Mechanism of electroacupuncture and herb-partitioned moxibustion on ulcerative colitis animal model: A study based on proteomics
Qin Qi, Rui Zhong, Ya-Nan Liu, Chen Zhao, Yan Huang, Yuan Lu, Zhe Ma,Han-Dan Zheng, Lu-Yi Wu
Abstract
Key Words: Proteomics; Ulcerative colitis; Moxibustion; Electroacupuncture; Differential proteins
INTRODUCTION
Ulcerative colitis (UC) is a chronic, nonspecific inflammatory disease, with lesions are mainly confined to the large intestinal mucosa and submucosal layer. Mucosal inflammation at the affected site is characterized by a diffuse distribution and its clinical manifestations mainly include abdominal pain,diarrhoea, mucus pus and blood in stool[1]. UC has a prolonged disease course, which can lead to the development of colorectal cancer, accounting for 10% to 15% of deaths among patients with UC, and is listed as one of the refractory diseases by the World Health Organization. Epidemiological data show that the prevalence of UC in Asia has increased exponentially in recent years, and the growth rate of UC in some East Asian countries including China has increased in multiples over the past decade[2,3].
The pathogenesis of UC is not yet clear, but it is believed to be related to genetic susceptibility,epithelial barrier deficiency, immune disorders, and environmental factors[4]. The modern treatment methods for UC mainly include glucocorticoids, immunosuppressants, 5-aminosalicylic acid, and biological agents (monoclonal antibodies), but it is prone to relapse after drug withdrawal, and multiple drugs have adverse side effects. Therefore, active exploration of the pathogenesis of UC, identification of disease biomarkers, and screening of specific therapeutic targets are important for the diagnosis and treatment of UC. As one of the unique treatments of traditional Chinese medicine, acupuncture and moxibustion have significant efficacy and advantages in the treatment of UC[5-7], but the specific mechanism requires further clarification.
In recent years, proteomic technology has been widely used in the study of UC[8,9], revealing a variety of biological markers related to immunity and inflammation in UC[10]. The activity of mitochondrial oxidative phosphorylation complex in intestinal tissues of UC patients decreases, and the dysfunction of mitochondrial oxidative phosphorylation may be involved in the pathogenesis of UC,but it is not clear whether acupuncture and moxibustion have a regulatory effect on it. Therefore, our study used isotope-labeled relative and absolute quantification (iTRAQ) proteomics technology to quantitatively analyse proteins in the colon tissues of UC rats, to analyse the biological functions of differentially expressed proteins (DEPs) and observe changes of immune and oxidative phosphorylation-associated protein expression profiles in colon tissues of UC rats and regulatory effect of acupuncture and moxibustion, in order to identify UC-relevant protein targets and further explore the mechanism of effects of acupuncture and moxibustion on UC.
MATERIALS AND METHODS
Animals
Healthy clean grade naïve male Sprague-Dawley (SD) rats, aged 6 wk, with body weight of 180 ± 20 g were purchased from Shanghai Slac Laboratory Animal Co., Ltd. [Laboratory Animal Use Permit No.SYXK (Shanghai) 2014-0008] and housed in the Experimental Animal Center of Shanghai University of Traditional Chinese Medicine. The animal protocol was designed to minimize pain or discomfort to the animals. The housing environment was a 12 h circadian rhythm, with a room temperature of 20 ± 2 °C and 50%-70% indoor humidity, 4 rats per cage, and rats were free to standard food and pure water. This study was approved by the Ethics Committee of the Experimental Animal Center of Shanghai University of Traditional Chinese Medicine. We tried all efforts to minimize animal suffering.
Dextran sulfate sodium-induced UC model preparation
For preparing the UC rat model, the rats received 3% dextran sulfate sodium (DSS, molecular weight:36000-50000, MP Biomedicals, United States) in drinking water by oral for 7 d, and then were switched to pure water for 7 d, and the same procedures were repeated once (drinking 3% DSS solution for 7 d,followed by drinking pure water for 7 d)[11]. After modeling, rats were anaesthetizedviaintraperitoneal injection with 2% pentobarbital sodium (30-40 mg/kg) (P3761, Sigma, United States) for tissue collection, the gross injury and hematoxylin and eosin (HE) staining were used to evaluate whether the model was successfully established.
Grouping and interventions
After one week of adaptive feeding, 32 rats were randomly divided into the normal (N) group, the DSSinduced UC model (M) group, the herb-partitioned moxibustion (HM) group, and the electroacupuncture (EA) group, with 8 rats in each group. UC rat model was prepared by DSS except for the N group. After model establishment, the rats in the HM and EA group were treated with corresponding treatment at Tianshu (ST25, bilateral) and Qihai (CV6) acupoints[12]. In the HM group, Chinese medicine powder containing Coptis chinensis, Radix aconiti lateralis, Cortex Cinnamomi, Radix aucklandiae, Flos carthami, Salvia miltiorrhiza, and Angelica sinensis was mixed and stirred with yellow wine to make herbal cake, then an herbal cake was prepared with a thickness of 0.5 cm and a diameter of 1 cm using a specific mould, and conical moxa cones weighing approximately 90 mg were made with moxa (Nanyang Hanyi Moxa Co., Ltd., Nanyang, China) by a mould. The rats were supine immobilized on a self-made fixator, the prepared moxa cone was placed on the top of herbal cake, and the herbal cake was placed at the ST25 and CV6 acupoints, then the moxa cone was ignited using line incense, 2 moxa cones per acupoint per time, once a day for 7 d (Figure 1)[13]. In the EA group, the rats were also supine immobilized on the self-made fixator, a 0.25 mm × 25 mm acupuncture needle was inserted into the ST25 and CV6 acupoints for 5 mm, then the needle handle was connected to a Han’s-200 acupoint nerve stimulator, with a frequency of 2/100 Hz and a current of 1 mA. The needle was kept for 10 min per time, once a day for a total of 7 d[14]. The rats in the N group and the M group did not receive any treatment, but only the same grasping fixation as that in the treatment groups at 10 o’clock every day for a total of 7 d.

Figure 1 Schematic diagram of herb-partitioned moxibustion at the rat Tianshu (ST25, bilateral) and Qihai (CV6) acupoints.
Collection of colon tissue and macroscopic scoring of colon injury
After the intervention, rats were anaesthetized with intraperitoneal injection of 2% pentobarbital sodium. The abdominal cavity was exposed and the entire colon from the pubic symphysis to the distal cecum was collected, and the length of the colon was recorded. The distal colon with a length of 6-8 cm was collected and cut longitudinally along the mesentery, observed the gross morphology, the macroscopic scoring of the colon was performed according to the following parameters: Hyperemia,wall thickening, ulceration, inflammation extension, and damage no damage, score 0; hyperemia without ulcers, score 1; hyperemia and wall thickening without ulcers, score 2; one ulceration site without wall thickening, score 3; two or more ulceration sites, score 4; 0.5 cm extension of inflammation or major damage, score 5; 1 cm extension of inflammation or severe damage, score 6[15]. Then was cut into two parts transversely, one part was fixed in 4% paraformaldehyde for morphological observation,and the remaining part was placed in liquid nitrogen for 1 h and transferred to a -80 °C freezer for future detection.
