X chromosome inactivation [15].Therefore,research on qualitative and quantitative detection of such modifications is essential to understand the epigenetic functions of the formylated bases.
Chemistry-based innovative detection techniques have greatly promoted the research of this field [16,17].Among them,reactionbased fluorescent probes are considered as efficient methods to study aldehyde groups in DNA because of high sensitivity,good selectivity,intuitive phenomenon and simple operation.To date,focus has been on the reactive aldehyde group,such as 2-amino(thio)phenol [18,19],hydroxylamine [20–23],hydrazine[24,25],1,2-phenylenediamine [26,27],hydrazide [28],amine [29],benzothiazole [30],2,3,3-trimethylindole [31],-CH2- reagents [32–34] and Wittig derivatives [35–37].They can selectively and effectively label one of the modified bases of 5fC,5fU or AP,mainly to study the role of a certain modified base in epigenetics,while ignoring other aldehyde modified bases or the conditions for selective labeling are more stringent.The biological functions produced by the aldehydes present in DNA have many similar characteristics.They are all regulators of gene expression and highly involved in the process of DNA methylation and demethylation [38–40].Therefore,introducing a simple and efficient compound which can calculate the whole contents of the formylated bases is useful and meaningful.In 2018,Zhouet al.reported a naphthalimide hydroxylamine probe that can selectively label all natural aldehydes in DNA instead of hydroxymethyl and methyl [41].They discussed and compared the reactivity of hydroxylamine and amine groups toward aldehydes in DNA,whereas multistep synthesis is a problem in this method.
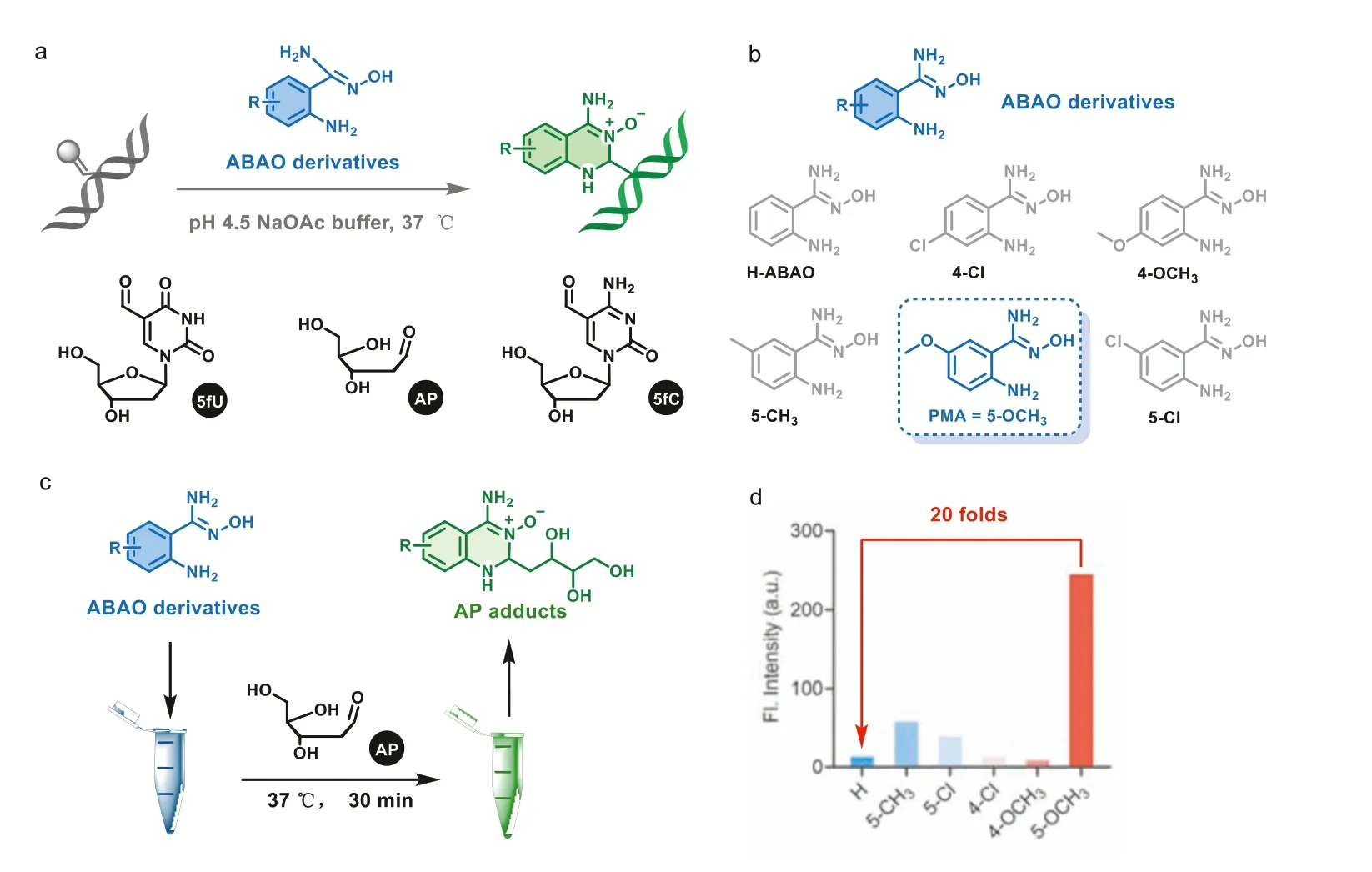
Fig.1.(a) The reaction scheme of ABAO derivatives fluorescent labeling 5fU,5fC and AP.(b) Chemical structures of ABAO derivatives.(c) Flowchart of fluorescent screening ABAO derivatives (Blue tube represents ABAO derivatives in NaOAc buffer (100 mmol/L,pH 4.50),green tube represents AP after incubation with ABAO derivatives for 30 min at 37 °C).(d) Fluorescence emission intensity (λem=535 nm) of AP adducts (X-axis is the species of ABAO substituents,the test concentration of AP and ABAO derivatives are 20 μmol/L).
