Comparative multi-locus assessment of modern Asian newts (Cynops, Paramesotriton, and Pachytriton:Salamandridae) in southern China suggests a shared biogeographic history
Zhi-Yong Yuan, Yun-Ke Wu, Fang Yan, Robert W. Murphy, Theodore J. Papenfuss, David B. Wake,Ya-Ping Zhang, Jing Che
1 State Key Laboratory of Genetic Resources and Evolution & Yunnan Key Laboratory of Biodiversity and Ecological Security of Gaoligong Mountain, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China
2 Key Laboratory of Freshwater Fish Reproduction and Development (Ministry of Education), School of Life Sciences, Southwest University, Chongqing 400715, China
3 Department of Ecology and Evolutionary Biology, Cornell University, Ithaca 14850, USA
4 College of Life Science, Yunnan University, Kunming, Yunnan 650091, China
5 Centre for Biodiversity and Conservation Biology, Royal Ontario Museum, Toronto, Ontario M5S 2C6, Canada
6 Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720, USA
ABSTRACT Evolutionary biologists are always interested in deciphering the geographic context of diversification,therefore they introduced the concept of comparative phylogeography, which helps to identify common mechanisms that contribute to shared genetic structures among organisms from the same region.Here, we used multi-locus genetic data along with environmental data to investigate shared phylogeographic patterns among three Asianendemic newt genera, Cynops, Paramesotriton and Pachytriton, which occurred in montane/submontane streams or ponds in southern China. Our 222 samples from 78 localities covered the entire range of the three genera and represented the largest dataset of this group to date. We reconstructed matrilineal genealogies from two protein-coding,mitochondrial genes, and gene network from two nuclear genes. We also estimated divergence times of major cladogenetic events and used occurrence data to evaluate niche difference and similarity between lineages. Our results revealed a common basal split in all three genera that corresponds to the separation of two geographic terrains of southern China. Those ancient divergence occurred during middle to late Miocene and likely correlate with paleoclimatic fluctuations caused by the uplift of the Qinghai-Xizang (Tibet) Plateau (QTP). Particularly,the strengthening and weakening of Asian summer monsoons during the Miocene may have profoundly impacted southern China and led to repeatedly vicariance in those newts. However, despite differences in realized niches between lineages,there is no evidence for divergence of fundamental niches. Preservation of old newt matriline lineages in mountains of southern China suggests that the region acts as both museums and cradles of speciation. Based on those results, we advocate a multi-pronged protection strategy for newts in the three genera.
Keywords: Amphibian; Comparative phylogeography; Qinghai-Xizang (Tibet) Plateau;East Asian monsoons; Ecological niche modeling;Species museums and cradles
lNTRODUCTlON
Comparison of biogeographic patterns of co-distributed organisms may offer insights into a shared evolutionary history(Bermingham & Moritz, 1998; Lapointe & Rissler, 2005). This phylogeographic approach may reveal how various species responded to the same palaeogeographical and palaeoclimatic changes across a shared landscape (Bell et al.,2010; Castoe et al., 2009; Myers et al., 2019). For example,range contraction/expansion and dispersal from/into refugia during the Quaternary glaciation episodes often resulted in deep genealogical lineages among continental taxa from the northern hemisphere (Riddle, 2016). Moreover, marine organisms distributed in the vast Indo-Polynesian tropical seas shows little genetic structure across this broad region due to the continuity of shallow habitats (Bowen et al., 2016).
Southern China is characterized by a mosaic of mountains with a highly rugged and dissected topology. Elevation decreases from the “Roof of the World”, the Qinghai-Xizang(Tibet) Plateau (QTP) in the west, all the way to nearly zero at eastern coastal areas. Such dramatic change in elevations divides southern China into three stepwise geographical terrains from west to east (Figure 1) (Jiang & Yang, 2009;Zhao et al., 1995). Terrain I corresponds to the mighty QTP with an average elevation over 4 000 m a.s.l.; Terrain II is a transitional belt east to Terrain III with an average of elevation around 2 000 m a.s.l.; and Terrain III, which has the lowest elevation (generally less than 1 000 m a.s.l.), encompasses the eastern plains such as the Twain-Hu Plain and those relatively low-elevation rolling hills (Hu et al., 2010; Figure 1).There are also a few prominent mountain ranges in Terrain III,including the Nanling Mountains and Wuyi Mountains that can reach 2 000 m a.s.l.