Morphological observation of the colon
Colon tissues fixed in 4% paraformaldehyde fixative solution were dehydrated, embedded in paraffin,and sliced into 4 μm sections for HE staining with the following steps: Dewaxing in xylene I and II solutions for 20 min each; elution in 100%, 95%, 90%, 80%, and 70% ethanol for 5 min each; rinsing with double distilled water for 5 min × 2 times; haematoxylin staining for 2-3 min; rinsing with running water for 10 min; 1% hydrochloric acid ethanol differentiation solution for 1-2 s; rinsing with running water for 5 min; eosin dye staining for 2-3 min; dehydration in 70%, 80%, 90%, and 100% ethanol for 1-2 s each; xylene I and II solutions for 15 min each. After drying and sealing, the colon sections were observed under a light microscope and microscopic scoring of the colon was performed according to the following parameters: Damage/necrosis, inflammatory cell infiltration, submucosal edema, and hemorrhage of mucosa. Colonic gross damage scores were recorded according to the severity of changes: No change, score 0; mild, score 1; moderate, score 2; severe, score 3[16].
Proteomic analysis
Protein extraction and enzymatic digestion: Colon samples from 3 rats in each group were randomly collected and ground into powder, and an appropriate amount of radioimmunoprecipitation assay(RIPA) protein lysis buffer (R0010, Solarbio, United States) was added (150 μL RIPA was add to 20 mg colon tissue), high-abundance protein in the sample was removed, a 5-fold volume of 10% trichloroacetic acid/cold acetone was added to the sample and precipitated at -20 °C overnight. The precipitated proteins were centrifuged at 4 °C, 15000 g for 20 min, the supernatant was removed, and samples were air-dried and precipitated after repeating the above steps several times. After lysis, samples were centrifuged at 25000 g for 20 min at 4 °C, and the supernatant was the protein solution. A total of 100 μg of protein solution was collected from each sample, and trypsin (2.5 μg) was added to the protein solution at a ratio of 40: 1 of protein: Enzyme, enzymolysis at 37 °C for 4 h. Trypsin was then added according to the above ratio and continue enzymolysis at 37 °C for 8 h.
Peptide labelling and separation: The digested peptide fragments were desalted using a Strata X column and vacuum-dried. The peptide samples were dissolved in 0.5 M triethylammonium bicarbonate, and then were were labeled with iTRAQ (iTRAQ8-plexreagent kit, Sigma, United States)and maintained at room temperature for 2 h. The Shimadzu LC-20AB liquid chromatography system(separation column: 5 μm, 4.6 mm × 250 mm Gemini C18) was used for liquid phase separation of samples. Dried peptide samples were redissolved in 2 mL of mobile phase A (5% ACN, pH 9.8), the solution was loaded, and gradient elution was performed at a flow rate of 1 mL/min as follows: 5%mobile phase B (95% ACN, pH 9.8) for 10 min, 5%-35% mobile phase B for 40 min, 35%-95% mobile phase B for 1 min, mobile phase B for 3 min, and equilibration with 5% mobile phase B for 10 min. The elution peak was monitored at a wavelength of 214 nm, and 1 fractionated peptide was collected every minute. The chromatographic elution peaks were combined with the samples to obtain 20 fractionated peptides, which were then freeze-dried.
High-performance liquid chromatography: The dried peptide samples were redissolved using mobile phase A (2% ACN, 0.1% FA) and centrifuged at 20000 g for 10 min. The supernatant was collected and loaded onto the column, and samples were separated by a Prominence nano LC (LC-20AD, Shimadzu,Japan). The sample was first enriched in a trap column and desalted, and the tandem self-assembled C18 column (75-μm inner diameter, 3.6-μm pore size, 15-cm length) was used. Separation was performed at a flow rate of 300 nL/min through an effective gradient: at 0-8 min, 5% mobile phase B(98% ACN, 0.1% FA); at 8-43 min, mobile phase B increased linearly from 8% to 35%; at 43-48 min,mobile phase B increased from 35% to 60%; at 48-50 min, mobile phase B increased from 60% to 80%; at 50-55 min, 80% mobile phase B; and at 55-65 min, 5% mobile phase B.
Mass spectrometric detection: The separated peptides were transferred into an electrospray ionization tandem mass spectrometer: TripleTOF 5600 (SCIEX, Framingham, United States) and theion source was Nanospray III source (SCIEX, Framingham, United States). During data collection, the parameters of the mass spectrometer were set as follows: An ion source spray voltage of 2300 V, a nitrogen pressure of 30 psi, atomizer 15, and a spray interface temperature of 150 °C. High-sensitivity mode was used for scanning. The cumulative time of primary mass spectrometry scanning was 250 ms, and the cumulative of secondary mass spectrometry scanning time was 100 ms.
iTRAQ quantification and bioinformatics analysis
ITRAQ quantification was performed using IQuant software. The selection of significantly DEPs in a single experiment were as follows: Fold change (FC) ≥ 1.2 was set as up-regulation and FC ≤ 0.83 as down-regulation, as well as Q-value < 0.05. The Gene Ontology (GO) database was used for functional annotation analysis of DEPs. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database was used for pathway enrichment analysis of DEPs. The protein-protein interaction network was performed using STRING software with an interaction confidence of 0.7 (the maximum confidence was 1). Cytoscape (v.3.7.1) software was used to visualize the enrichment analysis results. Figure 2 showed the process of proteomics detection and analysis.
Western blot analysis
We selected DEPs in UC that were reversed by HM and EA for verification by using western blot method. Total proteins were extracted from colon tissues using RIPA buffer supplemented with protease and phosphatase inhibitors. Protein concentration was measured with the bicinchoninic acid protein concentration assay kit. The proteins were separated by polyacrylamide gelelectrophoresis method, with the voltage of concentrated glue was 90 V and that of separating glue was 120 V (BIORAD, 1645050), then the separated proteins were transferred to polyvinylidene fluoride membranes.The modified membranes were blocked with 5% bovine serum albumin in TBST, and then was incubated overnight at 4 °C with primary antibody. The antibody dilutions were prepared according to the instructions [Synaptic vesicle glycoprotein 2A (Sv2a), 1: 1000; nuclear cap binding protein subunit 1(Ncbp1), 1: 800; Cps, 1: 5000; cytochrome c oxidase subunit 4 isoform 1 (Cox4i1), 1: 2000; ATP synthase beta subunit precursor 1 (Atp5f1), 1: 1000; doublecortin like kinase 3 (Dclk3), 1: 800)]. The membranes were washed with western detergent at room temperature for 6 times, 5 min each and then were incubated with secondary antibodies at room temperature for 1 h. Followed by washed with western detergent at room temperature for 6 times, 5 min each. The membranes were stained with enhanced chemiluminescence solution and visualized in a gel imager system (Tanon, 4600).