In 2014,Kitovet al.studied the rapid cyclization reaction of 2-amino benzamidoxime (ABAO) derivatives with aldehyde in water to form dihydroquinazoline products to detect aldehyde-containing proteins and study protein functionalization [42].para–Methoxy ABAO (PMA) in the tested analogues exhibited a high absorption coefficient and fluorescent properties.In 2019,Ressmannet al.further studied this reaction and adapted it to a universal method for detecting various chemically different aldehydes produced from enzyme-mediated whole cell transformation [43].Given the excellent properties of the reaction in protein functionalization of aldehyde groups,we will further challenge its application in nucleic acid labeling.In this work,we first try to react ABAO with aldehydes in DNA to explore the feasibility between them.Next,we synthesized a series of ABAO derivatives and selected the probe with the best signal for subsequent experimental researches (Fig.1a).The direct condensation of aldehydes and PMA is very effective and highly selective.Other nucleobase modifications that do not involve an aldehyde group cannot proceed through intramolecular cyclization with PMA.This method allowed for labeling the whole aldehyde in DNA based on fluorescence and it can be quantified.
In order to explore the reactivity of ABAO derivatives with the formylated nucleobases,the commercially available ABAO firstly tried to react with 5-formyl-2′-deoxyuridine (5fU),5-formyl-2′-deoxycytidine (5fC) and 2-deoxy-D-ribose (AP).All three of them can generate corresponding adduct in acetonitrile and pH 4.5 sodium acetate buffer at room temperature.Next,we synthesized a series of ABAO derivatives (5-Cl/5-CH3/5–OCH3/4-Cl/4–OCH3) to select the fluorescent probe with the best response to aldehydes in DNA (Fig.1b).All of them were readily synthesized in one step (Scheme S1 in Supporting information).Next,2-deoxy-D-ribose was selected as the substrate for preliminary model reactions due to rapid response (Fig.1c).According to fluorescence spectra of generated adducts (Fig.1d),fluorescence of ABAO (H)and 4-position substituted derivatives were weak,however,fluorescence intensity of 5-position substituted derivatives significantly increased.Especially,5–OCH3(PMA) had a nearly 20-fold enhancement at the peak of fluorescence emission at 535 nm,which might be due to electronic effect on the probe.In 2014,Ratmiret al.first reported that ABAO can be used for the modification of aldehyde-terminated proteins,and revealed the change inπ-conjugation upon reaction providing opportunities for designing fluorogenic ABAO derivatives [42],which was suggested to be aldehydes in DNA reason why fluorescence intensity of PMA was higher than other synthetic derivatives.As a result,we identified that PMA may be a candidate for aldehydes in DNA under certain conditions.
From the synthesis part,we already know that PMA was used to derivatize the formylated nucleobases to form the corresponding adducts in acetonitrile and sodium acetate buffer at room temperature.However,the reaction time was too long and the conversion rate was not ideal.Therefore,we try to improve derivative efficiency of PMA through more systematic condition selection.Specifically,we took 5fU (20 mmol/L,100 μL) as substrate to optimize derivatization conditions,including reaction solvent,reaction temperature,reaction time and the amount of PMA.For the choice of reaction solvent,the most ideal is pure water or nearly neutral aqueous solution.But it turns out that pH 4.50 NaOAc buffer was a necessary condition for the reaction (Fig.S1a in Supporting information).Notably,the temperature range was between 25°C and 60°C or the reaction time was reduced to 0.5 h or the equivalent ratio reaction can achieve nearly 99% conversion (Figs.S1b-d in Supporting information).It is worth noting that 5fC exists intramolecular hydrogen bond formed between the aldehyde oxygen atom and the amino group,5fC is less reactive than 5fU and AP[44].Taking into account the inertness of 5fC,it requires longer reaction time under optimized conditions.It is delightful that we can make appropriate adjustments according to the actual sample situation because of wide selection range of reaction temperature,time and the amount of PMA.This catalyst-free labeling reaction is efficient and environmentally friendly,and will have great potential in the formylate nucleobases analysis.