Figure 1 Topographic map of southern China, showing the locations of major mountain ranges, plateau, and plains
The complex landscape of southern China can lead toin situdiversification of taxa due to habitat discontinuity, which separated once-connected populations that ultimately became independently evolving lineages. The mountains of southern China not only act as museums that sheltered biota at times when environmental conditions were harsh but also as cradles for speciation (López-Pujol et al., 2011a, 2011b; Li et al.2021a; Rahbek et al., 2019a, 2019b). For example, the lowlands of the Twain-Hu Plain may constitute a major geographic barrier for montane species to disperse between Terrain I and II, while the Nanling Mountains could act as a dispersal corridor for those montane organisms. The impact of geological complexity on local taxa is further exacerbated by distinctive climatic factors in southern China. Particularly, the unique East Asian monsoon was developed with warm, rainy summers and cold, dry winters after the Late Oligocene, in response to orogenesis of the Himalayas and the QTP(Farnsworth et al., 2019; Harris, 2006; Sun & Wang, 2005).Monsoon systems often strongly associate with turnover of components of terrestrial ecosystems (Chen et al., 2018; Guo et al., 2008; Li et al., 2021b; Spicer, 2017). From the beginning of the Neogene, the East Asian summer monsoon further intensified but also underwent several unusually dry periods (Clift et al., 2008; Li et al., 2021b; Sun & Wang, 2005).Such climatic fluctuations would play an important role in shaping the distribution of organisms and left hallmarks in their genetics (Chen et al., 2018, 2020). One example involves aquatic montane newts that likely underwent range expansion followed by lineage diversification with the aid of the heavy summer rains (Wu et al., 2013).
Amphibians are ideal for investigating organismal responses to historical environmental changes, because precipitation and temperature govern their life history and species richness(Buckley & Jetz, 2007; Vieites et al., 2009). Their relatively low vagility, high philopatry, and low physiological tolerance to higher temperatures and dry environmental conditions suggest that they are often subject to allopatric speciation (Zeisset &Beebee, 2008). In East Asia there is a unique group of salamanders (Cynops,Paramesotriton,Pachytriton, andLaotriton) often referred as the modern Asian newts (Zhang et al., 2008), most of which are cold-adapted, montane species inhabiting streams or ponds at elevations between 500-2 400 m a.s.l. (Fei et al., 2006). Interestingly, 29 out of the extant 34 species are endemic to southern China, suggesting this region as a diversity center of this group. Fossil record and molecular dating indicate that these newts have a long evolutionary history extending back to early Miocene to Oligocene (Zhang et al., 2008). The combination of physiological features and old age makes this group an ideal system to investigate external environmental factors that have shaped their biogeographic patterns.
Here we use mitochondrial DNA (mtDNA) and nuclear DNA(nDNA) sequence data along with environmental data to investigate whether a shared biogeographic pattern can be recognized among the three genera of modern Asian newts and to identify likely external drivers for such pattern if exists.This is the first large-scale, comparative study that include most species from mainlandCynops,ParamesotritonandPachytriton, whereas previous work focused mainly on description of new species (i.e., Gu et al., 2012; Nishikawa et al., 2011, 2012; Wu et al., 2009, 2010a; Yuan et al., 2013).We further suggest the role of “museum and cradle” of southern China for those newts. Finally, based on the results we recommend conservation priorities and management as some newts are under threatened status.
MATERlALS AND METHODS
Sampling
We followed the classification of AmphibiaWeb (2022) and Frost (2022) while recognizing that alternative taxonomies exist (e.g., Dubois & Raffaëlli, 2009). Our ingroup sampling included 27 of 34 described species of modern Asian newts inCynops,Paramesotriton, andPachytriton(Supplementary Table S1). In total, 222 specimens were collected from 78 localities from 2010 to 2014. This sampling covered the entire known distributional range of the three genera on the mainland and our samples represented the largest dataset for this group to date (Supplementary Table S1; Figure 1).Cynops chenggongensisandC. wolterstorffihave not been collected since the 1990s and thus tissue samples were not available.Pachytrion changiwas described from a Japanese pet market so its type locality remains unknown (Nishikawa et al., 2012). Four recently described species ofParamesotritonandPachytritonwere not included here. However, omitting those taxa should not affect the backbone of the reconstructed phylogeny (see results).Calotriton asperandEuproctus platycephaluswere chosen as outgroup taxa based on Weisrock et al. (2006) and Zhang et al. (2008).
DNA extraction, sequencing and alignment
Genomic DNA was extracted using the phenol-chloroform method as described in Sambrook et al. (1989). Partial fragments of mitochondrial NADH dehydrogenase subunit 2(ND2) and cytochromeb(cytb) were sequenced for all specimens. In addition, 39ND2sequences and 35 cytbsequences were downloaded from GenBank. Data from the closely related, monotypicLaotritonwere also downloaded.For the nuclear genes, fragments of the genes encoding the brain-derived neurotrophic factor (BDNF) and proopiomelanocortin (POMC) were sequenced from a subset of samples consisting of 83 individuals (Supplementary Table S1). These samples were selected to represent all major mitochondrial matrilines. PCR conditions of these gene fragments were performed as in Vieites et al. (2007) and Wu et al. (2010b).