Statistical analysis
The western blot analysis data were analyzed with SPSS 20.0 statistical software. Data that consistent with the normal distribution were presented as mean ± SD, and one-way analysis of variance was used for comparison between groups, followed by the Fisher’s Least significant difference method if data met the homogeneity of variance. For data that did not meet the normal distribution were presented as medians (P25,P75), and nonparametric test was used for comparison.P< 0.05 was considered statistically significance.
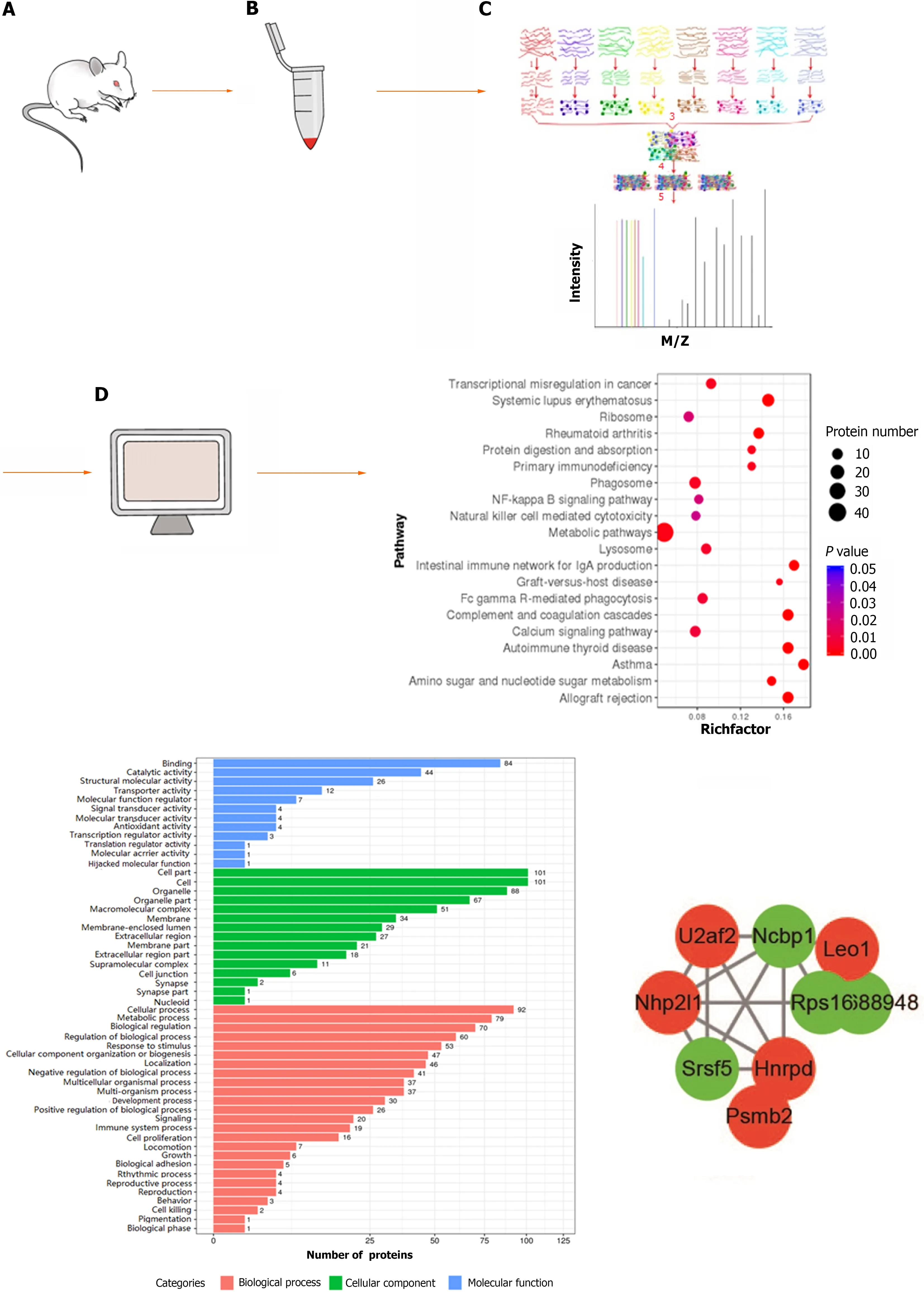
Figure 2 Flow chart of proteomics detection and analysis. A: Sprague-Dawley rat; B: Protein extraction of colon tissue; C: Protein enzymatic digestion,peptide labelling, separation, and high-performance liquid chromatography analysis; D: Bioinformatics analysis.
RESULTS
Effects of HM and EA on DSS induced UC rats
In the N group, rats were active and energetic, and their feces were moderately hard and soft. The rats in the M group had loose, bloody stools and decreased activities. Compared with the M group, the activity of rats in the HM and EA groups was obviously improved, and the stool gradually took shape without obvious mucus and blood. We found that the colon in the N group did not exhibit adhesion or bleeding points; the colonic mucosal surfaces were clean and smooth. In contracts, the colon in the M group exhibited significant adhesion and multiple bleeding points; the inner wall was not smooth, and partially visible scattered ulcers and thickened intestinal walls were observed. In the HM and EA groups, the adhesion and the bleeding points were decreased (Figure 3A).
The morphological changes of colon tissues were observed using HE staining. The colonic mucosal epithelium of the normal group was intact, with clear tissue structure, regular arranged glands, and no congestion, oedema or ulcers were evident. While the mucosal epithelium of UC rats was absent, ulcers had formed, with a loss or reduction of glands and goblet cells, and substantial inflammatory cells were infiltrated in mucosa and submucosa. After HM and EA interventions, the mucosal epithelium was restored, with healed ulcer, increased glands and goblet cells, decreased mucosal and submucosal inflammatory cell infiltration and congestion (Figure 3B).