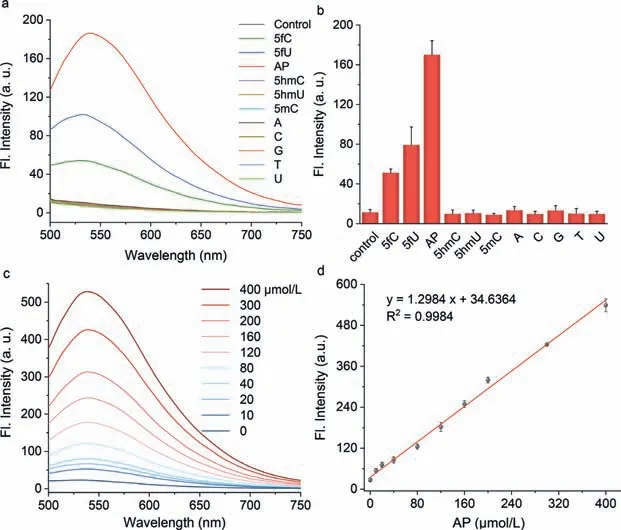
Fig.2.Selectivity and fluorescence titration of PMA to aldehydes in DNA.(a) Fluorescence spectra and (b) emission intensity of PMA after incubation with aldehydes in DNA and other nucleobases in NaOAc buffer (100 mmol/L,pH 4.50) for 24 h at 37 °C,including A,T,C,G,U,5mC,5hmC,5hmU (10 μmol/L).PMA without any nucleoside serving as the control.(c) Fluorescence spectrum changes after PMA (20 μmol/L) reaction with different concentrations of AP (0–400 μmol/L) in NaOAc buffer (100 mmol/L,pH 4.50)for 15 min at 37 °C and (d) liner relationship of emission intensity toward AP (λex/λem=405/535 nm).
We subsequently evaluated the feasibility of PMA in fluorescence detection of aldehyde groups.we first measured the selectivity of PMA for aldehyde groups in order to eliminate the interference of other base modifications.As shown in Figs.2a and b,both five common nucleobases and their natural modifications (5mC,5hmC,5hmU) observed no obvious spectral changes for PMA,and only aldehyde-modified nucleosides (5fU,5fC,and AP) induced a new fluorescence emission at 535 nm (λex=405 nm) and were accompanied by a yellow visible change in color (Fig.S2 in Supporting information),indicating the specific recognition of PMA toward 5fU,5fC and AP.This result again proved that PMA can selectively condense with 5fU,5fC,and AP bearing a reactive aldehyde group to yield fluorescent adductsviaintermolecular cyclization,whereas other nucleobases without this group do not affect the analysis of the formylated bases at all.Subsequently,fluorescence titration experiments were performed to investigate quantification of PMA for 5fU,5fC and AP.As depicted in Fig.2c,with the increase of aldehydes,fluorescence emission at 535 nm gradually increased and fluorescence intensity was linearly positive correlation with the AP concentration range of 0–400 μmol/L (Fig.2d).The detection limits of PMA to single base (5fU,5fC and AP) are 1.05,1.22 and 1.75 μmol/L,respectively (Fig.S3 in Supporting information),which suggest PMA is promising for qualitative and quantitative detection the formylated modifications under mild conditions.Besides,reaction progress of PMA with 5fU,5fC and AP was monitoredviafluorescence kinetics (Fig.S4 in Supporting information).
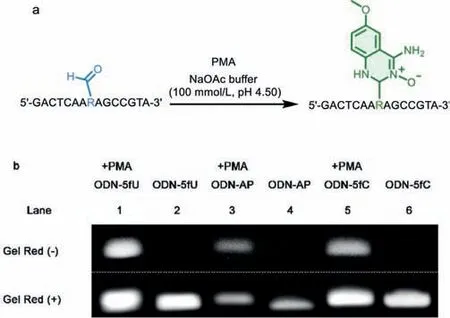
Fig.3.(a) Reaction scheme of ODN-5fU/5fC/AP with PMA to give adducts (R-CHO:ODN-5fU/5fC/AP),conditions: NaOAc buffer (100 mmol/L,pH 4.50),37 °C,24 h.(b)Denaturing PAGE analysis of ODN-5fU (lane 1),ODN-AP (lane 3),ODN-5fC (lane 5)after incubation with PMA.ODN-5fU (lane 2),ODN-AP (lane 4),ODN-5fC (lane 6)without PMA serving as the control.The image before and after SuperRed staining is above and below the dashed line,respectively.