Purifications were performed using the Gel Extraction Mini Kit (Watson BioTechnologies, China) and sequencing was performed in both directions using the BigDye Terminator Cycle Sequencing Kit (v.2.0; Applied Biosystems, USA) by an ABI PRISM 3730 automated DNA sequencer (Applied Biosystems, USA). Nucleotide sequences were aligned using Clustal X v.1.81 (Thompson et al., 1997) with default parameters and manually checked in mega v.6.0.6 (Kumar et al., 2018).
Matrilineal genealogy constructions
Considering that all mtDNA gene sequences are inherited as one linkage group, mtDNA gene segments were concatenated into a single partition for subsequent analyses. Bayesian Inference (BI), Maximum Likelihood (ML) and Maximum Parsimony (MP) methods were used to reconstruct the matrilineal relationships. Best-fitting nucleotide substitution model for each codon position and tRNA was selected independently using the Akaike Information Criterion in MrModeltest v.2.3 (Nylander, 2004).
Bayesian inference were conducted using MrBayes v.3.1.2(Ronquist & Huelsenbeck, 2003). Four Markov chain Monte Carlo (MCMC) were run for 4 000 000 generations using the default heating value. Trees were sampled every 100 generations. Stationarity was determined when the potential scale reduction factor (PSRF) reached 1 and the log-likelihood(-lnL) scores plotted against generation time stabilized(Huelsenbeck & Ronquist, 2001). The first 25% of the trees were discarded as burnin. The frequency of nodal resolution,termed a Bayesian posterior probability (BPP), was assumed to indicate confidence. Maximum likelihood analyses were conducted using RAxML v.7.0.4 (Stamatakis et al., 2008).Bootstrap analyses with 1 000 nonparametric bootstrap replicates were performed to assess nodal support. Maximum parsimony analyses were conducted using paup* v.4.0b10a(Swofford, 2003). Heuristic searches with tree bisection and reconnection (TBR) were executed in 1 000 random addition replicates with all characters treated as unordered and equally weighted. Bootstrap support analyses were performed with 1 000 pseudo-replicates. We consider BPP ≥ 95% and BS ≥ 70% as evidence of high statistical support for the node.
Nuclear data analyses
We used phase v.2.1.1 (Stephens et al., 2001) to resolve haplotypes from heterozygous individuals. Five independent runs of 20 000 iterations were implemented from each locus of each genus, discarding the first 5 000 as burnin and allowing recombination. Allelic combinations phased with a BPP of 0.90 or higher were retained for further analyses. Allelic medianjoining networks (MJNs) forBDNFandPOMCwere constructed separately for each of the genera using network v.4.5 (Bandelt et al., 1999). To visualize the overall divergence pattern, we also constructed a multi-locus, individual-based network for each genus. We used paup* to generate matrices of uncorrected p-distances between phased haplotypes at each locus. We combined individual locus-matrices into one multi-locus distance matrix using pofad v.1.03 (Joly &Bruneau, 2006). Finally, networks were reconstructed in splitstree v.4.6 (Huson & Bryant, 2006) using Neighbor Net(Bryant & Moulton, 2004).
Divergence time estimation and biogeographic inference
Divergence dates were estimated using the mtDNA data under a Bayesian molecular clock framework as implemented in beast v.1.6.1 (Drummond & Rambaut, 2007). Because we were mostly interested in major cladogeneses, we selected one individual per species (Supplementary Table S1).We took two secondary calibration points from Zhang et al. (2008),which evaluated divergence times with mitogenomic data of salamandrids. The first calibration prior (Supplementary Figure S1 and Table S2, C1) was placed at the split between the modern Asian newts and the modern European newts at 27 million years ago (Ma) with a normal-distributed 95%confidence interval (CI) between 25.6 and 28.7 Ma. The second calibration point (Supplementary Figure S1 and Table S2, C2) was the most recent common ancestor of the modern Asian newts at 19.5 Ma with a normal-distributed 95% CI between 16.9 and 21.5 Ma. We assumed a relaxed,uncorrelated lognormal clock for our rate-variation model and a pure birth model (Yule process) for the tree prior. A test run of 6 000 000 generations was performed to optimize the scale factors of the prior-function. The final MCMC was run for 30 000 000 generations with a sampling frequency of 1 000.Burnin and convergence of the chains were determined by tracer v.1.5 (Rambaut & Drummond, 2007). Further, the measures of effective sample sizes (ESS) estimated in beast v.1.6.1 were used to reflect the efficiency of chain mixing.Values greater than 200 were considered good mixing.
Historical biogeography analyses were carried out using RASP v.3.2 (Yu et al., 2015). We used the parsimony-based statistical-dispersal-vicariance (S-DIVA; Yu et al., 2010) and the likelihood-based divergence-extinction-cladogenesis(DEC; Ree & Smith, 2008) model to reconstruct the possible ancestral ranges and biogeographical divergences of the modern Asian newts. Two geographic areas corresponding to Terrains II (A) and III (B) were determined. The maximum credibility tree from the beast analysis with all outgroups removed were used for the analysis.