The macroscopic colon injury scores and histopathology scores in the M group were significantly increased than that in the N group (P< 0.01). Conversely, the macroscopic colon injury scores and histopathology scores in the HM and EA groups were significantly decreased compared to the rats in the M group (P< 0.01) (Figures 3C and D). The colonic length of the rats in the M group were significantly decreased compared with the N group, the colonic length of the rats in the HM and EA groups were significantly increased compared with the M group (P< 0.01) (Figure 3E).
iTRAQ quantification of DEPs
The volcano plot in Figure 4 described the DEPs between the groups. The quantitative results showed that 202 DEPs were identified in DSS-induced UC rats compared with the N group, of which 111 were up-regulated and 91 were down-regulated; 117 DEPs were identified in HM group compared with the M group, of which 41 were up-regulated and 76 were down-regulated; 145 DEPs were identified in EA group compared with the M model group, of which 59 were up-regulated and 86 were down-regulated(Table 1, Figure 5).
Comparison of the M/N and HM/M groups showed that among these proteins, 25 DEPs were common to N, M and HM groups, with 17 [Sv2a, Igkv13-85, Ncbp1, Aldo-keto reductase family 1 member B8, Rathemoglobin beta-chain, Ribosomal protein S8 (RpS8), Fga, Rps2-ps6, carbamoyl phosphate synthetase 1 (Cps1), Txn, ATP synthase subunit g (ATP5L), Cox4i1, Atp5h, Necap2, Atp5f1,Papss2, Acly] up-regulated in UC but were down-regulated by HM, and 8 [Spout1, calcium-activated chloride channel regulator 1 (CLCA1), Sval1, Hspb7, Peptidyl-prolyl cis-trans isomerase, Aldh1a1, Lyz2,Dclk3] down-regulated in UC but were up-regulated by HM (Figure 5A, Table 2). Comparison of the M/N and EA/M groups showed that 15 DEPs were common to N, M and EA groups, with 9 (Sv2a,Ncbp1, Rat hemoglobin beta-chain, Fga, Cps1, ATP5L, Cox4i1, Atp5f1, Lmod1) up-regulated in UC but were down-regulated by EA, and 6 (Spout1, Sh3bgrl3, Peptidyl-prolyl cis-trans isomerase, MHC class I RT1.Aw3 protein, Tkt, Dclk3) down-regulated in UC but were up-regulated by EA (Figure 5B, Table 3).
GO enrichment analysis for DEPs
GO annotation of DEPs between groups describes the characteristics of genes and gene products in terms of molecular functions, cellular components and biological processes. 53 GO functions were significantly differentiated between the M and the N group, 52 GO functions are significantly differentiated between the M group and the HM group, and 51 GO functions are significantly differentiated between the M group and the EA group. The GO functional annotations of DEPs in the M/N, HM/M,and EA/M comparisons were basically similar. Biological process was the most favorable enrichment component, in which DEGs were significantly enriched in cellular process, metabolic process, biological regulation and so on. Cellular component analysis was enriched in cell part, cell, organelles, macromolecular complexes,etc.Molecular functions mainly included binding, synaptic activity, structural molecule activity, molecular function regulation, molecular sensor activity, and transcriptional regulation activity (Figure 6).
KEGG pathway enrichment analysis for DEPs
KEGG pathway enrichment analysis was performed for DEPs between the groups, we found that these DEPs were mainly enriched in inflammation responses and immune-related pathways. The pathways in which DEPs between the N and M groups mainly enriched included primary immunodeficiency, the noncanonical nuclear factor-kappaB (NF-κB) signalling pathway, natural killer cells mediated cytotoxicity, intestinal immune network for immunoglobulin A (IgA) production, FcγR-mediated phagocytosis, and complement and coagulation cascades (Figure 7A, Table 4). The pathways in which DEPs between the HM and the M groups mainly enriched included primary immunodeficiency,oxidative phosphorylation, nitrogen metabolism, natural killer cells mediated cytotoxicity, the intestinal immune network for IgA production, FcγR-mediated phagocytosis, complement and coagulation cascades, and B cell receptor signalling pathway (Figure 7B, Table 5). The pathways in which DEPs between the EA and the M groups mainly enriched included the phosphoinositol-3 kinase (PI3K)-protein kinase B (Akt) signalling pathway, oxidative phosphorylation, intestinal immune network for IgA production, and the calcium signalling pathway (Figure 7C, Table 6).

Table 1 List of differential protein numbers
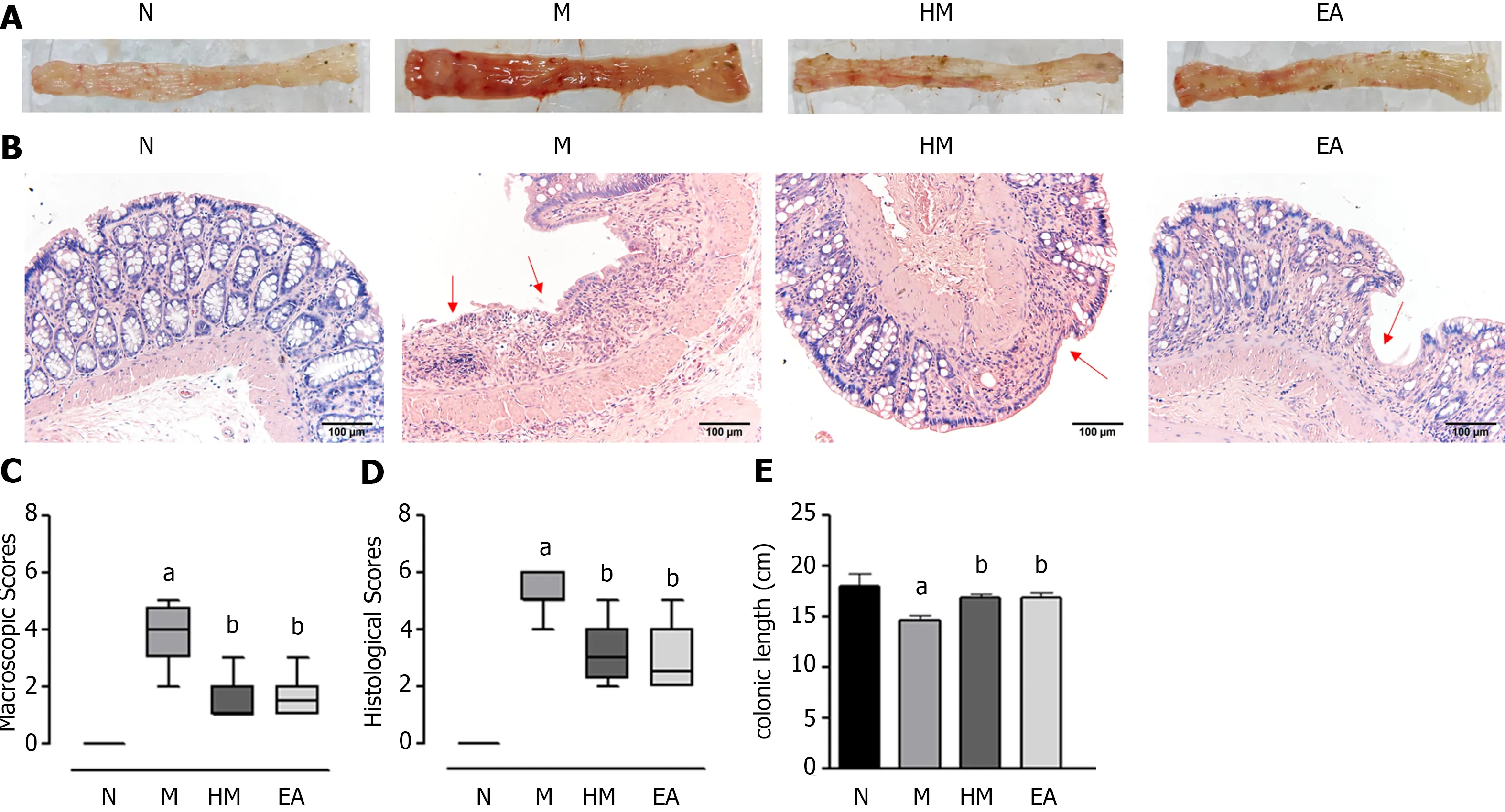
Figure 3 Colon tissue injuries in each group. A: Gross structure; B: Histopathological observation (hematoxylin and eosin, × 200); C: Macroscopic scores; D:Histopathological scores; E: Colonic length. aP < 0.01 vs N group; bP < 0.01 vs M group. N: Normal group; M: Dextran sulfate sodium-induced ulcerative colitis model group; HM: Herb-partitioned moxibustion group; EA: Electroacupuncture group.