Our original intention of designing the probe is to detect aldehydes group in DNA.Therefore,we further tested the reactivity of PMA to label aldehyde-modified DNA.Three 15-mer oligodeoxynucleotides containing one 5fU (ODN-5fU),5fC (ODN-5fC),AP (ODNAP) were incubated with PMA in NaOAc buffer,respectively(Fig.3a).The whole was separated by electrophoresis without further purification after the reaction.Only experimental groups with probe were observed the bands and migrated slowly due to the increase of molecular weight,while no signal was found in the blank lanes (Fig.3b).Given the high reactivity of PMA to aldehyde groups,we proceed to study its specificity.ODN-C,ODN-T (C,T is in the position of 5fU,respectively) showed negative results,which contained no aldehyde group cannot be identified by probe (Fig.S5 in Supporting information).All lanes were obtained after staining with 3×SuperRed to ensure the complete existence of the control DNAs.The PAGE test proved that PMA had higher reactivity and selectivity in aldehyde than methyl,hydroxymethyl modified DNAs again.
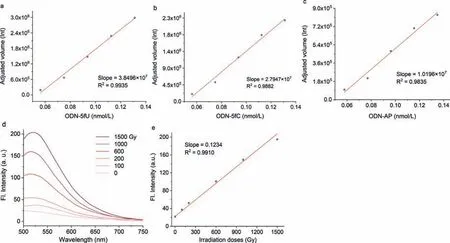
Fig.4.Correlation between various concentration of ODN-5fU (a),ODN-5fC (b),or ODN-AP (c) and adjusted volume after incubation with PMA in NaOAc buffer (100 mmol/L,pH 4.50) for 24 h at 37 °C.(d) Fluorescence spectra of γ-irradiated calf thymus DNA (50 μL) after incubation with PMA (λex=405 nm,the total volume was 71 μL).(e)Quantification of aldehyde mutations in γ-irradiated ctDNA (the test volume was 5 μL) at different irradiation doses (0–1500 Gy).
To confirm whether PMA can directly quantity ODNs.PMA was incubated with different concentrations of ODNs (including one 5fU/5fC/AP,respectively)viaPAGE analysis.The final products were imaged with BIO-RAD Gel DocTMXR+ (Fig.S6 in Supporting information).Between volume of bands and concentration of ODNs exhibited an apparent linear correlation and suggested that PMA can chemoselective quantification of aldehydes in DNAs and was sensitive to the nmol/L level (Figs.4a-c).Subsequently,in order to prove whether our probe can be used for detection of complex biological samples,we preparedγ-irradiated calf thymus DNA (ctDNA) with various doses (0–1500 Gy).γ-Irradiation was reported to cause more DNA damages,like 5mC to 5fC,5hmU to 5fU [45].After incubation with PMA,a satisfactory linear fit equation (R2=0.9910)was obtained through direct fluorescence readouts to estimate the overall aldehydes abundance (Figs.4d and e).Taken together,the above findings verified that PMA,5–methoxy-ABAO,is also reliable and practical for the analysis of aldehydes in DNA.
In summary,we successfully introduced ABAO derivatives in nucleic acid labeling.5-Methoxy-ABAO (PMA),the amino benzamidoxime framework,combines high reactivity,specific selectivity,long-term stability and intriguing changes in fluorescence upon reaction.PMA can be used to quantify 5fU,5fC and AP within the low μmol/L range applying fluorescence detection and quantify aldehyde in ODNs down to nmol/L concentrations by PAGE analysis.Moreover,our method does not need tedious synthesis of reference probe and can estimate the overall aldehydes abundance on complex biological samples such asγ-irradiated calf thymus DNA.These features broaden the application of nucleophile PMA and provide a potential candidate for marking aldehyde instead of methyl,hydroxymethyl-modified DNAs in epigenome research.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported by National Natural Science Foundation of China (Nos.22077088 and 21877082) and Foundation from Science and Technology Department of Sichuan Province(Nos.2020JDJQ0017,2021YFH0132).
Supplementary materials
Supplementary material associated with this article can be found,in the online version,at doi:10.1016/j.cclet.2022.04.069.
 Chinese Chemical Letters2023年1期
Chinese Chemical Letters2023年1期
- Chinese Chemical Letters的其它文章
- Diabetic wound healing activated by supramolecular cascade reaction
- MBenes: Two-dimensional transition-metal borides with ordered metal vacancies
- Wet-adhesive materials of oral and maxillofacial region: From design to application
- Diverse catalytic systems for nitrogen-heterocycle formation from O-acyl ketoximes
- Fluorine-containing drugs approved by the FDA in 2021
- The development and application of dual-comb spectroscopy in analytical chemistry