Ecological niche comparison
In order to determine whether genetic groupings are associated with differences in their realized ecological niches,we used occurrence data and environmental layers to statistically assess niche identity and divergence between genetic groups within each of the three genera. To increase the number of occurrence data, we obtained additional GPS coordinates from the VertNet database (http://vertnet.org/index.html, Supplementary Table S3). Nineteen environmental layers of temperature and precipitation with a 30 arc-sec resolution (~1 km2) were downloaded from WorldClim(http://www.worldclim.org/) (Hijmans et al., 2005). We clipped this coverage to a region that encompassed southern China(N20-33°, E100-122°) and only retained one layer if multiple layers had correlation efficiencies >0.9, preferentially choosing layers that measured averages over those that measured extremes or seasonality (Kozak & Wiens, 2006; Shepard &Burbrink, 2008). Correlation efficiency was determined using functionraster.cor.matrixin the R package ENMTools v 1.0.6(Warren et al., 2019, 2021). We further used the ENMTools package along with Maxent model to determine niche identity(the identity test, functionidentity.test) and background similarity (the background test, functionbackground.test). The former test statistically assesses whether a pair of taxa exhibit identical niches, and the latter test asks to what extent the observed differences in niches of a pair of taxa can be effectively attributed to their background differences, which is suitable for comparing allopatric taxa (Warren et al., 2008).The original background test described in Warren (Warren et al., 2008) involved two asymmetric tests, whereas the R package introduced a modified symmetric background test to avoid generating confusing results (http://enmtools.blogspot.com/2016/07/backgroundsimilarity-tests-inenmtools.html) and thus is used here. Background buffer raster for each taxon was created from occurrence data with a circular radius of 50 km, from which 1 000 background points were drawn. We further tested for an abrupt environmental transition defined by a linear line between lineages (functionrangebreak.linear). This analysis can provide a more biologically meaningful result than the prior two tests if a steep biogeographic boundary occurs between two taxa/lineages on either side (Glor & Warren, 2011). Lastly, for all three tests we used the two conventional metrics, Hellinger’sIand Schoener’sD, calculated in environmental space (E-space) as opposed to geographic space (G-space), as the former measurement is continuous in space and not limited by discrete units or predetermined maximum or minimum values that each predictor variable may take (Warren et al., 2019). All tests were run with 99 replicates and statistical significance was determined by nonparametric Monte Carlo method(Warren et al., 2021).
RESULTS
Sequence characteristics
We obtained 1 128 bp of cytband 1 132 bp ofND2from 222 specimens, representing the densest sampling of the modern Asian newts. Newly generated sequences are deposited in GenBank (accession numbers: ON793668-ON793756,ON793810-ON793941, ON793995-ON794124, ON793648,Supplementary Table S1). No premature stop codon or indel was detected in protein-coding regions. Six indels were found in tRNATrp. The concatenated dataset contained 174 haplotypes with 1 200 variable sites, of which 1 112 were potentially parsimony informative. The haplotypes were highly restricted geographically, as no haplotype was shared among locations. ForND2, the best-fit model was TRN+G for the first codon position, HKY+G for the second codon position, and GTR+G for the third codon position. GTR+G was selected the best model for tRNA. For cytb, the best model was F81 for the first and second codon positions and GTR+G for the third position.
The nDNA data included 609 bp forBDNFand 479 bp forPOMCfrom 83 representative specimens. No indel was detected.BDNFhad 13 haplotypes from mainlandCynops, 13 fromParamesotriton, and 19 fromPachytriton.POMChad 13 haplotypes in mainlandCynops, 13 inParamesotriton, and 14 inPachytriton.All sequences were deposited in GenBank(accession numbers: ON793650-ON793667, ON793757-ON793809, ON793942-ON793994, ON794125-ON794160,ON793647, ON793649, Supplementary Table S1).