Protein-protein interactions analysis
The STRING database (v.11.0) was used to construct the protein-protein interactions (PPIs) network for the transcription factors with an interaction score > 0.7 (high confidence) among the groups. Cytoscape(v.3.7.1) software was used to visualize the enrichment results. The statistical significance was set asP<0.05. The analysis showed extensive interactions among the DEPs between groups (Figure 8). A total of 100 DEPs (63 up-regulated proteins and 37 down-regulated proteins) were included in the PPI network constructed by DEPs between the N and M groups, and Nme2 served as the junction point of two pathways, indicating that it was important for screening key candidate proteins. 38 DEPs (8 upregulated proteins and 30 down-regulated proteins) were included in the PPI network constructed from DEPs between the M and HM groups, in which ribosomal pathway-related proteins were all downregulated, and oxidative phosphorylation pathway-related proteins, including Atp5o, ATP5L, Atp5f,Atp5h, Cox4i1 were also down-regulated. 55 DEPs (19 up-regulated proteins and 36 down-regulated proteins) were included in the PPI network constructed from DEPs between the EA and M groups. In addition, proteins involved in inflammation regulation such as serpins were identified (serpinb6 in M/N comparisons, serpinc1 in HM/M and EA/M comparisons).

Table 2 Differentially expressed proteins in dextran sulfate sodium-induced ulcerative colitis model group rats that regulated by herbpartitioned moxibustion
Western blot verification
We verified the expression of some DEPs that could be regulated by HM and EA. The western blot results showed that, compared with the N group, the expression of Sv2a, Ncbp1, Cps1, Cox4i1, Atp5f1 proteins were increased in the colon of UC rats, while the expression of Dclk3 protein was decreased (P< 0.01). HM and EA interventions can significantly down-regulate the expression of Sv2a, Ncbp1, Cps1,Cox4i1, Atp5f1 proteins and up-regulate Dclk3 protein (P< 0.01). It indicated that the western blotverification results were consistent with the iTRAQ results (Figure 9).

Table 3 Differentially expressed proteins in dextran sulfate sodium-induced ulcerative colitis model group rats that regulated by electroacupuncture
DISCUSSION
UC is a type of inflammatory bowel disease (IBD) characterized by chronic recurrent, and the specific aetiology and pathogenesis of UC are still not clear. Proteomics is a discipline that systematically quantifies all the proteins in cells or organisms and elucidates their biological functions, it has enormous potential for early diagnosis, prevention, and prognosis prediction of diseases[17,18]. In recent years,proteomics has gradually been applied to the study of UC, providing strong support for the research of pathogenesis, clinical diagnosis, and treatment of UC. A study has been found that 7 protein genes were differentially expressed in the intestinal tissues of UC patients using proteomics, including prohibitin,heat shock proteins, alpha-1 antitrypsin, ventralis intermedius, caspase-1, cytokeratin 20, Filamin Ainteracting protein 1, FLNa[19]. Another study analysed the transcriptome and proteome characteristics of the colon of UC patients, and found that the expression of genes and proteins related to immune and inflammatory responses in UC patients were dysregulated. At the same time, the complement cascade, metabolic processes, and peroxisome proliferator-activated receptor signal transduction were inhibited[20]. Therefore, iTRAQ proteomic technology can also be a feasible method to reveal potential targets for UC drug therapy[8].
In the present study, we performed proteomic analysis using iTRAQ labelling combined with mass spectrometry technology to identify DEPs in colon of DSS-induced UC rats, and selected DEPs that could be reversed by HM and EA. DSS has been widely used to prepare experimental colitis to study the new drug therapy, immunity and mechanism of action[21-23]. Our results showed that HM and EA improved the colonic mucosal injury of UC rats. At the same time, we have compared the normal andDSS-induced UC rats using iTRAQ labelling and found that 202 protein genes were differentially expressed between the two group. Among them, galectin-3, an up-regulated protein, is an endogenous lectin with extensive immunomodulatory functions, and can promote the inflammatory response in DSS-induced colitis by activating the NLR family pyrin domain containing 3 inflammasome[24].Moreover, galectin-3 has also been shown to be a key regulator of inflammation and can be a potential biomarker for IBD[25]. s100a9 is a subunit of calprotectin with pro-inflammatory activity, and the level of s100a9 was significantly increased in the faeces of DSS-induced colitis mice compared with normal mice[26,27]. This is consistent with the upregulation of s100a9 in UC colon tissue in our study.Guanylate cyclase activator 2A (GUCA2A) is the ligand of guanylate cyclase C, which is mainly expressed in intestinal epithelial cells, and can regulate intestinal barrier function and intestinal homeostasis through the cyclic guanosine monophosphate-dependent signalling pathway[28]. The down-regulation of GUCA2A can injure the intestinal mucosa barrier and affect the growth and repair of intestinal epithelial cells in IBD patients[29]. Interestingly, we found that the expression of GUCA2A was down-regulated in UC colon tissue, which was consistent with previous findings. These results suggest that galectin-3, s100a9 and GUCA2A may be potential biomarkers of UC.