Matrilineal genealogy and nuclear gene networks
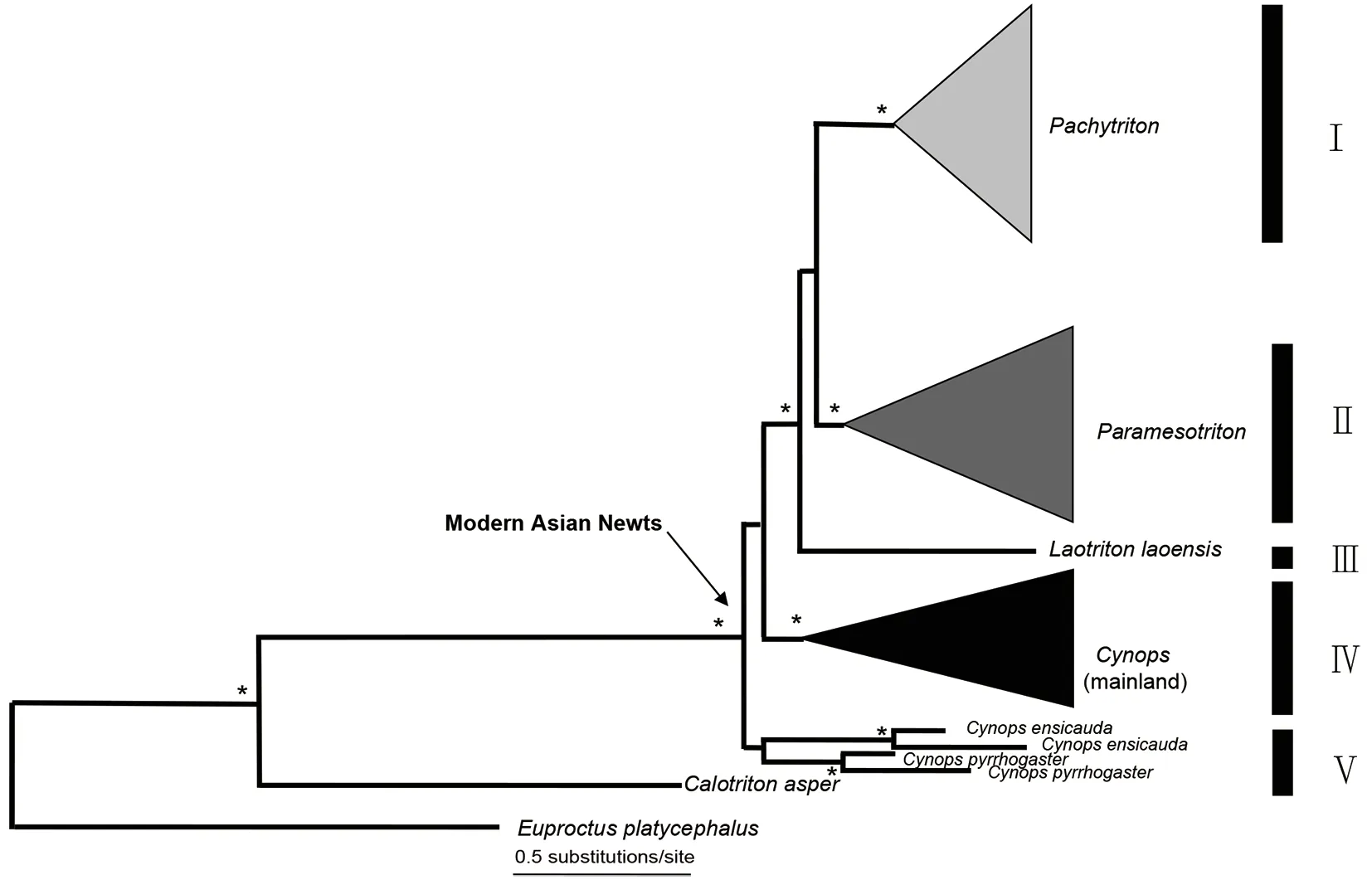
Figure 2 Bayesian inference tree for the 174 sampled haplotypes of modern Asian newts based on concatenated sequence data from cyt b and ND2

Figure 3 Sampling sites and relationships for mainland Cynops
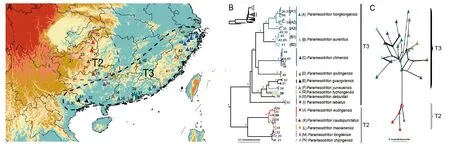
Figure 4 Sampling sites and relationships for Paramesotriton

Figure 5 Sampling sites and relationships for Pachytriton
The BI, ML and MP trees were nearly identical and thus, only the BI tree was shown (Figures 2, 3B, 4B, 5B). Monophyly of the modern Asian newts was strongly supported (Figure 2,lineages I-V), same as the monophyly ofPachytriton(Figure 2, lineage I),Paramesotriton(Figure 2, lineage II), and mainlandCynops(Figure 2, lineage IV). The monophyly ofCynopsremained ambiguous because Japanese species were recovered as the sister group to all other mainland modern Asian newts, albeit with weak nodal support (BI<95%and ML/MP bootstrap support <70%). The monotypic genusLaotritonformed the sister taxon toPachytritonandParamesotriton. Phylogenetic relationships within each genus were identical or congruent with latest studies (Li et al., 2018;Wu et al., 2010c; Wu & Murphy, 2015; Yuan et al., 2013,2014, 2016a, 2016b).
Comparison among mtDNA genealogies of the three genera showed that they all shared a basal split between lineages that inhabit Terrain II and III in southern China (referred as the T2 and T3 lineage, Figures 3-5; Supplementary Figure S2).The nDNA networks were in strong congruence with the mtDNA results in all three genera (Figures 3C, 4C, 5C;Supplementary Figure S2). Specifically, within mainlandCynops,C. cyanurusrepresented the only T2 species from the Yunnan-Guizhou Plateau of Terrain II, whereas four T3 species occurred in the eastern coastal regions of Terrain III.Paramesotritonwas comprised of five T2 species from the Dalou Mountain to the middle of the Nanling Mountains, and nine T3 species from the middle of the Nanling Mountains eastward to coastal southern China and northernmost Vietnam (Figure 4). InPachytriton, there were two T2 species that occur from the Guizhou Plateau to Dayao Mountain and the Miaoling Mountain; and six T3 species were distributed from the middle of the Nanling Mountains eastward to the coast of southern China (Figure 5).