Table 4 The Kyoto Encyclopedia of Genes and Genomes pathway related to immunity and inflammation with differentially expressed proteins between normal group and dextran sulfate sodium-induced ulcerative colitis model group

Table 5 The Kyoto Encyclopedia of Genes and Genomes pathway related to immunity, inflammation and oxidative phosphorylation with differentially expressed proteins between herb-partitioned moxibustion group and dextran sulfate sodium-induced ulcerative colitis model group

Table 6 The Kyoto Encyclopedia of Genes and Genomes pathway related to immunity, inflammation and oxidative phosphorylation with differentially expressed proteins between electroacupuncture group and dextran sulfate sodium-induced ulcerative colitis model group

Figure 4 Volcano plot of differentially expressed proteins between groups. A: Volcano plot of differentially expressed proteins (DEPs) between the normal and dextran sulfate sodium-induced ulcerative colitis model group; B: Volcano plot of DEPs between the dextran sulfate sodium-induced ulcerative colitis model and herb-partitioned moxibustion group; C: Volcano plot of DEPs between the dextran sulfate sodium-induced ulcerative colitis model and electroacupuncture group.

Figure 5 Venn diagram of differentially expressed proteins among groups. A: Venn diagram of differentially expressed proteins (DEPs) in dextran sulfate sodium-induced ulcerative colitis model group that can be regulated by herb-partitioned moxibustion; B: Venn diagram of DEPs in dextran sulfate sodiuminduced ulcerative colitis model group that can be regulated by electroacupuncture. N: Normal group; M: Dextran sulfate sodium-induced ulcerative colitis model group; HM: Herb-partitioned moxibustion group; EA: Electroacupuncture group.


Figure 6 Biological function annotation of differentially expressed proteins between groups. A: Biological function annotation of differentially expressed proteins (DEPs) between the normal and dextran sulfate sodium-induced ulcerative colitis model group; B: Biological function annotation of DEPs between the dextran sulfate sodium-induced ulcerative colitis model and herb-partitioned moxibustion group; C: biological function annotation of DEPs between the dextran sulfate sodium-induced ulcerative colitis model and electroacupuncture group.
After receiving the HM intervention, a total of 117 DEPs were identified, of which 25 proteins were reversely regulated by HM, including 17 proteins up-regulated in UC but were down-regulated by HM,and 8 down-regulated in UC but were up-regulated by HM. Among the 17 proteins down-regulated by HM, ATP5L, Atp5f, and Atp5h are the subunits of ATP synthase that involved in energy metabolism and oxidative phosphorylation; Cox4i1 is an important regulatory subunit of cytochrome c oxidase and is mainly associated with mitochondrial oxidative phosphorylation; Cps1, the carbamyl phosphate synthetase 1, was over-expressed in the non-dysplastic tissue of UC progressors[30]; RpS8 is a small subunit protein of ribosomes that plays a critical role in regulation of protein translation. Although it is unclear whether these proteins play a role in UC, ATP-induced energy metabolism disorders and intestinal epithelial mitochondrial damage are important pathogenesis of UC[31]. Therefore, changes in the expression of these proteins may be related to the occurrence of UC and the therapeutic effect of HM. Further studies should be performed to clarify the exact mechanisms of these oxidative phosphorylation proteins. Among the 8 proteins up-regulated by HM, CLCA1 is one of the major nonmucinous proteins in intestinal mucus, which is normally expressed in the gastrointestinal tract and is most abundant in the intestinal mucosa. Studies have shown that CLCA1 regulates the structural arrangement of mucus, participate in the mucus processing, and plays an important role in regulating intestinal homeostasis and intestinal inflammation[32-34]. CLCA1 can also exert a tumor suppressor effect in colorectal cancer (CRC) by inhibiting the Wnt/β-catenin signalling pathway and epithelialmesenchymal transition process[35]. The serum concentration and mRNA expression of CLCA1 in CRC tissues were negatively correlated with metastasis and tumor staging[36,37]. These findings suggest that CLCA1 may play an important role in moxibustion in inhibiting the development and carcinogenesis of UC. While 15 proteins were reversely regulated by EA, of which 9 up-regulated in UC but were downregulated by EA, and 6 down-regulated in UC but were up-regulated by EA. We found EA could also regulate the expression of Atp5f, Cox4i1, Cps1 that explained above, but the functions of other reversely regulated proteins in UC and immune inflammation remain unknown and require further validation and study.


Figure 7 Kyoto Encyclopedia of Genes and Genomes pathway enrichment of differentially expressed proteins between groups. A: Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of differentially expressed proteins (DEPs) between the normal and dextran sulfate sodiuminduced ulcerative colitis model group; B: KEGG pathway enrichment of DEPs between the dextran sulfate sodium-induced ulcerative colitis model and herbpartitioned moxibustion group; C: KEGG pathway enrichment of DEPs between the dextran sulfate sodium-induced ulcerative colitis model and electroacupuncture group. Each point in the KEGG pathway enrichment plot represents a KEGG pathway, with the left axis showing the pathway name and the abscissa showing the enrichment factor, which represents the ratio of the number of DEPs annotated to that pathway to the number of proteins annotated to that pathway for that species’protein. A larger enrichment factor indicates more reliable enrichment of DEPs in the pathway.
We further explored the potential functions of DEPs by GO and KEGG functional enrichment analyses. GO analysis revealed that the majority of the DEPs were primarily involved in cellular process, metabolic process, biological regulation in biological processes, cell part, cell, organelles,macromolecular complexes in cellular component, and binding, synaptic activity, structural molecule activity, molecular function regulation, molecular sensor activity, and transcriptional regulation activity in molecular function. The results indicated that most differentially abundant proteins were associated with cell structure and catalytic activity which play an important role in cell function. For KEGG analysis, we found that these DEPs were mainly enriched in inflammation responses and immunerelated pathways, such as primary immunodeficiency, NF-κB signalling pathway, natural killer cells mediated cytotoxicity, intestinal immune network for IgA production and so on. It has been confirmed that inflammatory response and immune response play an important role in UC. Numerous literature evidence that NF-κB pathway plays an essential role in pathogenic development of UC[38]. In addition,PPI network analysis showed that DEPs related to ribosomal pathways and oxidative phosphorylation pathways were all down-regulated by HM. As we all know that classical signalling pathways, such as the NF-κB signalling pathway, PI3K-Akt signalling pathway, have been confirmed to be associated with UC[39-42], and IgG receptor FcγR also plays an important role in UC intestinal immunity and inflammation[43,44]. The intestinal immune network for IgA production and Fc epsilon RI signalling pathway may also the UC-specific signalling pathway. A study on pathway enrichment analysis of differentially expressed genes between IBD and healthy individuals revealed that primary immunodeficiency,complement and coagulation cascades, and nitrogen metabolism pathways may also play a key role in the progression of IBD[45]. Multiple algorithm analysis found that calcium signalling pathway was involved in the process of UC[46]. This is consistent with our findings at the protein level. Therefore, the abnormality of above pathways may be the key to the pathogenesis of UC, and HM and EA may play the therapeutic role in UC through the regulation of key molecules in these pathways. In addition, we found that proteins involved in inflammation regulation such as serpins were identified (serpinb6 in M/N comparisons, serpinc1 in HM/M and EA/M comparisons). Serpins are the largest known family of serine proteinase inhibitors, which regulate innate immunity by inhibiting the serine proteinase cascades that initiate immune responses such as melanization and antimicrobial peptide production[47]. Several human serpins have been shown to regulate serine proteases associated with processes such as inflammation and immune responses[48]. And increased activity of serine proteases is demonstrated in IBD patients and may contribute to the onset and the maintenance of the disease[49].This appears to be a novel finding that will require additional research.