Divergence times and biogeographic inference
The beast tree (Supplementary Figure S1 and Table S2) was largely congruent with those of BI, ML and MP based on the complete sampling, except thatCynopswas resolved as monophyletic. The modern Asian newts were estimated to have diverged around the beginning of the Miocene (ca. 19.6 Ma; CI 18.6-20.5 Ma) (Supplementary Figure S1 and Table S2). Splits between the T2 and T3 matrilines of mainlandCynops,ParamesotritonandPachytritondiffered in their estimated ages. In mainlandCynopsthe divergence occurred about 15.5 Ma, inParamesotritonit occurred around 13.2 Ma and inPachytritonapproximately 9.2 Ma. Most speciation events that gave rise to extant taxa occurred before the Quaternary except for five closely related species in theParamesotritoncaudopunctatusgroup. The ancestral ranges of modern Asian newts were likely either in Terrains III (SDIVA, Figure 6) or across Terrains II and III (DEC,Supplementary Figure S3). Both DEC and S-DIVA suggested that vicariances between Terrains II and III were vital in shaping the current distribution pattern ofCynops,ParamesotritonandPachytriton(Figure 6).
Ecological niche comparison
We retained 11 environmental layers after omitting those that were highly correlated. Maxent ecological niche models were built for T2 and T3 lineages of each genus with AUC statistics>0.9. The identity tests in all three genera strongly rejected the null hypothesis that environmental distributions of T2 and T3 lineages represent random draws from the same underlying distribution (allP<0.05; Table 1; Supplementary Figure S4),suggesting that T2 and T3 lineages occupy significantly different realized niches. However, the symmetric background tests cannot reject the null hypothesis that lineages are more(or less) similar in their environmental distributions than expected given their respective ranges (Table 1;Supplementary Figure S5). This result indicated that significance in the identity test was mostly caused by distinct environmental conditions between the Terrains II and III instead of by differential niche preference between T2 and T3 lineages. Additionally, range-break tests also did not identify a single abrupt environmental transition between distributional ranges of T2 and T3 lineages, as all tests were non-significant except for theIstatistics inParamesotriton(Table 1;Supplementary Figure S6).
DlSCUSSlON
We used genetic and environmental data to reveal a shared T2-T3 phylogeographic divergence within mainlandCynops,Paramesotriton, andPachytritonfrom montane and submontane regions of continental East Asia. Our results are based on the most comprehensive samplings of the modern Asian newts to date regarding both species diversity (27 of 34 described species) and sampling range (78 localities).Phylogenetic relationships and divergence times are largely congruent with previous studies. Importantly, this work represents the first study to analyze the modern Asian newts under a comparative phylogeographic framework and provides insights into the regional biodiversity that likely impacted by the serial orogeneses of the QTP system. We want to highlight the necessity that some vulnerable newt species warrant state protection status, because they not only exemplify the long evolutionary history of this group but also occupy a very restricted range and thus are threatened with extinction.
Phylogenetic relationships of the modern Asian newts
Phylogenetic relationships among the three genera inferred in this study are similar to those from earlier works based on either mitogenomic data or exclusively nuclear data (Veith et al., 2018; Zhang et al., 2008):ParamesotritonandPachytritonare recovered as sister genera to the exclusion ofCynops.However, the position of the enigmatic, monotypic genusLaotritonis questionable, which could be placed as the sister taxon to (1)ParamesotritonandPachytritonas shown in this study, (2) just toPachytriton(Zhang et al., 2008), or (3) just toParamesotriton(Veith et al., 2018). None of those placements received robust nodal supports. The other fundamental question is the monophyly ofCynops, which is comprised of the mainland lineage and the Japanese lineage. Our result suggests a likely paraphyletic relationship with respect to the two lineages, which are found to form a low-support monophyletic group by mitogenomic data (Zhang et al., 2008).However, the mitogenomic data also cannot reject a paraphyly hypothesis ofCynops(Zhang et al., 2008).More recently, a comprehensive transcriptomic dataset corroborated a paraphyleticCynopswith the Japanese lineage being the first split among modern Asian newts, and the spurious monophyly is probably caused by introgression between the mainland and Japanese lineage (Rancilhac et al., 2021). These results would support the separation ofCynopsinto two genera and the revival ofHypselotritonfor all mainland species ofCynopsas suggested by Dubois & Raffaëlli (2009). In regard to intrageneric relationships, our mtDNA-based results are identical or congruent with those most recent studies on respective genus (Li et al., 2018; Wu et al., 2010c; Wu &Murphy, 2015; Yuan et al., 2013, 2014, 2016a, 2016b). Unlike mtDNA genealogies, the nuclear networks are less resolved.The most prominent divergence is the basal split between T2 and T3 lineages in all three genera.