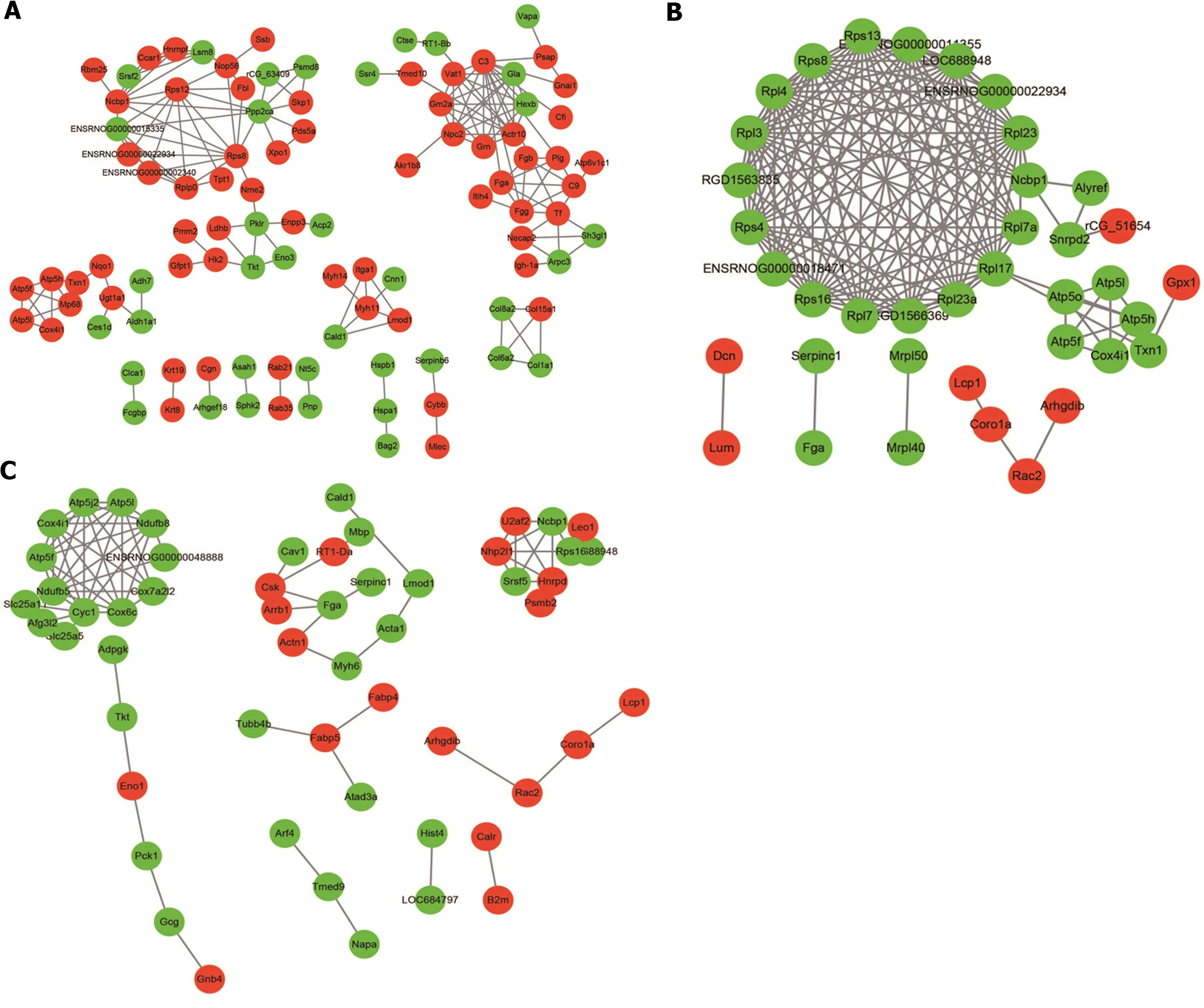
Figure 8 Protein-protein interaction network of differentially expressed proteins between groups. A: Protein-protein interaction (PPI) network of differentially expressed proteins (DEPs) between the normal and dextran sulfate sodium-induced ulcerative colitis model group; B: PPI network of DEPs between the dextran sulfate sodium-induced ulcerative colitis model and herb-partitioned moxibustion group; C: PPI network of DEPs between the dextran sulfate sodium-induced ulcerative colitis model and electroacupuncture group. Circle colours indicate changes in protein expression, with red indicating up-regulation and green indicating down-regulation, and line thickness indicates interaction intensity.
We also found that both HM and EA reversed the expression of Sv2a, Ncbp1, Rat hemoglobin betachain (Fragment), Fga, Cps1, ATP5L, Cox4i1, Atp5f1, Spout1, Peptidyl-prolyl cis-trans isomerase, Dclk3 proteins, while the rest is what they can reverse individually. The KEGG pathways of DEPs between HM and M groups, EA and M groups were also partly different. These results may reflect the commonness and characteristics of the therapeutic effects of HM and EA. So, the effects of HM and EA on UC may involve the differently regulating proteins and pathways. In addition, based on the results of proteomics, we selected 6 DEPs that could be reversed by HM and EA for verification. We found that both HM and EA decreased the expression of Sv2a, Ncbp1, Cps1, Cox4i1, Atp5f1 and increased the expression of Dclk3, which were consistent with the iTRAQ results, confirming the reliability of the iTRAQ results. Cox4i1 is the main subunit of cytochrome c oxidase, a key enzyme in energy metabolism,and plays an important role in mitochondrial oxidative phosphorylation. Defects in oxidative phosphorylation leads to a decrease in cellular ATP production. Studies have reported that the activity of mitochondrial respiratory chain complexes in UC patients is reduced, and mucosal ATP is absent[50,51]. Atp5f1 is a nuclear gene responsible for encoding the F0 subunit of ATP synthase, which is closely related to energy metabolism. Studies have found that the high expression of Cps1 plays an important role in the formation stage from UC to colitis associated cancer[52]. The intestinal mucosa of UC patients has been under chronic stress for a long time. The dynamic balance between oxidation and antioxidant defense mechanisms in the body is broken, causing mitochondrial damage, weakened oxidative phosphorylation, and reduced energy supply, which in turn causes cell apoptosis and colonic mucosal barrier dysfunction. The above results showed that HM and EA play a therapeutic effect on UC for mitochondrial function and energy metabolism. However, the role of Sv2a, Ncbp1 and Dclk3 in UC or mitochondrial oxidative phosphorylation has not been reported yet, and further research is needed.