Figure 6 Ancestral distributions reconstructed by the statistical dispersal-vicariance analysis (S-DlVA)

Table 1 Nonparametric Monte Carlo P-values of Schoener’s D and Hellinger’s l metrics in environmental space (E-space) from the three tests in ENMTools
Split between T2 and T3 lineage in southern China
For all three genera of modern Asian newts, the genetic divergence of T2 and T3 lineages correspond to spatial separation in Terrains II and III. Mitochondrial matrilines,nDNA networks, and ancestral range analysis all strongly support this result. Previous phylogeographical studies from other species also identified a similar T2-T3 divergence pattern (Guo et al., 2016; Lin et al., 2014; Yan et al., 2021).
Ecological niche modeling and the associated identity tests revealed significant differences between the realized ecological niches of T2 and T3 lineages in all three genera.However, the symmetric background test cannot reject the null hypothesis in any genus, suggesting that it is background environmental differences, rather than the divergence of fundamental ecological niches, that contributes to the observed ecological differences between T2 and T3 lineages.Combined results of the two tests suggest that the T2 and T3 lineage of each genus likely have retained similar niche preferences (e.g., preferring cool montane streams), but their realized niches appear different because they occupy environmentally heterogeneous, allopatric ranges. It must be noted that the background test can be flawed by problematic definition of environmental background, which may not necessarily reflect where the species would indeed occur(Glor & Warren, 2011). In our case, we also tested for background buffer raster with a circular radius of 100 km and 200 km (results not shown) and the conclusion remained the same.
Most species of the three genera only occur in montane or submontane aquatic habitats, which are particularly fragmentated at the boundary between Terrain II and III due to rugged and dissected topology (e.g., near the west end of the Nanling Mountains). It is highly likely that countless valleys among those mountains form multiple dispersal barriers for the T2 and T3 lineages, because they are restricted to cooltemperature, high-elevation streams or ponds and cannot cross lowland valleys under current climate model. Results from the range-break tests are in line with this hypothesis as a single biogeographic barrier between the ranges of T2 and T3 lineages was not supported by most tests. Divergence times between T2 and T3 lineages varied among the three genera,again suggesting that the observed split was not caused by a single vicariance but occurred repeatedly through the evolutionary history (Figure 6).
Orogenesis of the QTP system can be a factor that shaped the T2-T3 splits in modern Asian newts. This series of geological events had driven cladogenesis in many taxa via vicariance (e.g., Che et al., 2010; Yan et al., 2021; Zhou et al.,2012). Allopatric divergence could be driven and then maintained by distinct topological landscapes and climatological patterns between Terrains II and III (Guo et al.,2016; Yan et al., 2021). The uplift of QTP not only created a range of distinct habitats that differ in elevations over thousands of meters, but also led to changes in climatic variables, especially the development of Asian monsoons.Intermittent uplifting of the QTP caused southern China to experience cyclical wet-dry environments driven by oscillating Asian monsoons (Farnsworth et al., 2019; Li et al., 2021b; Sun& Wang, 2005). Substantially intensified summer monsoons formed waterways that connected neighboring stream systems, resulting in dispersal corridors that probably facilitated the colonization of new habitats, as it had happened in North American salamanders (Keen, 1984; Schalk &Luhring, 2010). In geological episodes when summer monsoons weakened, habitats became fragmentated and thus newt populations were isolated from each other. Summer monsoon activity may have facilitated speciation, for example inPachytriton(Wu et al., 2013). Three periods of drastic decrease in strength of monsoons and increased dryness occurred at around 16.5-15.0 Ma (Clift et al., 2008;Farnsworth et al., 2019; Wei et al., 2006), 14.25-11.35 Ma(Jiang et al., 2008; Jiang & Ding, 2009;) and again 10.0-3.5 Ma (Barry et al., 2002; Clift et al., 2008; Fan et al., 2007). Our estimated splits between T2 and T3 of mainlandCynops(Supplementary Figure S1 and Table S2: node 7; 15.5 Ma),Paramesotriton(Supplementary Figure S1 and Table S2:node 10; 13.2 Ma) andPachytriton(Supplementary Figure S1 and Table S2: node 11; 9.2 Ma) largely coincide with these weak summer monsoon episodes, indicating their influence on basal cladogenesis in those newts.
Museums and cradles
Our results indicate that mountains of southern China not only acted as cradles of speciation for modern Asian newts but also as museums in preserving old genetic lineages. Similar phenomenon is also observed in endemic Chinese flora, as previous studies suggested that those mountains harbor high species diversity (the cradle) as well as shelter biota at inhospitable times (the museum) (López-Pujol et al., 2011a,2011b; Li et al., 2021a; Lu et al., 2018).
The modern Asian newts are microhabitat-specialists adapted to cool, montane water bodies. When climate became less favorable, montane areas provide an array of sheltered habitats with mild and moist conditions (Bell et al., 2010;Rahbek et al., 2019a, 2019b) at which newts could survive.The mtDNA data demonstrated that newts have been geographically restricted to local montane areas, as haplotypes are rarely shared between populations from different mountains. The lack of shared haplotypes generated profound geographic structures with deeply divergent matrilines. Limited or no gene flow can lead to radiations of range-restricted species such as in the case of Appalachian lungless salamanders of North America (Ruben & Boucot,1989). Except for theParamesotritoncaudopunctatusgroup,all other species in southern China originated before the Quaternary, some of which dated back to the late Miocene.These old taxa are often characterized by small ranges. For example,Cynops glaucus(14.8 Ma) is restricted to Lianhua Mountain,Paramesotriton labiatus(8.1 Ma) is just known from Dayao Mountain, andPachytriton moi(7.2 Ma) can only be found at high elevations of Mao’er Mountain. The contrast between old age and restricted distribution indicates long-termin situpersistence, an outcome of physiographical and climatological heterogeneity of southern China (Qian &Ricklefs, 2000). Our results support the mountains of southern China as both museums and cradles for modern Asian newts.
Conservation implications
Following the principle of identifying evolutionary significant units (Moritz, 1994; Ryder, 1986), our study can provide a guide to the assessment of conservation status of modern Asian newts, especially for species that deserve more imminent protection. Some endemic, old-aged species only occur on a single mountain or a small area, such asCynops glaucus,C. fudingensis,Pachytriton moi, andParamesotriton labiatus. They are vulnerable to habitat disturbance and the entire population can be easily extirpated by environmental fluctuation or human activities (Wu et al., 2013). For example,the type localities ofC. glaucusandC. fudingensisare undergoing development for tourism (Zhou J., personal communication). In addition, many species of the modern Asian newts, such asPachytriton granulosus,Paramesotriton hongkongensis, andC. orientalis, are overharvested for the pet trade or used in traditional medicine in an unsustainable manner (IUCN, 2021; Xie et al., 2007). Fortunately, the recently updated List of Wild Animals under Special State Protection in China has given all members ofParamesotritonandC. orphicusthe Class II status (http://www.forestry.gov.cn/main/586/20210208/095403793167571.html), which imposes heavy penalty if people destroy their habitat or illegally collect them. However, the new list still overlooks other taxa as it remains unclear whyPachytriton, which is similar toParamesotritonin many ways, was not given any protection status (Wang et al., 2021). Protection of these species must be multi-pronged. Education and public awareness are essential. There is an urgent need to launch a conservation agenda that can ensure the preservation of the landscape.Genetically informed management plans can maximize the maintenance of diversity. Despite recent government effort,there is still a need to strengthen both the legal system and enforcement of existing laws.
SClENTlFlC FlELD SURVEY PERMlSSlON lNFORMATlON
Field collections followed the rules of the Wildlife Protection Law of the People’s Republic of China. All procedures and handing of animals were approved by the Ethics Committee of the Kunming Institute of Zoology, Chinese Academy of Sciences (approval No. SYDW-20110105-28).
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETlNG lNTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRlBUTlONS
J.C. and Y.P.Z. conceived and supervised the study. Z.Y.Y.,Y.K.W., F.Y. collected samples. Z.Y.Y. and Y.K.W. performed the analyses. Z.Y.Y., Y.K.W., J.C. and Y.P.Z. wrote the manuscript. F.Y., R.W.M., T.J.P., and D.B.W. provided valuable suggestions and revisions. All authors read and approved the final version of the manuscript.
ACKOWLEDGEMENTS
We thank Ke Jiang, Li-Min Ding, Shun-Qing Lu, Jun-Xiao Yang, Jie-Qiong Jin, Hong-Man Chen, Hai-Peng Zhao, Miao Hou, Liang Zhang, Ping-Fan Wei, Xiao-Long Liu, Ying-Yong Wang and Hai-Tao Zhao for providing tissues and helping during collecting. We are grateful to Wei-Wei Zhou, Dong-Yi Wu and Pin-Fan Wei for invaluable comments and analysis on the manuscript preparation. We thank the local staff of the nature reserves for the permit to collect a limited number of newts.
- Zoological Research的其它文章
- Diversity of reptile sex chromosome evolution revealed by cytogenetic and linked-read sequencing
- Coevolutionary insights between promoters and transcription factors in the plant and animal kingdoms
- Deficiency of transmembrane AMPA receptor regulatory protein γ-8 leads to attention-deficit hyperactivity disorder-like behavior in mice
- Global cold-chain related SARS-CoV-2 transmission identified by pandemic-scale phylogenomics
- The Hippo pathway and its correlation with acute kidney injury
- Genomics and morphometrics reveal the adaptive evolution of pikas