In summary, iTRAQ proteomics was used in this study to identify DEPs in the colon of DSS-induced UC rats, as well as proteins that could be regulated by EA and HM, and analysed the function of these DEPs. However, the specific mechanisms of these DEPs were still unclear. They may be involved in one or more regulatory pathways, thus participate in the pathogenesis of UC and the mechanism of EA and HM treatments. The underlying mechanisms may require further study through animal and cell experiments.
CONCLUSION
In conclusion, our study used proteomics to provide evidence that EA and HM might regulate immunerelated pathways by regulating the expression of ATP5L, Atp5f1, Cox4i1 that associated with oxidative phosphorylation, thereby alleviating colonic inflammation of DSS-induced UC rats. Further investigations at the role of these proteins in UC will be helpful for revealing the pathogenesis and the mechanism of acupuncture and moxibustion on UC.
ARTICLE HIGHLIGHTS


Research motivation
The mechanisms of UC and the therapeutic targets of acupuncture and moxibustion treatment are complicated, and whether acupuncture and moxibustion play a therapeutic role in UC by regulating proteome changes remains unclear.
Research objectives
The present study aims to investigate the underlying mechanism of electroacupuncture (EA) and moxibustion on UC rats by using isotope-labeled relative and absolute quantification (iTRAQ)proteomics technology.
Research methods
Male Sprague-Dawley rats were randomly divided into the normal (N) group, the DSS-induced UC model (M) group, the herb-partitioned moxibustion (HM) group, and the EA group. 3% DSS was used to establish the UC rat model except for the N group, and HM and EA at the Tianshu (bilateral) and Qihai acupoints were performed respectively. Haematoxylin and eosin staining was used for morphological evaluation of colon tissues. iTRAQ and liquid chromatography-tandem mass spectrometry were performed for proteome analysis of the colon tissues, followed by bioinformatics analysis and proteinprotein interaction networks establishment of differentially expressed proteins (DEPs) between groups.Then western blot was used for verification of selected DEPs.
Research results
Our study revealed that HM and EA could regulate the expression of multiple proteins in colon of DSSinduced UC model rat. The DEPs were involved in various biological processes such as biological regulation, immune system progression and in multiple pathways including natural killer cell mediated cytotoxicity, intestinal immune network for immunoglobulin A production, and FcγR-mediated phagocytosis. Network analysis revealed that multiple pathways for the DEPs of each group were involved in protein-protein interactions. Subsequent verification of selected DEPs [synaptic vesicle glycoprotein 2A, nuclear cap binding protein subunit 1, carbamoyl phosphate synthetase 1, cytochrome c oxidase subunit 4 isoform 1 (Cox4i1), ATP synthase beta subunit precursor (Atp5F1), doublecortin like kinase 3] by western blot confirmed the reliability of the iTRAQ data.
Research conclusions
HM and EA might regulate immune-related pathways by regulating the expression of ATP5L, Atp5f1,Cox4i1 that associated with oxidative phosphorylation pathways, thereby alleviating colonic inflammation of DSS-induced UC rats.
Research perspectives
The present study revealed the possible molecular mechanisms of acupuncture and moxibustion treatment on UC, it may provide new light on clinical therapy of acupuncture and moxibustion treatment of UC.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the Experimental Animal Center of Shanghai University of Traditional Chinese Medicine for providing the animals, acknowledge the Beijing Genomics Institute for technical services and support of proteomics testings.
FOOTNOTES
Author contributions:Wu LY, Huang Y and Qi Q conceived and designed this study; Zhong R, Liu YN, Ma Z and Zheng HD performed the animal experiments, acquired and analyzed the data; Qi Q wrote the main manuscript; Liu YN and Ma Z prepared the figures and tables; Lu Y gave guidance to the manuscript writing; Zhao C, Huang Y and Wu LY revised and improved the manuscript; and all authors reviewed and approved the final version of this manuscript.
Supported bythe National Natural Science Foundation of China No. 81973955, 82004475 and 82174501; Shanghai Clinical Research Center for Acupuncture and Moxibustion No. 20MC1920500; Clinical Key Specialty Construction Foundation of Shanghai No. shslczdzk04701; Natural Science Foundation of Shanghai No. 21ZR1460200.
Institutional animal care and use committee statement:All animal experiments were performed according to the protocols approved by the Animal Ethics Committee of the Experimental Animal Center of Shanghai University of Traditional Chinese Medicine.
Conflict-of-interest statement:All the authors report no relevant conflicts of interest for this article.
Data sharing statement:No additional data are available.
ARRIVE guidelines statement:The authors have read the ARRIVE guidelines, and the manuscript has been prepared and revised according to the ARRIVE guidelines.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BYNC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is noncommercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin:China
ORCID number:Qin Qi 0000-0002-0563-1560; Rui Zhong 0000-0002-5136-335X; Ya-Nan Liu 0000-0002-3290-9206; Chen Zhao 0000-0002-2872-6626; Yan Huang 0000-0002-5588-0274; Yuan Lu 0000-0001-7584-5727; Zhe Ma 0000-0003-0956-4438; Han-Dan Zheng 0000-0002-8640-9194; Lu-Yi Wu 0000-0001-8888-1698.
S-Editor:Wang JJ
L-Editor:A
P-Editor:Cai YX
 World Journal of Gastroenterology2022年28期
World Journal of Gastroenterology2022年28期
- World Journal of Gastroenterology的其它文章
- Peroxisome proliferator-activated receptor gamma as a therapeutic target for hepatocellular carcinoma:Experimental and clinical scenarios
- Gut microbiota alteration and modulation in hepatitis B virus-related fibrosis and complications:Molecular mechanisms and therapeutic inventions
- Combination approaches in hepatocellular carcinoma: How systemic treatment can benefit candidates to locoregional modalities
- Update on endoscopic ultrasound-guided liver biopsy
- Non-alcoholic fatty liver disease-related hepatocellular carcinoma: Is there a role for immunotherapy?
- Potassium-competitive acid blockers and gastroesophageal reflux disease
